Please be patient as the page loads
|
PDCL2_HUMAN
|
||||||
| SwissProt Accessions | Q8N4E4 | Gene names | PDCL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosducin-like protein 2. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PDCL2_HUMAN
|
||||||
| θ value | 6.63862e-128 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8N4E4 | Gene names | PDCL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosducin-like protein 2. | |||||
|
PDCL2_MOUSE
|
||||||
| θ value | 6.24247e-110 (rank : 2) | NC score | 0.992038 (rank : 2) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q78Y63 | Gene names | Pdcl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosducin-like protein 2. | |||||
|
PDCL3_HUMAN
|
||||||
| θ value | 2.03155e-68 (rank : 3) | NC score | 0.967221 (rank : 4) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9H2J4 | Gene names | PDCL3, VIAF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosducin-like protein 3 (Viral IAP-associated factor 1) (VIAF-1) (HTPHLP). | |||||
|
PDCL3_MOUSE
|
||||||
| θ value | 4.23606e-66 (rank : 4) | NC score | 0.969714 (rank : 3) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BVF2, Q3TH06, Q99JX2, Q9D0W3 | Gene names | Pdcl3, Viaf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosducin-like protein 3 (Viral IAP-associated factor 1) (VIAF-1). | |||||
|
PHLP_HUMAN
|
||||||
| θ value | 1.86753e-13 (rank : 5) | NC score | 0.630551 (rank : 5) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q13371, Q96AF1, Q9UEW7, Q9UFL0 | Gene names | PDCL | |||
|
Domain Architecture |

|
|||||
| Description | Phosducin-like protein (PHLP). | |||||
|
PHLP_MOUSE
|
||||||
| θ value | 4.59992e-12 (rank : 6) | NC score | 0.617909 (rank : 6) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9DBX2, Q3TKI0 | Gene names | Pdcl | |||
|
Domain Architecture |
|
|||||
| Description | Phosducin-like protein (PHLP). | |||||
|
TXND9_HUMAN
|
||||||
| θ value | 3.29651e-10 (rank : 7) | NC score | 0.547257 (rank : 9) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O14530, Q53RV8, Q6NSF5, Q9BRU6 | Gene names | TXNDC9, APACD | |||
|
Domain Architecture |
|
|||||
| Description | Thioredoxin domain-containing protein 9 (Protein 1-4) (ATP-binding protein associated with cell differentiation). | |||||
|
PHOS_HUMAN
|
||||||
| θ value | 7.34386e-10 (rank : 8) | NC score | 0.602501 (rank : 7) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P20941, Q14816 | Gene names | PDC | |||
|
Domain Architecture |

