Please be patient as the page loads
|
PHEX_HUMAN
|
||||||
| SwissProt Accessions | P78562, O00678, Q13646, Q93032, Q99827 | Gene names | PHEX, PEX | |||
|
Domain Architecture |

|
|||||
| Description | Phosphate-regulating neutral endopeptidase (EC 3.4.24.-) (Metalloendopeptidase homolog PEX) (X-linked hypophosphatemia protein) (HYP) (Vitamin D-resistant hypophosphatemic rickets protein). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PHEX_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P78562, O00678, Q13646, Q93032, Q99827 | Gene names | PHEX, PEX | |||
|
Domain Architecture |

|
|||||
| Description | Phosphate-regulating neutral endopeptidase (EC 3.4.24.-) (Metalloendopeptidase homolog PEX) (X-linked hypophosphatemia protein) (HYP) (Vitamin D-resistant hypophosphatemic rickets protein). | |||||
|
PHEX_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999779 (rank : 2) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P70669, P97439 | Gene names | Phex, Hyp, Pex | |||
|
Domain Architecture |

|
|||||
| Description | Metalloendopeptidase homolog PEX (EC 3.4.24.-) (Phosphate regulating neutral endopeptidase) (X-linked hypophosphatemia protein) (HYP) (Vitamin D-resistant hypophosphatemic rickets protein). | |||||
|
MMEL1_MOUSE
|
||||||
| θ value | 2.8663e-147 (rank : 3) | NC score | 0.976017 (rank : 4) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9JLI3, Q3U495, Q9ERK2, Q9ERK3, Q9QZV6, Q9QZV7 | Gene names | Mmel1, Nep2, Nl1, Sep | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane metallo-endopeptidase-like 1 (EC 3.4.24.11) (Neprilysin-2) (Neprilysin II) (NL2) (NEPII) (NEP2(m)) (Neprilysin-like peptidase) (NEPLP) (Neprilysin-like 1) (NL-1) (Soluble secreted endopeptidase) [Contains: Membrane metallo-endopeptidase-like 1, soluble form (Neprilysin-2 secreted) (NEP2(s))]. | |||||
|
MMEL1_HUMAN
|
||||||
| θ value | 4.28312e-143 (rank : 4) | NC score | 0.976123 (rank : 3) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q495T6, Q495T7, Q495T8, Q5SZS6, Q96PH9 | Gene names | MMEL1, MELL1, MMEL2, NEP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane metallo-endopeptidase-like 1 (EC 3.4.24.11) (Membrane metallo-endopeptidase-like 2) (Neprilysin-2) (Neprilysin II) (NL2) (NEPII) (NEP2(m)) [Contains: Membrane metallo-endopeptidase-like 1, soluble form (Neprilysin-2 secreted) (NEP2(s))]. | |||||
|
ECE1_HUMAN
|
||||||
| θ value | 5.80223e-132 (rank : 5) | NC score | 0.971854 (rank : 8) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P42892, Q14217, Q58GE7, Q5THM5, Q5THM7, Q5THM8, Q9UJQ6, Q9UPF4, Q9UPM4, Q9Y501 | Gene names | ECE1 | |||
|
Domain Architecture |

|
|||||
| Description | Endothelin-converting enzyme 1 (EC 3.4.24.71) (ECE-1). | |||||
|
ECE1_MOUSE
|
||||||
| θ value | 3.76091e-131 (rank : 6) | NC score | 0.972078 (rank : 7) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q4PZA2, Q4PZ99, Q4PZA1, Q6P9Q9 | Gene names | Ece1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endothelin-converting enzyme 1 (EC 3.4.24.71) (ECE-1). | |||||
|
NEP_HUMAN
|
||||||
| θ value | 2.52267e-127 (rank : 7) | NC score | 0.973773 (rank : 5) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P08473 | Gene names | MME, EPN | |||
|
Domain Architecture |

