Please be patient as the page loads
|
SPAT2_HUMAN
|
||||||
| SwissProt Accessions | Q9UM82, O94857 | Gene names | SPATA2, KIAA0757, PD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spermatogenesis-associated protein 2 (Spermatogenesis-associated protein PD1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SPAT2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9UM82, O94857 | Gene names | SPATA2, KIAA0757, PD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spermatogenesis-associated protein 2 (Spermatogenesis-associated protein PD1). | |||||
|
TRI42_HUMAN
|
||||||
| θ value | 0.125558 (rank : 2) | NC score | 0.084725 (rank : 2) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8IWZ5, Q8N832, Q8NDL3 | Gene names | TRIM42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 42. | |||||
|
DPOLA_MOUSE
|
||||||
| θ value | 0.163984 (rank : 3) | NC score | 0.071602 (rank : 4) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P33609 | Gene names | Pola1, Pola | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase alpha catalytic subunit (EC 2.7.7.7). | |||||
|
TXLNB_HUMAN
|
||||||
| θ value | 0.365318 (rank : 4) | NC score | 0.043834 (rank : 7) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 912 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8N3L3, Q76L25, Q86T52, Q8N3S2 | Gene names | TXLNB, C6orf198, MDP77 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-taxilin (Muscle-derived protein 77) (hMDP77). | |||||
|
GOGA4_MOUSE
|
||||||
| θ value | 0.47712 (rank : 5) | NC score | 0.022835 (rank : 21) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
RN169_HUMAN
|
||||||
| θ value | 0.47712 (rank : 6) | NC score | 0.081604 (rank : 3) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8NCN4, Q6N015 | Gene names | RNF169, KIAA1991 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 169. | |||||
|
RBY1A_HUMAN
|
||||||
| θ value | 1.38821 (rank : 7) | NC score | 0.030579 (rank : 16) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15414, Q15377, Q8NHR0 | Gene names | RBMY1A1, RBM1, YRRM1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding motif protein, Y chromosome, family 1 member A1 (RNA- binding motif protein 1). | |||||
|
ZN238_HUMAN
|
||||||
| θ value | 1.38821 (rank : 8) | NC score | 0.014958 (rank : 28) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 836 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99592, Q13397, Q5VU40, Q8N463 | Gene names | ZNF238, RP58, TAZ1, ZBTB18 | |||
|
Domain Architecture |
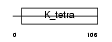
|
|||||
| Description | Zinc finger protein 238 (Transcriptional repressor RP58) (58 kDa repressor protein) (Zinc finger protein C2H2-171) (Translin-associated zinc finger protein 1) (TAZ-1) (Zinc finger and BTB domain-containing protein 18). | |||||
|
ZN238_MOUSE
|
||||||
| θ value | 1.81305 (rank : 9) | NC score | 0.014538 (rank : 30) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 837 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9WUK6 | Gene names | Znf238, Rp58, Zfp238 | |||
|
Domain Architecture |
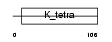
|
|||||
| Description | Zinc finger protein 238 (Zfp-238) (Transcriptional repressor RP58) (58 kDa repressor protein). | |||||
|
SRCH_HUMAN
|
||||||
| θ value | 2.36792 (rank : 10) | NC score | 0.052219 (rank : 6) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P23327, Q504Y6 | Gene names | HRC, HCP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcoplasmic reticulum histidine-rich calcium-binding protein precursor. | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 11) | NC score | 0.032947 (rank : 12) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
MLL3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 12) | NC score | 0.035643 (rank : 8) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
LIPA2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 13) | NC score | 0.016622 (rank : 25) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 1282 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BSS9, Q8BN73 | Gene names | Ppfia2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-2) (PTPRF-interacting protein alpha-2). | |||||