|
|||||
| Description | Phosducin (PHD) (33 kDa phototransducing protein) (MEKA protein). | |||||
|
PHOS_MOUSE
|
||||||
| θ value | 6.21693e-09 (rank : 9) | NC score | 0.589145 (rank : 8) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9QW08 | Gene names | Pdc, Rpr1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosducin (PHD) (33 kDa phototransducing protein) (Rod photoreceptor 1) (RPR-1). | |||||
|
TXND9_MOUSE
|
||||||
| θ value | 4.0297e-08 (rank : 10) | NC score | 0.527619 (rank : 10) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9CQ79, Q3TKD2 | Gene names | Txndc9, Apacd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 9 (ATP-binding protein associated with cell differentiation). | |||||
|
TXNL2_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 11) | NC score | 0.208640 (rank : 11) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9CQM9, Q9JLZ2 | Gene names | Txnl2, Picot | |||
|
Domain Architecture |
|
|||||
| Description | Thioredoxin-like protein 2 (PKC-interacting cousin of thioredoxin) (PKC-theta-interacting protein) (PKCq-interacting protein). | |||||
|
TXNL2_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 12) | NC score | 0.187440 (rank : 12) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O76003, Q96CE0, Q9P1B0, Q9P1B1 | Gene names | TXNL2, PICOT | |||
|
Domain Architecture |
|
|||||
| Description | Thioredoxin-like protein 2 (PKC-interacting cousin of thioredoxin) (PKC-theta-interacting protein) (PKCq-interacting protein). | |||||
|
RB6I2_HUMAN
|
||||||
| θ value | 0.163984 (rank : 13) | NC score | 0.023569 (rank : 25) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1584 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8IUD2, Q6NVK2, Q8IUD3, Q8IUD4, Q8IUD5, Q8NAS1, Q9NXN5, Q9UIK7, Q9UPS1 | Gene names | ERC1, ELKS, KIAA1081, RAB6IP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1). | |||||
|
UBP8_HUMAN
|
||||||
| θ value | 0.47712 (rank : 14) | NC score | 0.021711 (rank : 26) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 751 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P40818, Q7Z3U2, Q86VA0, Q8IWI7 | Gene names | USP8, KIAA0055, UBPY | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 8 (EC 3.1.2.15) (Ubiquitin thioesterase 8) (Ubiquitin-specific-processing protease 8) (Deubiquitinating enzyme 8) (hUBPy). | |||||
|
GLE1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 15) | NC score | 0.047170 (rank : 19) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q53GS7, O75458, Q53GT9, Q5VVU1, Q8NCP6, Q9UFL6 | Gene names | GLE1L, GLE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin GLE1 (GLE1-like protein) (hGLE1). | |||||
|
K0831_MOUSE
|
||||||
| θ value | 1.06291 (rank : 16) | NC score | 0.051086 (rank : 17) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8CDJ3, Q69ZY1, Q6PFY6, Q8C6N0, Q8R3M3 | Gene names | Kiaa0831, D14Ertd436e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0831. | |||||
|
MYO10_HUMAN
|
||||||
| θ value | 1.06291 (rank : 17) | NC score | 0.012002 (rank : 36) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 920 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9HD67, Q9NYM7, Q9P110, Q9P111, Q9UHF6 | Gene names | MYO10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin X). | |||||
|
IF2P_HUMAN
|
||||||
| θ value | 1.38821 (rank : 18) | NC score | 0.025247 (rank : 24) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O60841, O95805, Q9UF81, Q9UMN7 | Gene names | EIF5B, IF2, KIAA0741 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5B (eIF-5B) (Translation initiation factor IF-2). | |||||
|
CD008_HUMAN
|
||||||
| θ value | 1.81305 (rank : 19) | NC score | 0.048217 (rank : 18) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P78312, O43607, P78311, P78313, Q9UEG8 | Gene names | C4orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C4orf8 (Protein IT14). | |||||
|
THIO_HUMAN
|
||||||
| θ value | 1.81305 (rank : 20) | NC score | 0.071769 (rank : 13) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P10599, Q96KI3 | Gene names | TXN, TRDX, TRX, TRX1 | |||
|
Domain Architecture |
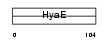
|
|||||
| Description | Thioredoxin (Trx) (ATL-derived factor) (ADF) (Surface-associated sulphydryl protein) (SASP). | |||||
|
DYXC1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 21) | NC score | 0.020946 (rank : 29) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R368 | Gene names | Dyx1c1, Ekn1 | |||
|
Domain Architecture |

