Please be patient as the page loads
|
SAMD8_HUMAN
|
||||||
| SwissProt Accessions | Q96LT4 | Gene names | SAMD8 | |||
|
Domain Architecture |
|
|||||
| Description | Sphingomyelin synthase-related protein 1 (Sterile alpha motif domain- containing 8). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SAMD8_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96LT4 | Gene names | SAMD8 | |||
|
Domain Architecture |

|
|||||
| Description | Sphingomyelin synthase-related protein 1 (Sterile alpha motif domain- containing 8). | |||||
|
SAMD8_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999077 (rank : 2) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9DA37 | Gene names | Samd8 | |||
|
Domain Architecture |
|
|||||
| Description | Sphingomyelin synthase-related protein 1 (Sterile alpha motif domain- containing 8). | |||||
|
SMS1_HUMAN
|
||||||
| θ value | 1.04096e-72 (rank : 3) | NC score | 0.931520 (rank : 5) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q86VZ5 | Gene names | TMEM23, MOB | |||
|
Domain Architecture |
|
|||||
| Description | Phosphatidylcholine:ceramide cholinephosphotransferase 1 (EC 2.7.-.-) (Transmembrane protein 23) (Sphingomyelin synthase 1) (Mob protein). | |||||
|
SMS1_MOUSE
|
||||||
| θ value | 6.7473e-72 (rank : 4) | NC score | 0.930586 (rank : 6) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8VCQ6, Q8C464, Q8C583, Q8C652 | Gene names | Tmem23 | |||
|
Domain Architecture |
|
|||||
| Description | Phosphatidylcholine:ceramide cholinephosphotransferase 1 (EC 2.7.-.-) (Transmembrane protein 23) (Sphingomyelin synthase 1) (Mob protein). | |||||
|
SMS2_HUMAN
|
||||||
| θ value | 4.52582e-68 (rank : 5) | NC score | 0.932876 (rank : 3) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NHU3 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylcholine:ceramide cholinephosphotransferase 2 (EC 2.7.-.-) (Sphingomyelin synthase 2). | |||||
|
SMS2_MOUSE
|
||||||
| θ value | 2.24614e-67 (rank : 6) | NC score | 0.932721 (rank : 4) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9D4B1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylcholine:ceramide cholinephosphotransferase 2 (EC 2.7.-.-) (Sphingomyelin synthase 2). | |||||
|
DGKD_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 7) | NC score | 0.086137 (rank : 7) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q16760, Q14158, Q6PK55, Q8NG53 | Gene names | DGKD, KIAA0145 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Diacylglycerol kinase delta (EC 2.7.1.107) (Diglyceride kinase delta) (DGK-delta) (DAG kinase delta) (130 kDa diacylglycerol kinase). | |||||
|
DGKH_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 8) | NC score | 0.085350 (rank : 8) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q86XP1, Q5VZW0, Q6PI56, Q86XP2, Q8N3N0, Q8N7J9 | Gene names | DGKH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Diacylglycerol kinase eta (EC 2.7.1.107) (Diglyceride kinase eta) (DGK-eta) (DAG kinase eta). | |||||
|
LIPB2_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 9) | NC score | 0.067483 (rank : 9) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 599 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8ND30, O75337, Q8WW26 | Gene names | PPFIBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-beta-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein-binding protein 2) (PTPRF-interacting protein-binding protein 2). | |||||
|
LIPB2_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 10) | NC score | 0.064605 (rank : 10) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O35711, Q7TMG4, Q8CBS6, Q99KX6 | Gene names | Ppfibp2, Cclp1 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-beta-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein-binding protein 2) (PTPRF-interacting protein-binding protein 2) (Coiled-coil-like protein 1). | |||||
|
CSKI1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 11) | NC score | 0.029635 (rank : 17) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8WXD9, Q9P2P0 | Gene names | CASKIN1, KIAA1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
CSKI1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 12) | NC score | 0.030805 (rank : 16) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 1031 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6P9K8, Q6ZPU2, Q8BWU2, Q8BX99, Q9CXH0 | Gene names | Caskin1, Kiaa1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
SARM1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 13) | NC score | 0.059876 (rank : 11) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6PDS3, Q5SYG5, Q5SYG6, Q6A054, Q6SZW0, Q8BRI9 | Gene names | Sarm1, Kiaa0524 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sterile alpha and TIR motif-containing protein 1 (Tir-1 homolog). | |||||
|
TLR13_MOUSE
|
||||||
| θ value | 1.81305 (rank : 14) | NC score | 0.013659 (rank : 20) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6R5N8, Q3TDS2 | Gene names | Tlr13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Toll-like receptor 13 precursor. | |||||
|
BFAR_HUMAN
|
||||||
| θ value | 3.0926 (rank : 15) | NC score | 0.034084 (rank : 14) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NZS9 | Gene names | BFAR, BAR, RNF47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional apoptosis regulator (RING finger protein 47). | |||||
|