|
MYH3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 14) | NC score | 0.010176 (rank : 33) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 1660 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P11055, Q15492 | Gene names | MYH3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, fast skeletal muscle, embryonic (Muscle embryonic myosin heavy chain) (SMHCE). | |||||
|
TRI42_MOUSE
|
||||||
| θ value | 4.03905 (rank : 15) | NC score | 0.064614 (rank : 5) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9D2H5 | Gene names | Trim42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 42. | |||||
|
UBP53_HUMAN
|
||||||
| θ value | 4.03905 (rank : 16) | NC score | 0.028899 (rank : 18) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q70EK8, Q68DA5, Q8WVQ5, Q9P2J7 | Gene names | USP53, KIAA1350 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inactive ubiquitin carboxyl-terminal hydrolase 53 (Inactive ubiquitin- specific peptidase 53). | |||||
|
ZF64B_HUMAN
|
||||||
| θ value | 4.03905 (rank : 17) | NC score | 0.005922 (rank : 37) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 740 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NTW7, Q8WU98, Q9H9P1 | Gene names | ZFP64, ZNF338 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 64, isoforms 3 and 4 (Zinc finger protein 338). | |||||
|
ATE1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 18) | NC score | 0.033093 (rank : 11) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95260, O95261, Q8WW04 | Gene names | ATE1 | |||
|
Domain Architecture |

|
|||||
| Description | Arginyl-tRNA--protein transferase 1 (EC 2.3.2.8) (R-transferase 1) (Arginyltransferase 1) (Arginine-tRNA--protein transferase 1). | |||||
|
DAG1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 19) | NC score | 0.031054 (rank : 15) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q62165, Q61094, Q61141, Q61497 | Gene names | Dag1, Dag-1 | |||
|
Domain Architecture |

|
|||||
| Description | Dystroglycan precursor (Dystrophin-associated glycoprotein 1) [Contains: Alpha-dystroglycan (Alpha-DG); Beta-dystroglycan (Beta- DG)]. | |||||
|
JIP2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 20) | NC score | 0.025470 (rank : 19) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13387, Q96G62, Q99771, Q9NZ59, Q9UKQ4 | Gene names | MAPK8IP2, IB2, JIP2 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 2 (JNK-interacting protein 2) (JIP-2) (JNK MAP kinase scaffold protein 2) (Islet-brain-2) (IB-2) (Mitogen-activated protein kinase 8-interacting protein 2). | |||||
|
KCND1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 21) | NC score | 0.010207 (rank : 32) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q03719, Q8CC68 | Gene names | Kcnd1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 1 (Voltage-gated potassium channel subunit Kv4.1) (mShal). | |||||
|
MRCKG_HUMAN
|
||||||
| θ value | 5.27518 (rank : 22) | NC score | 0.007516 (rank : 34) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 1519 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6DT37, O00565 | Gene names | CDC42BPG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK gamma (EC 2.7.11.1) (CDC42- binding protein kinase gamma) (Myotonic dystrophy kinase-related CDC42-binding kinase gamma) (Myotonic dystrophy protein kinase-like alpha) (MRCK gamma) (DMPK-like gamma). | |||||
|
ZN142_HUMAN
|
||||||
| θ value | 5.27518 (rank : 23) | NC score | 0.007488 (rank : 35) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P52746, Q92510 | Gene names | ZNF142, KIAA0236 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 142 (HA4654). | |||||
|
HRX_HUMAN
|
||||||
| θ value | 6.88961 (rank : 24) | NC score | 0.028927 (rank : 17) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
JADE2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 25) | NC score | 0.016079 (rank : 26) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6ZQF7, Q3UHD5, Q6IE83 | Gene names | Phf15, Jade2, Kiaa0239 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
K1802_MOUSE
|
||||||