|
|||||
| Description | Dyslexia susceptibility 1 candidate gene 1 protein homolog. | |||||
|
THIO_MOUSE
|
||||||
| θ value | 2.36792 (rank : 22) | NC score | 0.069774 (rank : 14) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P10639, Q52KC4, Q9D8R0 | Gene names | Txn, Txn1 | |||
|
Domain Architecture |
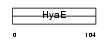
|
|||||
| Description | Thioredoxin (Trx) (ATL-derived factor) (ADF). | |||||
|
NFM_HUMAN
|
||||||
| θ value | 3.0926 (rank : 23) | NC score | 0.006790 (rank : 45) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
RB6I2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 24) | NC score | 0.018723 (rank : 30) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q99MI1, Q80TK7, Q8BPL1, Q8C7Y1, Q99MI2 | Gene names | Erc1, Cast2, Kiaa1081, Rab6ip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1) (ERC1) (CAZ-associated structural protein 2) (CAST2). | |||||
|
SLK_HUMAN
|
||||||
| θ value | 3.0926 (rank : 25) | NC score | 0.004449 (rank : 48) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1775 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9H2G2, O00211, Q6P1Z4, Q86WU7, Q86WW1, Q92603, Q9NQL0, Q9NQL1 | Gene names | SLK, KIAA0204, STK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (hSLK) (Serine/threonine-protein kinase 2) (CTCL tumor antigen se20-9). | |||||
|
VATE_MOUSE
|
||||||
| θ value | 3.0926 (rank : 26) | NC score | 0.029842 (rank : 22) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P50518 | Gene names | Atp6v1e1, Atp6e | |||
|
Domain Architecture |
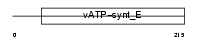
|
|||||
| Description | Vacuolar ATP synthase subunit E (EC 3.6.3.14) (V-ATPase E subunit) (Vacuolar proton pump E subunit) (V-ATPase 31 kDa subunit) (P31). | |||||
|
CCD25_MOUSE
|
||||||
| θ value | 4.03905 (rank : 27) | NC score | 0.028334 (rank : 23) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q78PG9, Q9CSQ8, Q9CUF8 | Gene names | Ccdc25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 25. | |||||
|
GAS8_HUMAN
|
||||||
| θ value | 4.03905 (rank : 28) | NC score | 0.017455 (rank : 32) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 819 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O95995 | Gene names | GAS8, GAS11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Growth-arrest-specific protein 8 (Growth arrest-specific 11). | |||||
|
CNGB1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 29) | NC score | 0.009943 (rank : 40) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14028, Q14029 | Gene names | CNGB1, CNCG4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel 4 (CNG channel 4) (CNG-4) (CNG4) (Cyclic nucleotide-gated cation channel modulatory subunit). | |||||
|
IF3A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 30) | NC score | 0.020948 (rank : 28) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 875 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q14152, O00653 | Gene names | EIF3S10, KIAA0139 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 10 (eIF-3 theta) (eIF3 p167) (eIF3 p180) (eIF3 p185) (eIF3a). | |||||
|
IF3A_MOUSE
|
||||||
| θ value | 5.27518 (rank : 31) | NC score | 0.021503 (rank : 27) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 871 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P23116, Q60697, Q62162 | Gene names | Eif3s10, Csma, Eif3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 10 (eIF-3 theta) (eIF3 p167) (eIF3 p180) (eIF3 p185) (eIF3a) (p162 protein) (Centrosomin). | |||||
|
ITSN2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 32) | NC score | 0.011544 (rank : 37) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1371 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Z0R6, Q9Z0R5 | Gene names | Itsn2, Ese2, Sh3d1B | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (EH and SH3 domains protein 2) (EH domain and SH3 domain regulator of endocytosis 2). | |||||
|
KIFC3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 33) | NC score | 0.009659 (rank : 42) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 619 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9BVG8, O75299, Q8IUT3, Q96HW6 | Gene names | KIFC3 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIFC3. | |||||
|
STABP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 34) | NC score | 0.018452 (rank : 31) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95630, Q3MJE7 | Gene names | STAMBP, AMSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STAM-binding protein (EC 3.1.2.15) (Associated molecule with the SH3 domain of STAM). | |||||
|
SWP70_MOUSE
|
||||||
| θ value | 5.27518 (rank : 35) | NC score | 0.015643 (rank : 33) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1097 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6A028, O88443, Q3TQR6, Q3UCA3, Q6P1D0 | Gene names | Swap70, Kiaa0640 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Switch-associated protein 70 (SWAP-70). | |||||
|
VASP_MOUSE
|
||||||
| θ value | 5.27518 (rank : 36) | NC score | 0.012568 (rank : 35) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P70460, Q3TAP0, Q3TCD2, Q3U0C2, Q3UDF1, Q91VD2, Q9R214 | Gene names | Vasp | |||
|
Domain Architecture |