|
|||||
| Description | Neprilysin (EC 3.4.24.11) (Neutral endopeptidase) (NEP) (Enkephalinase) (Neutral endopeptidase 24.11) (Atriopeptidase) (Common acute lymphocytic leukemia antigen) (CALLA) (CD10 antigen). | |||||
|
NEP_MOUSE
|
||||||
| θ value | 4.75757e-126 (rank : 8) | NC score | 0.973436 (rank : 6) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q61391, Q6NXX5, Q8K251 | Gene names | Mme | |||
|
Domain Architecture |

|
|||||
| Description | Neprilysin (EC 3.4.24.11) (Neutral endopeptidase) (NEP) (Enkephalinase) (Neutral endopeptidase 24.11) (Atriopeptidase) (CD10 antigen). | |||||
|
ECE2_HUMAN
|
||||||
| θ value | 1.29561e-123 (rank : 9) | NC score | 0.970166 (rank : 9) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O60344, Q96NX3, Q96NX4 | Gene names | ECE2, KIAA0604 | |||
|
Domain Architecture |

|
|||||
| Description | Endothelin-converting enzyme 2 (EC 3.4.24.71) (ECE-2). | |||||
|
ECEL1_MOUSE
|
||||||
| θ value | 1.94503e-103 (rank : 10) | NC score | 0.963492 (rank : 10) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9JMI0 | Gene names | Ecel1, Dine, Xce | |||
|
Domain Architecture |

|
|||||
| Description | Endothelin-converting enzyme-like 1 (EC 3.4.24.-) (Xce protein) (Damage-induced neuronal endopeptidase). | |||||
|
ECEL1_HUMAN
|
||||||
| θ value | 9.98939e-100 (rank : 11) | NC score | 0.963305 (rank : 11) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O95672, Q45UD9, Q53RF9, Q6UW86, Q86TH4, Q9NY95 | Gene names | ECEL1, XCE | |||
|
Domain Architecture |

|
|||||
| Description | Endothelin-converting enzyme-like 1 (EC 3.4.24.-) (Xce protein). | |||||
|
KELL_HUMAN
|
||||||
| θ value | 8.28633e-54 (rank : 12) | NC score | 0.938420 (rank : 12) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P23276, Q96RS8, Q99885 | Gene names | KEL | |||
|
Domain Architecture |

|
|||||
| Description | Kell blood group glycoprotein (EC 3.4.24.-) (CD238 antigen). | |||||
|
C8AP2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 13) | NC score | 0.010776 (rank : 14) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 548 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UKL3, Q6PH76, Q7LCQ7, Q86YD9, Q9NUQ4, Q9NZV9, Q9P2N1, Q9Y563 | Gene names | CASP8AP2, FLASH, KIAA1315, RIP25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CASP8-associated protein 2 (FLICE-associated huge protein). | |||||
|
FKB11_MOUSE
|
||||||
| θ value | 3.0926 (rank : 14) | NC score | 0.018653 (rank : 13) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D1M7, Q9CRE4 | Gene names | Fkbp11 | |||
|
Domain Architecture |
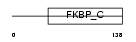
|
|||||
| Description | FK506-binding protein 11 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (19 kDa FK506-binding protein) (FKBP-19). | |||||
|
AN30A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 15) | NC score | 0.000410 (rank : 17) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 1119 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BXX3, Q5W025 | Gene names | ANKRD30A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 30A (NY-BR-1 antigen). | |||||
|
SYMM_MOUSE
|
||||||
| θ value | 6.88961 (rank : 16) | NC score | 0.008659 (rank : 15) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q499X9, Q3T9W7, Q8BWR9, Q8BXY1 | Gene names | Mars2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methionyl-tRNA synthetase, mitochondrial precursor (EC 6.1.1.10) (Methionine--tRNA ligase 2) (Mitochondrial methionine--tRNA ligase) (MtMetRS). | |||||
|
UTP20_MOUSE
|
||||||
| θ value | 8.99809 (rank : 17) | NC score | 0.006626 (rank : 16) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5XG71, Q80V22, Q8BXH9, Q8CHL6, Q99K11 | Gene names | Utp20, Drim | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small subunit processome component 20 homolog (Down-regulated in metastasis protein). | |||||
|
PHEX_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P78562, O00678, Q13646, Q93032, Q99827 | Gene names | PHEX, PEX | |||
|
Domain Architecture |