| θ value | 6.88961 (rank : 26) | NC score | 0.014805 (rank : 29) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
KTNB1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 27) | NC score | 0.007194 (rank : 36) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BG40, Q8CD18, Q8R1J0, Q9CWV2 | Gene names | Katnb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p80 WD40-containing subunit B1 (Katanin p80 subunit B1) (p80 katanin). | |||||
|
LATS2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 28) | NC score | 0.004982 (rank : 39) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 884 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NRM7, Q9P2X1 | Gene names | LATS2, KPM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase LATS2 (EC 2.7.11.1) (Large tumor suppressor homolog 2) (Serine/threonine-protein kinase kpm) (Kinase phosphorylated during mitosis protein) (Warts-like kinase). | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | 6.88961 (rank : 29) | NC score | 0.015516 (rank : 27) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
RHG04_HUMAN
|
||||||
| θ value | 6.88961 (rank : 30) | NC score | 0.011411 (rank : 31) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P98171, Q14144 | Gene names | ARHGAP4, KIAA0131, RGC1, RHOGAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 4 (Rho-GAP hematopoietic protein C1) (p115). | |||||
|
RNF31_MOUSE
|
||||||
| θ value | 6.88961 (rank : 31) | NC score | 0.022482 (rank : 22) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q924T7 | Gene names | Rnf31 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 31. | |||||
|
TRIM1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 32) | NC score | 0.035349 (rank : 9) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UJV3, Q5JYF5, Q8WWK1, Q9UJR9 | Gene names | MID2, FXY2, RNF60, TRIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Midline-2 (Midline defect 2) (Tripartite motif-containing protein 1) (Midin-2) (RING finger protein 60). | |||||
|
DGLA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 33) | NC score | 0.017353 (rank : 24) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y4D2, Q6WQJ0 | Gene names | NSDDR, C11orf11, KIAA0659 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sn1-specific diacylglycerol lipase alpha (EC 3.1.1.-) (DGL-alpha) (Neural stem cell-derived dendrite regulator). | |||||
|
PIGN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 34) | NC score | 0.020479 (rank : 23) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95427, Q7L8F8, Q8TC01, Q9NT05 | Gene names | PIGN, MCD4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GPI ethanolamine phosphate transferase 1 (EC 2.-.-.-) (Phosphatidylinositol-glycan biosynthesis class N protein) (PIG-N) (MCD4 homolog). | |||||
|
TRI18_HUMAN
|
||||||
| θ value | 8.99809 (rank : 35) | NC score | 0.031544 (rank : 14) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O15344, O75361, Q9BZX5 | Gene names | MID1, FXY, RNF59, TRIM18, XPRF | |||
|
Domain Architecture |

|
|||||
| Description | Midline-1 (EC 6.3.2.-) (Tripartite motif-containing protein 18) (Putative transcription factor XPRF) (Midin) (RING finger protein 59) (Midline 1 RING finger protein). | |||||
|
TRI18_MOUSE
|
||||||
| θ value | 8.99809 (rank : 36) | NC score | 0.032239 (rank : 13) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O70583, O35418, Q7TPT6 | Gene names | Mid1, Fxy, Trim18 | |||
|
Domain Architecture |

|
|||||
| Description | Midline-1 (EC 6.3.2.-) (Tripartite motif-containing protein 18) (Midin). | |||||
|
TRI32_HUMAN
|
||||||
| θ value | 8.99809 (rank : 37) | NC score | 0.023202 (rank : 20) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13049, Q9NQP8 | Gene names | TRIM32, HT2A | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 32 (EC 6.3.2.-) (Zinc finger protein HT2A) (72 kDa Tat-interacting protein). | |||||
|
TRIM1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 38) | NC score | 0.033955 (rank : 10) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9QUS6 | Gene names | Mid2, Fxy2, Trim1 | |||
|
Domain Architecture |

|
|||||
| Description | Midline-2 (Midline defect 2) (Tripartite motif-containing protein 1). | |||||