|
|||||
| Description | Vasodilator-stimulated phosphoprotein (VASP). | |||||
|
ZN326_HUMAN
|
||||||
| θ value | 5.27518 (rank : 37) | NC score | 0.032576 (rank : 20) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5BKZ1, Q5VW93, Q5VW94, Q5VW96, Q5VW97, Q6NSA2, Q7Z638, Q7Z6C2 | Gene names | ZNF326 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 326. | |||||
|
ZN326_MOUSE
|
||||||
| θ value | 5.27518 (rank : 38) | NC score | 0.031977 (rank : 21) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O88291, Q8BSJ5, Q8K1X9, Q9CYG9 | Gene names | Znf326, Zan75, Zfp326 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 326 (Zinc finger protein-associated with nuclear matrix of 75 kDa). | |||||
|
ACHB3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 39) | NC score | 0.004300 (rank : 49) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q05901, Q15827 | Gene names | CHRNB3 | |||
|
Domain Architecture |
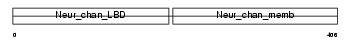
|
|||||
| Description | Neuronal acetylcholine receptor protein subunit beta-3 precursor. | |||||
|
GBP2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 40) | NC score | 0.009775 (rank : 41) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P32456, Q86TB0 | Gene names | GBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced guanylate-binding protein 2 (GTP-binding protein 2) (Guanine nucleotide-binding protein 2) (GBP-2) (HuGBP-2). | |||||
|
NEGR1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 41) | NC score | 0.008492 (rank : 44) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80Z24, Q3UHQ8, Q80T70 | Gene names | Negr1, Kiaa3001, Ntra | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal growth regulator 1 precursor (Kilon protein) (Kindred of IgLON) (Neurotractin). | |||||
|
AT10D_HUMAN
|
||||||
| θ value | 8.99809 (rank : 42) | NC score | 0.004614 (rank : 47) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9P241, Q8NC70, Q96SR3 | Gene names | ATP10D, KIAA1487 | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase VD (EC 3.6.3.1) (ATPVD). | |||||
|
HS90A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 43) | NC score | 0.009110 (rank : 43) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P07900, Q2PP14, Q9BVQ5 | Gene names | HSP90AA1, HSP90A, HSPC1, HSPCA | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock protein HSP 90-alpha (HSP 86) (NY-REN-38 antigen). | |||||
|
ITSN2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 44) | NC score | 0.010876 (rank : 38) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1173 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NZM3, O95062, Q15812, Q9HAK4, Q9NXE6, Q9NYG0, Q9NZM2, Q9ULG4 | Gene names | ITSN2, KIAA1256, SH3D1B, SWAP | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (SH3P18) (SH3P18-like WASP-associated protein). | |||||
|
MAT1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 45) | NC score | 0.014863 (rank : 34) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P51949, Q3TZP0, Q9D8A0, Q9D8D2 | Gene names | Mnat1, Mat1 | |||
|
Domain Architecture |
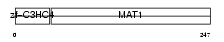
|
|||||
| Description | CDK-activating kinase assembly factor MAT1 (RING finger protein MAT1) (Menage a trois) (CDK7/cyclin H assembly factor) (p36) (p35). | |||||
|
NEGR1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 46) | NC score | 0.005775 (rank : 46) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7Z3B1, Q5VT21, Q6UY06, Q8NAQ3 | Gene names | NEGR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal growth regulator 1 precursor. | |||||
|
TAF3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 47) | NC score | 0.010674 (rank : 39) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 463 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5HZG4, Q3U490, Q3UWX2, Q8BIU8, Q99JH4 | Gene names | Taf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
THIOM_HUMAN
|
||||||
| θ value | 8.99809 (rank : 48) | NC score | 0.063001 (rank : 15) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99757, Q5JZA0, Q9UH29 | Gene names | TXN2, TRX2 | |||
|
Domain Architecture |
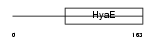
|
|||||
| Description | Thioredoxin, mitochondrial precursor (Mt-Trx) (MTRX) (Thioredoxin-2). | |||||
|
THIOM_MOUSE
|
||||||
| θ value | 8.99809 (rank : 49) | NC score | 0.062862 (rank : 16) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P97493, Q545D5 | Gene names | Txn2 | |||
|
Domain Architecture |
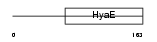
|
|||||
| Description | Thioredoxin, mitochondrial precursor (Mt-Trx) (MTRX) (Thioredoxin-2). | |||||
|
PDCL2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 6.63862e-128 (rank : 1) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8N4E4 | Gene names | PDCL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosducin-like protein 2. | |||||
|
PDCL2_MOUSE
|
||||||
| NC score | 0.992038 (rank : 2) | θ value | 6.24247e-110 (rank : 2) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q78Y63 | Gene names | Pdcl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosducin-like protein 2. | |||||
|
PDCL3_MOUSE
|
||||||
| NC score | 0.969714 (rank : 3) | θ value | 4.23606e-66 (rank : 4) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BVF2, Q3TH06, Q99JX2, Q9D0W3 | Gene names | Pdcl3, Viaf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosducin-like protein 3 (Viral IAP-associated factor 1) (VIAF-1). | |||||
|
PDCL3_HUMAN
|
||||||
| NC score | 0.967221 (rank : 4) | θ value | 2.03155e-68 (rank : 3) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9H2J4 | Gene names | PDCL3, VIAF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosducin-like protein 3 (Viral IAP-associated factor 1) (VIAF-1) (HTPHLP). | |||||
|
PHLP_HUMAN
|
||||||
| NC score | 0.630551 (rank : 5) | θ value | 1.86753e-13 (rank : 5) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q13371, Q96AF1, Q9UEW7, Q9UFL0 | Gene names | PDCL | |||
|
Domain Architecture |