|
|||||
| Description | Phosphate-regulating neutral endopeptidase (EC 3.4.24.-) (Metalloendopeptidase homolog PEX) (X-linked hypophosphatemia protein) (HYP) (Vitamin D-resistant hypophosphatemic rickets protein). | |||||
|
PHEX_MOUSE
|
||||||
| NC score | 0.999779 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P70669, P97439 | Gene names | Phex, Hyp, Pex | |||
|
Domain Architecture |

|
|||||
| Description | Metalloendopeptidase homolog PEX (EC 3.4.24.-) (Phosphate regulating neutral endopeptidase) (X-linked hypophosphatemia protein) (HYP) (Vitamin D-resistant hypophosphatemic rickets protein). | |||||
|
MMEL1_HUMAN
|
||||||
| NC score | 0.976123 (rank : 3) | θ value | 4.28312e-143 (rank : 4) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q495T6, Q495T7, Q495T8, Q5SZS6, Q96PH9 | Gene names | MMEL1, MELL1, MMEL2, NEP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane metallo-endopeptidase-like 1 (EC 3.4.24.11) (Membrane metallo-endopeptidase-like 2) (Neprilysin-2) (Neprilysin II) (NL2) (NEPII) (NEP2(m)) [Contains: Membrane metallo-endopeptidase-like 1, soluble form (Neprilysin-2 secreted) (NEP2(s))]. | |||||
|
MMEL1_MOUSE
|
||||||
| NC score | 0.976017 (rank : 4) | θ value | 2.8663e-147 (rank : 3) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9JLI3, Q3U495, Q9ERK2, Q9ERK3, Q9QZV6, Q9QZV7 | Gene names | Mmel1, Nep2, Nl1, Sep | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane metallo-endopeptidase-like 1 (EC 3.4.24.11) (Neprilysin-2) (Neprilysin II) (NL2) (NEPII) (NEP2(m)) (Neprilysin-like peptidase) (NEPLP) (Neprilysin-like 1) (NL-1) (Soluble secreted endopeptidase) [Contains: Membrane metallo-endopeptidase-like 1, soluble form (Neprilysin-2 secreted) (NEP2(s))]. | |||||
|
NEP_HUMAN
|
||||||
| NC score | 0.973773 (rank : 5) | θ value | 2.52267e-127 (rank : 7) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P08473 | Gene names | MME, EPN | |||
|
Domain Architecture |

|
|||||
| Description | Neprilysin (EC 3.4.24.11) (Neutral endopeptidase) (NEP) (Enkephalinase) (Neutral endopeptidase 24.11) (Atriopeptidase) (Common acute lymphocytic leukemia antigen) (CALLA) (CD10 antigen). | |||||
|
NEP_MOUSE
|
||||||
| NC score | 0.973436 (rank : 6) | θ value | 4.75757e-126 (rank : 8) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q61391, Q6NXX5, Q8K251 | Gene names | Mme | |||
|
Domain Architecture |

|
|||||
| Description | Neprilysin (EC 3.4.24.11) (Neutral endopeptidase) (NEP) (Enkephalinase) (Neutral endopeptidase 24.11) (Atriopeptidase) (CD10 antigen). | |||||
|
ECE1_MOUSE
|
||||||
| NC score | 0.972078 (rank : 7) | θ value | 3.76091e-131 (rank : 6) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q4PZA2, Q4PZ99, Q4PZA1, Q6P9Q9 | Gene names | Ece1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endothelin-converting enzyme 1 (EC 3.4.24.71) (ECE-1). | |||||
|
ECE1_HUMAN
|
||||||
| NC score | 0.971854 (rank : 8) | θ value | 5.80223e-132 (rank : 5) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P42892, Q14217, Q58GE7, Q5THM5, Q5THM7, Q5THM8, Q9UJQ6, Q9UPF4, Q9UPM4, Q9Y501 | Gene names | ECE1 | |||
|
Domain Architecture |