|
ZN575_HUMAN
|
||||||
| θ value | 8.99809 (rank : 39) | NC score | 0.005055 (rank : 38) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 739 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86XF7 | Gene names | ZNF575 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 575. | |||||
|
SPAT2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9UM82, O94857 | Gene names | SPATA2, KIAA0757, PD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spermatogenesis-associated protein 2 (Spermatogenesis-associated protein PD1). | |||||
|
TRI42_HUMAN
|
||||||
| NC score | 0.084725 (rank : 2) | θ value | 0.125558 (rank : 2) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8IWZ5, Q8N832, Q8NDL3 | Gene names | TRIM42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 42. | |||||
|
RN169_HUMAN
|
||||||
| NC score | 0.081604 (rank : 3) | θ value | 0.47712 (rank : 6) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8NCN4, Q6N015 | Gene names | RNF169, KIAA1991 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 169. | |||||
|
DPOLA_MOUSE
|
||||||
| NC score | 0.071602 (rank : 4) | θ value | 0.163984 (rank : 3) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P33609 | Gene names | Pola1, Pola | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase alpha catalytic subunit (EC 2.7.7.7). | |||||
|
TRI42_MOUSE
|
||||||
| NC score | 0.064614 (rank : 5) | θ value | 4.03905 (rank : 15) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9D2H5 | Gene names | Trim42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 42. | |||||
|
SRCH_HUMAN
|
||||||
| NC score | 0.052219 (rank : 6) | θ value | 2.36792 (rank : 10) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P23327, Q504Y6 | Gene names | HRC, HCP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcoplasmic reticulum histidine-rich calcium-binding protein precursor. | |||||
|
TXLNB_HUMAN
|
||||||
| NC score | 0.043834 (rank : 7) | θ value | 0.365318 (rank : 4) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 912 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8N3L3, Q76L25, Q86T52, Q8N3S2 | Gene names | TXLNB, C6orf198, MDP77 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-taxilin (Muscle-derived protein 77) (hMDP77). | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.035643 (rank : 8) | θ value | 3.0926 (rank : 12) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
TRIM1_HUMAN
|
||||||
| NC score | 0.035349 (rank : 9) | θ value | 6.88961 (rank : 32) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UJV3, Q5JYF5, Q8WWK1, Q9UJR9 | Gene names | MID2, FXY2, RNF60, TRIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Midline-2 (Midline defect 2) (Tripartite motif-containing protein 1) (Midin-2) (RING finger protein 60). | |||||
|
TRIM1_MOUSE
|
||||||
| NC score | 0.033955 (rank : 10) | θ value | 8.99809 (rank : 38) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9QUS6 | Gene names | Mid2, Fxy2, Trim1 | |||
|
Domain Architecture |

|
|||||
| Description | Midline-2 (Midline defect 2) (Tripartite motif-containing protein 1). | |||||
|
ATE1_HUMAN
|
||||||
| NC score | 0.033093 (rank : 11) | θ value | 5.27518 (rank : 18) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95260, O95261, Q8WW04 | Gene names | ATE1 | |||
|
Domain Architecture |

|
|||||
| Description | Arginyl-tRNA--protein transferase 1 (EC 2.3.2.8) (R-transferase 1) (Arginyltransferase 1) (Arginine-tRNA--protein transferase 1). | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.032947 (rank : 12) | θ value | 3.0926 (rank : 11) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
TRI18_MOUSE
|
||||||
| NC score | 0.032239 (rank : 13) | θ value | 8.99809 (rank : 36) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O70583, O35418, Q7TPT6 | Gene names | Mid1, Fxy, Trim18 | |||
|
Domain Architecture |

|
|||||
| Description | Midline-1 (EC 6.3.2.-) (Tripartite motif-containing protein 18) (Midin). | |||||
|
TRI18_HUMAN
|
||||||
| NC score | 0.031544 (rank : 14) | θ value | 8.99809 (rank : 35) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O15344, O75361, Q9BZX5 | Gene names | MID1, FXY, RNF59, TRIM18, XPRF | |||