|
|||||
| Description | Phosducin-like protein (PHLP). | |||||
|
PHLP_MOUSE
|
||||||
| NC score | 0.617909 (rank : 6) | θ value | 4.59992e-12 (rank : 6) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9DBX2, Q3TKI0 | Gene names | Pdcl | |||
|
Domain Architecture |

|
|||||
| Description | Phosducin-like protein (PHLP). | |||||
|
PHOS_HUMAN
|
||||||
| NC score | 0.602501 (rank : 7) | θ value | 7.34386e-10 (rank : 8) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P20941, Q14816 | Gene names | PDC | |||
|
Domain Architecture |

|
|||||
| Description | Phosducin (PHD) (33 kDa phototransducing protein) (MEKA protein). | |||||
|
PHOS_MOUSE
|
||||||
| NC score | 0.589145 (rank : 8) | θ value | 6.21693e-09 (rank : 9) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9QW08 | Gene names | Pdc, Rpr1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosducin (PHD) (33 kDa phototransducing protein) (Rod photoreceptor 1) (RPR-1). | |||||
|
TXND9_HUMAN
|
||||||
| NC score | 0.547257 (rank : 9) | θ value | 3.29651e-10 (rank : 7) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O14530, Q53RV8, Q6NSF5, Q9BRU6 | Gene names | TXNDC9, APACD | |||
|
Domain Architecture |

|
|||||
| Description | Thioredoxin domain-containing protein 9 (Protein 1-4) (ATP-binding protein associated with cell differentiation). | |||||
|
TXND9_MOUSE
|
||||||
| NC score | 0.527619 (rank : 10) | θ value | 4.0297e-08 (rank : 10) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9CQ79, Q3TKD2 | Gene names | Txndc9, Apacd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 9 (ATP-binding protein associated with cell differentiation). | |||||
|
TXNL2_MOUSE
|
||||||
| NC score | 0.208640 (rank : 11) | θ value | 0.0148317 (rank : 11) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9CQM9, Q9JLZ2 | Gene names | Txnl2, Picot | |||
|
Domain Architecture |