|
|||||
| Description | Endothelin-converting enzyme 1 (EC 3.4.24.71) (ECE-1). | |||||
|
ECE2_HUMAN
|
||||||
| NC score | 0.970166 (rank : 9) | θ value | 1.29561e-123 (rank : 9) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O60344, Q96NX3, Q96NX4 | Gene names | ECE2, KIAA0604 | |||
|
Domain Architecture |

|
|||||
| Description | Endothelin-converting enzyme 2 (EC 3.4.24.71) (ECE-2). | |||||
|
ECEL1_MOUSE
|
||||||
| NC score | 0.963492 (rank : 10) | θ value | 1.94503e-103 (rank : 10) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9JMI0 | Gene names | Ecel1, Dine, Xce | |||
|
Domain Architecture |

|
|||||
| Description | Endothelin-converting enzyme-like 1 (EC 3.4.24.-) (Xce protein) (Damage-induced neuronal endopeptidase). | |||||
|
ECEL1_HUMAN
|
||||||
| NC score | 0.963305 (rank : 11) | θ value | 9.98939e-100 (rank : 11) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O95672, Q45UD9, Q53RF9, Q6UW86, Q86TH4, Q9NY95 | Gene names | ECEL1, XCE | |||
|
Domain Architecture |

|
|||||
| Description | Endothelin-converting enzyme-like 1 (EC 3.4.24.-) (Xce protein). | |||||
|
KELL_HUMAN
|
||||||
| NC score | 0.938420 (rank : 12) | θ value | 8.28633e-54 (rank : 12) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P23276, Q96RS8, Q99885 | Gene names | KEL | |||
|
Domain Architecture |

|
|||||
| Description | Kell blood group glycoprotein (EC 3.4.24.-) (CD238 antigen). | |||||
|
FKB11_MOUSE
|
||||||
| NC score | 0.018653 (rank : 13) | θ value | 3.0926 (rank : 14) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D1M7, Q9CRE4 | Gene names | Fkbp11 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 11 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (19 kDa FK506-binding protein) (FKBP-19). | |||||
|
C8AP2_HUMAN
|
||||||
| NC score | 0.010776 (rank : 14) | θ value | 2.36792 (rank : 13) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 548 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UKL3, Q6PH76, Q7LCQ7, Q86YD9, Q9NUQ4, Q9NZV9, Q9P2N1, Q9Y563 | Gene names | CASP8AP2, FLASH, KIAA1315, RIP25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CASP8-associated protein 2 (FLICE-associated huge protein). | |||||
|
SYMM_MOUSE
|
||||||
| NC score | 0.008659 (rank : 15) | θ value | 6.88961 (rank : 16) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q499X9, Q3T9W7, Q8BWR9, Q8BXY1 | Gene names | Mars2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methionyl-tRNA synthetase, mitochondrial precursor (EC 6.1.1.10) (Methionine--tRNA ligase 2) (Mitochondrial methionine--tRNA ligase) (MtMetRS). | |||||
|
UTP20_MOUSE
|
||||||
| NC score | 0.006626 (rank : 16) | θ value | 8.99809 (rank : 17) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5XG71, Q80V22, Q8BXH9, Q8CHL6, Q99K11 | Gene names | Utp20, Drim | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small subunit processome component 20 homolog (Down-regulated in metastasis protein). | |||||
|
AN30A_HUMAN
|
||||||
| NC score | 0.000410 (rank : 17) | θ value | 5.27518 (rank : 15) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 1119 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BXX3, Q5W025 | Gene names | ANKRD30A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 30A (NY-BR-1 antigen). | |||||