|
Domain Architecture |

|
|||||
| Description | Midline-1 (EC 6.3.2.-) (Tripartite motif-containing protein 18) (Putative transcription factor XPRF) (Midin) (RING finger protein 59) (Midline 1 RING finger protein). | |||||
|
DAG1_MOUSE
|
||||||
| NC score | 0.031054 (rank : 15) | θ value | 5.27518 (rank : 19) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q62165, Q61094, Q61141, Q61497 | Gene names | Dag1, Dag-1 | |||
|
Domain Architecture |

|
|||||
| Description | Dystroglycan precursor (Dystrophin-associated glycoprotein 1) [Contains: Alpha-dystroglycan (Alpha-DG); Beta-dystroglycan (Beta- DG)]. | |||||
|
RBY1A_HUMAN
|
||||||
| NC score | 0.030579 (rank : 16) | θ value | 1.38821 (rank : 7) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15414, Q15377, Q8NHR0 | Gene names | RBMY1A1, RBM1, YRRM1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding motif protein, Y chromosome, family 1 member A1 (RNA- binding motif protein 1). | |||||
|
HRX_HUMAN
|
||||||
| NC score | 0.028927 (rank : 17) | θ value | 6.88961 (rank : 24) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
UBP53_HUMAN
|
||||||
| NC score | 0.028899 (rank : 18) | θ value | 4.03905 (rank : 16) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q70EK8, Q68DA5, Q8WVQ5, Q9P2J7 | Gene names | USP53, KIAA1350 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inactive ubiquitin carboxyl-terminal hydrolase 53 (Inactive ubiquitin- specific peptidase 53). | |||||
|
JIP2_HUMAN
|
||||||
| NC score | 0.025470 (rank : 19) | θ value | 5.27518 (rank : 20) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13387, Q96G62, Q99771, Q9NZ59, Q9UKQ4 | Gene names | MAPK8IP2, IB2, JIP2 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 2 (JNK-interacting protein 2) (JIP-2) (JNK MAP kinase scaffold protein 2) (Islet-brain-2) (IB-2) (Mitogen-activated protein kinase 8-interacting protein 2). | |||||
|
TRI32_HUMAN
|
||||||
| NC score | 0.023202 (rank : 20) | θ value | 8.99809 (rank : 37) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13049, Q9NQP8 | Gene names | TRIM32, HT2A | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 32 (EC 6.3.2.-) (Zinc finger protein HT2A) (72 kDa Tat-interacting protein). | |||||
|
GOGA4_MOUSE
|
||||||
| NC score | 0.022835 (rank : 21) | θ value | 0.47712 (rank : 5) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
RNF31_MOUSE
|
||||||
| NC score | 0.022482 (rank : 22) | θ value | 6.88961 (rank : 31) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q924T7 | Gene names | Rnf31 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 31. | |||||
|
PIGN_HUMAN
|
||||||
| NC score | 0.020479 (rank : 23) | θ value | 8.99809 (rank : 34) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95427, Q7L8F8, Q8TC01, Q9NT05 | Gene names | PIGN, MCD4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GPI ethanolamine phosphate transferase 1 (EC 2.-.-.-) (Phosphatidylinositol-glycan biosynthesis class N protein) (PIG-N) (MCD4 homolog). | |||||
|
DGLA_HUMAN
|
||||||
| NC score | 0.017353 (rank : 24) | θ value | 8.99809 (rank : 33) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y4D2, Q6WQJ0 | Gene names | NSDDR, C11orf11, KIAA0659 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sn1-specific diacylglycerol lipase alpha (EC 3.1.1.-) (DGL-alpha) (Neural stem cell-derived dendrite regulator). | |||||
|
LIPA2_MOUSE
|
||||||
| NC score | 0.016622 (rank : 25) | θ value | 4.03905 (rank : 13) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 1282 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BSS9, Q8BN73 | Gene names | Ppfia2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-2) (PTPRF-interacting protein alpha-2). | |||||
|
JADE2_MOUSE
|
||||||
| NC score | 0.016079 (rank : 26) | θ value | 6.88961 (rank : 25) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6ZQF7, Q3UHD5, Q6IE83 | Gene names | Phf15, Jade2, Kiaa0239 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.015516 (rank : 27) | θ value | 6.88961 (rank : 29) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
ZN238_HUMAN
|
||||||