|
|||||
| Description | Thioredoxin-like protein 2 (PKC-interacting cousin of thioredoxin) (PKC-theta-interacting protein) (PKCq-interacting protein). | |||||
|
TXNL2_HUMAN
|
||||||
| NC score | 0.187440 (rank : 12) | θ value | 0.0563607 (rank : 12) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O76003, Q96CE0, Q9P1B0, Q9P1B1 | Gene names | TXNL2, PICOT | |||
|
Domain Architecture |

|
|||||
| Description | Thioredoxin-like protein 2 (PKC-interacting cousin of thioredoxin) (PKC-theta-interacting protein) (PKCq-interacting protein). | |||||
|
THIO_HUMAN
|
||||||
| NC score | 0.071769 (rank : 13) | θ value | 1.81305 (rank : 20) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P10599, Q96KI3 | Gene names | TXN, TRDX, TRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Thioredoxin (Trx) (ATL-derived factor) (ADF) (Surface-associated sulphydryl protein) (SASP). | |||||
|
THIO_MOUSE
|
||||||
| NC score | 0.069774 (rank : 14) | θ value | 2.36792 (rank : 22) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P10639, Q52KC4, Q9D8R0 | Gene names | Txn, Txn1 | |||
|
Domain Architecture |

|
|||||
| Description | Thioredoxin (Trx) (ATL-derived factor) (ADF). | |||||
|
THIOM_HUMAN
|
||||||
| NC score | 0.063001 (rank : 15) | θ value | 8.99809 (rank : 48) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99757, Q5JZA0, Q9UH29 | Gene names | TXN2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Thioredoxin, mitochondrial precursor (Mt-Trx) (MTRX) (Thioredoxin-2). | |||||
|
THIOM_MOUSE
|
||||||
| NC score | 0.062862 (rank : 16) | θ value | 8.99809 (rank : 49) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P97493, Q545D5 | Gene names | Txn2 | |||
|
Domain Architecture |