PKCB1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 16) | NC score | 0.020318 (rank : 19) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 488 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ULU4, Q13517, Q4JJ94, Q4JJ95, Q5TH09, Q6MZM1, Q8WXC5, Q9H1F3, Q9H1F4, Q9H1F5, Q9H1L8, Q9H1L9, Q9H2G5, Q9NYN3, Q9UIX6 | Gene names | PRKCBP1, KIAA1125, RACK7, ZMYND8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein 1 (Rack7) (Cutaneous T-cell lymphoma- associated antigen se14-3) (CTCL tumor antigen se14-3) (Zinc finger MYND domain-containing protein 8). | |||||
|
SAMH1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 17) | NC score | 0.031488 (rank : 15) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y3Z3, Q5JXG8, Q9H004, Q9H005, Q9H3U9 | Gene names | SAMHD1, MOP5 | |||
|
Domain Architecture |
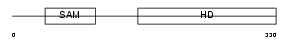
|
|||||
| Description | SAM domain and HD domain-containing protein 1 (Dendritic cell-derived IFNG-induced protein) (DCIP) (Monocyte protein 5) (MOP-5). | |||||
|
LIPB1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 18) | NC score | 0.048979 (rank : 12) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 640 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q86W92, O75336, Q86X70, Q9NY03, Q9ULJ0 | Gene names | PPFIBP1, KIAA1230 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-beta-1 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein-binding protein 1) (PTPRF-interacting protein-binding protein 1) (hSGT2). | |||||
|
LIPB1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 19) | NC score | 0.044358 (rank : 13) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 727 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8C8U0, Q69ZN5, Q6GQV3, Q80VB4, Q9CUT7 | Gene names | Ppfibp1, Kiaa1230 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-beta-1 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein-binding protein 1) (PTPRF-interacting protein-binding protein 1). | |||||
|
SLIK5_MOUSE
|
||||||
| θ value | 5.27518 (rank : 20) | NC score | 0.007788 (rank : 21) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 301 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q810B7, Q9CT21 | Gene names | Slitrk5 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT and NTRK-like protein 5 precursor. | |||||
|
BFAR_MOUSE
|
||||||
| θ value | 8.99809 (rank : 21) | NC score | 0.022013 (rank : 18) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8R079, Q8C1A7, Q9CXY3 | Gene names | Bfar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional apoptosis regulator. | |||||
|
SLIK5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 22) | NC score | 0.006953 (rank : 22) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O94991 | Gene names | SLITRK5, KIAA0918, LRRC11 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT and NTRK-like protein 5 precursor (Leucine-rich repeat-containing protein 11). | |||||
|
SAMD8_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96LT4 | Gene names | SAMD8 | |||
|
Domain Architecture |

|
|||||
| Description | Sphingomyelin synthase-related protein 1 (Sterile alpha motif domain- containing 8). | |||||
|
SAMD8_MOUSE
|
||||||
| NC score | 0.999077 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9DA37 | Gene names | Samd8 | |||
|
Domain Architecture |

|
|||||
| Description | Sphingomyelin synthase-related protein 1 (Sterile alpha motif domain- containing 8). | |||||
|
SMS2_HUMAN
|
||||||
| NC score | 0.932876 (rank : 3) | θ value | 4.52582e-68 (rank : 5) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NHU3 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylcholine:ceramide cholinephosphotransferase 2 (EC 2.7.-.-) (Sphingomyelin synthase 2). | |||||
|
SMS2_MOUSE
|
||||||
| NC score | 0.932721 (rank : 4) | θ value | 2.24614e-67 (rank : 6) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9D4B1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylcholine:ceramide cholinephosphotransferase 2 (EC 2.7.-.-) (Sphingomyelin synthase 2). | |||||
|
SMS1_HUMAN
|
||||||
| NC score | 0.931520 (rank : 5) | θ value | 1.04096e-72 (rank : 3) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q86VZ5 | Gene names | TMEM23, MOB | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylcholine:ceramide cholinephosphotransferase 1 (EC 2.7.-.-) (Transmembrane protein 23) (Sphingomyelin synthase 1) (Mob protein). | |||||
|
SMS1_MOUSE
|
||||||
| NC score | 0.930586 (rank : 6) | θ value | 6.7473e-72 (rank : 4) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8VCQ6, Q8C464, Q8C583, Q8C652 | Gene names | Tmem23 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylcholine:ceramide cholinephosphotransferase 1 (EC 2.7.-.-) (Transmembrane protein 23) (Sphingomyelin synthase 1) (Mob protein). | |||||
|
DGKD_HUMAN
|
||||||
| NC score | 0.086137 (rank : 7) | θ value | 0.00509761 (rank : 7) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q16760, Q14158, Q6PK55, Q8NG53 | Gene names | DGKD, KIAA0145 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Diacylglycerol kinase delta (EC 2.7.1.107) (Diglyceride kinase delta) (DGK-delta) (DAG kinase delta) (130 kDa diacylglycerol kinase). | |||||
|
DGKH_HUMAN
|
||||||