| NC score | 0.014958 (rank : 28) | θ value | 1.38821 (rank : 8) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 836 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99592, Q13397, Q5VU40, Q8N463 | Gene names | ZNF238, RP58, TAZ1, ZBTB18 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 238 (Transcriptional repressor RP58) (58 kDa repressor protein) (Zinc finger protein C2H2-171) (Translin-associated zinc finger protein 1) (TAZ-1) (Zinc finger and BTB domain-containing protein 18). | |||||
|
K1802_MOUSE
|
||||||
| NC score | 0.014805 (rank : 29) | θ value | 6.88961 (rank : 26) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
ZN238_MOUSE
|
||||||
| NC score | 0.014538 (rank : 30) | θ value | 1.81305 (rank : 9) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 837 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9WUK6 | Gene names | Znf238, Rp58, Zfp238 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 238 (Zfp-238) (Transcriptional repressor RP58) (58 kDa repressor protein). | |||||
|
RHG04_HUMAN
|
||||||
| NC score | 0.011411 (rank : 31) | θ value | 6.88961 (rank : 30) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P98171, Q14144 | Gene names | ARHGAP4, KIAA0131, RGC1, RHOGAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 4 (Rho-GAP hematopoietic protein C1) (p115). | |||||
|
KCND1_MOUSE
|
||||||
| NC score | 0.010207 (rank : 32) | θ value | 5.27518 (rank : 21) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q03719, Q8CC68 | Gene names | Kcnd1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 1 (Voltage-gated potassium channel subunit Kv4.1) (mShal). | |||||
|
MYH3_HUMAN
|
||||||
| NC score | 0.010176 (rank : 33) | θ value | 4.03905 (rank : 14) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 1660 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P11055, Q15492 | Gene names | MYH3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, fast skeletal muscle, embryonic (Muscle embryonic myosin heavy chain) (SMHCE). | |||||
|
MRCKG_HUMAN
|
||||||
| NC score | 0.007516 (rank : 34) | θ value | 5.27518 (rank : 22) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 1519 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6DT37, O00565 | Gene names | CDC42BPG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK gamma (EC 2.7.11.1) (CDC42- binding protein kinase gamma) (Myotonic dystrophy kinase-related CDC42-binding kinase gamma) (Myotonic dystrophy protein kinase-like alpha) (MRCK gamma) (DMPK-like gamma). | |||||
|
ZN142_HUMAN
|
||||||
| NC score | 0.007488 (rank : 35) | θ value | 5.27518 (rank : 23) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P52746, Q92510 | Gene names | ZNF142, KIAA0236 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 142 (HA4654). | |||||
|
KTNB1_MOUSE
|
||||||
| NC score | 0.007194 (rank : 36) | θ value | 6.88961 (rank : 27) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BG40, Q8CD18, Q8R1J0, Q9CWV2 | Gene names | Katnb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p80 WD40-containing subunit B1 (Katanin p80 subunit B1) (p80 katanin). | |||||
|
ZF64B_HUMAN
|
||||||
| NC score | 0.005922 (rank : 37) | θ value | 4.03905 (rank : 17) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 740 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NTW7, Q8WU98, Q9H9P1 | Gene names | ZFP64, ZNF338 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 64, isoforms 3 and 4 (Zinc finger protein 338). | |||||
|
ZN575_HUMAN
|
||||||
| NC score | 0.005055 (rank : 38) | θ value | 8.99809 (rank : 39) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 739 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86XF7 | Gene names | ZNF575 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 575. | |||||
|
LATS2_HUMAN
|
||||||
| NC score | 0.004982 (rank : 39) | θ value | 6.88961 (rank : 28) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 884 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NRM7, Q9P2X1 | Gene names | LATS2, KPM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase LATS2 (EC 2.7.11.1) (Large tumor suppressor homolog 2) (Serine/threonine-protein kinase kpm) (Kinase phosphorylated during mitosis protein) (Warts-like kinase). | |||||