|
|||||
| Description | Thioredoxin, mitochondrial precursor (Mt-Trx) (MTRX) (Thioredoxin-2). | |||||
|
K0831_MOUSE
|
||||||
| NC score | 0.051086 (rank : 17) | θ value | 1.06291 (rank : 16) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8CDJ3, Q69ZY1, Q6PFY6, Q8C6N0, Q8R3M3 | Gene names | Kiaa0831, D14Ertd436e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0831. | |||||
|
CD008_HUMAN
|
||||||
| NC score | 0.048217 (rank : 18) | θ value | 1.81305 (rank : 19) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P78312, O43607, P78311, P78313, Q9UEG8 | Gene names | C4orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C4orf8 (Protein IT14). | |||||
|
GLE1_HUMAN
|
||||||
| NC score | 0.047170 (rank : 19) | θ value | 1.06291 (rank : 15) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q53GS7, O75458, Q53GT9, Q5VVU1, Q8NCP6, Q9UFL6 | Gene names | GLE1L, GLE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin GLE1 (GLE1-like protein) (hGLE1). | |||||
|
ZN326_HUMAN
|
||||||
| NC score | 0.032576 (rank : 20) | θ value | 5.27518 (rank : 37) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5BKZ1, Q5VW93, Q5VW94, Q5VW96, Q5VW97, Q6NSA2, Q7Z638, Q7Z6C2 | Gene names | ZNF326 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 326. | |||||
|
ZN326_MOUSE
|
||||||
| NC score | 0.031977 (rank : 21) | θ value | 5.27518 (rank : 38) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O88291, Q8BSJ5, Q8K1X9, Q9CYG9 | Gene names | Znf326, Zan75, Zfp326 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 326 (Zinc finger protein-associated with nuclear matrix of 75 kDa). | |||||
|
VATE_MOUSE
|
||||||
| NC score | 0.029842 (rank : 22) | θ value | 3.0926 (rank : 26) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P50518 | Gene names | Atp6v1e1, Atp6e | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar ATP synthase subunit E (EC 3.6.3.14) (V-ATPase E subunit) (Vacuolar proton pump E subunit) (V-ATPase 31 kDa subunit) (P31). | |||||
|
CCD25_MOUSE
|
||||||
| NC score | 0.028334 (rank : 23) | θ value | 4.03905 (rank : 27) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q78PG9, Q9CSQ8, Q9CUF8 | Gene names | Ccdc25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 25. | |||||
|
IF2P_HUMAN
|
||||||
| NC score | 0.025247 (rank : 24) | θ value | 1.38821 (rank : 18) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O60841, O95805, Q9UF81, Q9UMN7 | Gene names | EIF5B, IF2, KIAA0741 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5B (eIF-5B) (Translation initiation factor IF-2). | |||||
|
RB6I2_HUMAN
|
||||||
| NC score | 0.023569 (rank : 25) | θ value | 0.163984 (rank : 13) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1584 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8IUD2, Q6NVK2, Q8IUD3, Q8IUD4, Q8IUD5, Q8NAS1, Q9NXN5, Q9UIK7, Q9UPS1 | Gene names | ERC1, ELKS, KIAA1081, RAB6IP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1). | |||||
|
UBP8_HUMAN
|
||||||
| NC score | 0.021711 (rank : 26) | θ value | 0.47712 (rank : 14) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 751 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P40818, Q7Z3U2, Q86VA0, Q8IWI7 | Gene names | USP8, KIAA0055, UBPY | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 8 (EC 3.1.2.15) (Ubiquitin thioesterase 8) (Ubiquitin-specific-processing protease 8) (Deubiquitinating enzyme 8) (hUBPy). | |||||
|
IF3A_MOUSE
|
||||||
| NC score | 0.021503 (rank : 27) | θ value | 5.27518 (rank : 31) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 871 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P23116, Q60697, Q62162 | Gene names | Eif3s10, Csma, Eif3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 10 (eIF-3 theta) (eIF3 p167) (eIF3 p180) (eIF3 p185) (eIF3a) (p162 protein) (Centrosomin). | |||||
|
IF3A_HUMAN
|
||||||
| NC score | 0.020948 (rank : 28) | θ value | 5.27518 (rank : 30) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 875 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q14152, O00653 | Gene names | EIF3S10, KIAA0139 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 10 (eIF-3 theta) (eIF3 p167) (eIF3 p180) (eIF3 p185) (eIF3a). | |||||
|
DYXC1_MOUSE
|
||||||
| NC score | 0.020946 (rank : 29) | θ value | 2.36792 (rank : 21) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R368 | Gene names | Dyx1c1, Ekn1 | |||
|
Domain Architecture |

|
|||||
| Description | Dyslexia susceptibility 1 candidate gene 1 protein homolog. | |||||
|
RB6I2_MOUSE
|
||||||
| NC score | 0.018723 (rank : 30) | θ value | 3.0926 (rank : 24) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q99MI1, Q80TK7, Q8BPL1, Q8C7Y1, Q99MI2 | Gene names | Erc1, Cast2, Kiaa1081, Rab6ip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1) (ERC1) (CAZ-associated structural protein 2) (CAST2). | |||||
|
STABP_HUMAN
|
||||||
| NC score | 0.018452 (rank : 31) | θ value | 5.27518 (rank : 34) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95630, Q3MJE7 | Gene names | STAMBP, AMSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STAM-binding protein (EC 3.1.2.15) (Associated molecule with the SH3 domain of STAM). | |||||
|
GAS8_HUMAN
|
||||||
| NC score | 0.017455 (rank : 32) | θ value | 4.03905 (rank : 28) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 819 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O95995 | Gene names | GAS8, GAS11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Growth-arrest-specific protein 8 (Growth arrest-specific 11). | |||||
|
SWP70_MOUSE
|
||||||
| NC score | 0.015643 (rank : 33) | θ value | 5.27518 (rank : 35) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1097 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6A028, O88443, Q3TQR6, Q3UCA3, Q6P1D0 | Gene names | Swap70, Kiaa0640 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Switch-associated protein 70 (SWAP-70). | |||||
|
MAT1_MOUSE
|
||||||
| NC score | 0.014863 (rank : 34) | θ value | 8.99809 (rank : 45) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P51949, Q3TZP0, Q9D8A0, Q9D8D2 | Gene names | Mnat1, Mat1 | |||
|
Domain Architecture |