| NC score | 0.085350 (rank : 8) | θ value | 0.00665767 (rank : 8) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q86XP1, Q5VZW0, Q6PI56, Q86XP2, Q8N3N0, Q8N7J9 | Gene names | DGKH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Diacylglycerol kinase eta (EC 2.7.1.107) (Diglyceride kinase eta) (DGK-eta) (DAG kinase eta). | |||||
|
LIPB2_HUMAN
|
||||||
| NC score | 0.067483 (rank : 9) | θ value | 0.0563607 (rank : 9) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 599 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8ND30, O75337, Q8WW26 | Gene names | PPFIBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-beta-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein-binding protein 2) (PTPRF-interacting protein-binding protein 2). | |||||
|
LIPB2_MOUSE
|
||||||
| NC score | 0.064605 (rank : 10) | θ value | 0.0961366 (rank : 10) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O35711, Q7TMG4, Q8CBS6, Q99KX6 | Gene names | Ppfibp2, Cclp1 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-beta-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein-binding protein 2) (PTPRF-interacting protein-binding protein 2) (Coiled-coil-like protein 1). | |||||
|
SARM1_MOUSE
|
||||||
| NC score | 0.059876 (rank : 11) | θ value | 0.813845 (rank : 13) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6PDS3, Q5SYG5, Q5SYG6, Q6A054, Q6SZW0, Q8BRI9 | Gene names | Sarm1, Kiaa0524 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sterile alpha and TIR motif-containing protein 1 (Tir-1 homolog). | |||||
|
LIPB1_HUMAN
|
||||||
| NC score | 0.048979 (rank : 12) | θ value | 5.27518 (rank : 18) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 640 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q86W92, O75336, Q86X70, Q9NY03, Q9ULJ0 | Gene names | PPFIBP1, KIAA1230 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-beta-1 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein-binding protein 1) (PTPRF-interacting protein-binding protein 1) (hSGT2). | |||||
|
LIPB1_MOUSE
|
||||||
| NC score | 0.044358 (rank : 13) | θ value | 5.27518 (rank : 19) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 727 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8C8U0, Q69ZN5, Q6GQV3, Q80VB4, Q9CUT7 | Gene names | Ppfibp1, Kiaa1230 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-beta-1 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein-binding protein 1) (PTPRF-interacting protein-binding protein 1). | |||||
|
BFAR_HUMAN
|
||||||
| NC score | 0.034084 (rank : 14) | θ value | 3.0926 (rank : 15) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NZS9 | Gene names | BFAR, BAR, RNF47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional apoptosis regulator (RING finger protein 47). | |||||
|
SAMH1_HUMAN
|
||||||
| NC score | 0.031488 (rank : 15) | θ value | 4.03905 (rank : 17) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y3Z3, Q5JXG8, Q9H004, Q9H005, Q9H3U9 | Gene names | SAMHD1, MOP5 | |||
|
Domain Architecture |

|
|||||
| Description | SAM domain and HD domain-containing protein 1 (Dendritic cell-derived IFNG-induced protein) (DCIP) (Monocyte protein 5) (MOP-5). | |||||
|
CSKI1_MOUSE
|
||||||
| NC score | 0.030805 (rank : 16) | θ value | 0.62314 (rank : 12) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 1031 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6P9K8, Q6ZPU2, Q8BWU2, Q8BX99, Q9CXH0 | Gene names | Caskin1, Kiaa1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
CSKI1_HUMAN
|
||||||
| NC score | 0.029635 (rank : 17) | θ value | 0.365318 (rank : 11) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8WXD9, Q9P2P0 | Gene names | CASKIN1, KIAA1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
BFAR_MOUSE
|
||||||
| NC score | 0.022013 (rank : 18) | θ value | 8.99809 (rank : 21) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8R079, Q8C1A7, Q9CXY3 | Gene names | Bfar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional apoptosis regulator. | |||||
|
PKCB1_HUMAN
|
||||||
| NC score | 0.020318 (rank : 19) | θ value | 4.03905 (rank : 16) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 488 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ULU4, Q13517, Q4JJ94, Q4JJ95, Q5TH09, Q6MZM1, Q8WXC5, Q9H1F3, Q9H1F4, Q9H1F5, Q9H1L8, Q9H1L9, Q9H2G5, Q9NYN3, Q9UIX6 | Gene names | PRKCBP1, KIAA1125, RACK7, ZMYND8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein 1 (Rack7) (Cutaneous T-cell lymphoma- associated antigen se14-3) (CTCL tumor antigen se14-3) (Zinc finger MYND domain-containing protein 8). | |||||
|
TLR13_MOUSE
|
||||||
| NC score | 0.013659 (rank : 20) | θ value | 1.81305 (rank : 14) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6R5N8, Q3TDS2 | Gene names | Tlr13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Toll-like receptor 13 precursor. | |||||
|
SLIK5_MOUSE
|
||||||
| NC score | 0.007788 (rank : 21) | θ value | 5.27518 (rank : 20) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 301 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q810B7, Q9CT21 | Gene names | Slitrk5 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT and NTRK-like protein 5 precursor. | |||||
|
SLIK5_HUMAN
|
||||||
| NC score | 0.006953 (rank : 22) | θ value | 8.99809 (rank : 22) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O94991 | Gene names | SLITRK5, KIAA0918, LRRC11 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT and NTRK-like protein 5 precursor (Leucine-rich repeat-containing protein 11). | |||||