|
|||||
| Description | CDK-activating kinase assembly factor MAT1 (RING finger protein MAT1) (Menage a trois) (CDK7/cyclin H assembly factor) (p36) (p35). | |||||
|
VASP_MOUSE
|
||||||
| NC score | 0.012568 (rank : 35) | θ value | 5.27518 (rank : 36) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P70460, Q3TAP0, Q3TCD2, Q3U0C2, Q3UDF1, Q91VD2, Q9R214 | Gene names | Vasp | |||
|
Domain Architecture |

|
|||||
| Description | Vasodilator-stimulated phosphoprotein (VASP). | |||||
|
MYO10_HUMAN
|
||||||
| NC score | 0.012002 (rank : 36) | θ value | 1.06291 (rank : 17) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 920 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9HD67, Q9NYM7, Q9P110, Q9P111, Q9UHF6 | Gene names | MYO10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin X). | |||||
|
ITSN2_MOUSE
|
||||||
| NC score | 0.011544 (rank : 37) | θ value | 5.27518 (rank : 32) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1371 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Z0R6, Q9Z0R5 | Gene names | Itsn2, Ese2, Sh3d1B | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (EH and SH3 domains protein 2) (EH domain and SH3 domain regulator of endocytosis 2). | |||||
|
ITSN2_HUMAN
|
||||||
| NC score | 0.010876 (rank : 38) | θ value | 8.99809 (rank : 44) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1173 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NZM3, O95062, Q15812, Q9HAK4, Q9NXE6, Q9NYG0, Q9NZM2, Q9ULG4 | Gene names | ITSN2, KIAA1256, SH3D1B, SWAP | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (SH3P18) (SH3P18-like WASP-associated protein). | |||||
|
TAF3_MOUSE
|
||||||
| NC score | 0.010674 (rank : 39) | θ value | 8.99809 (rank : 47) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 463 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5HZG4, Q3U490, Q3UWX2, Q8BIU8, Q99JH4 | Gene names | Taf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
CNGB1_HUMAN
|
||||||
| NC score | 0.009943 (rank : 40) | θ value | 5.27518 (rank : 29) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14028, Q14029 | Gene names | CNGB1, CNCG4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel 4 (CNG channel 4) (CNG-4) (CNG4) (Cyclic nucleotide-gated cation channel modulatory subunit). | |||||
|
GBP2_HUMAN
|
||||||
| NC score | 0.009775 (rank : 41) | θ value | 6.88961 (rank : 40) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P32456, Q86TB0 | Gene names | GBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced guanylate-binding protein 2 (GTP-binding protein 2) (Guanine nucleotide-binding protein 2) (GBP-2) (HuGBP-2). | |||||
|
KIFC3_HUMAN
|
||||||
| NC score | 0.009659 (rank : 42) | θ value | 5.27518 (rank : 33) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 619 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9BVG8, O75299, Q8IUT3, Q96HW6 | Gene names | KIFC3 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIFC3. | |||||
|
HS90A_HUMAN
|
||||||
| NC score | 0.009110 (rank : 43) | θ value | 8.99809 (rank : 43) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P07900, Q2PP14, Q9BVQ5 | Gene names | HSP90AA1, HSP90A, HSPC1, HSPCA | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock protein HSP 90-alpha (HSP 86) (NY-REN-38 antigen). | |||||
|
NEGR1_MOUSE
|
||||||
| NC score | 0.008492 (rank : 44) | θ value | 6.88961 (rank : 41) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80Z24, Q3UHQ8, Q80T70 | Gene names | Negr1, Kiaa3001, Ntra | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal growth regulator 1 precursor (Kilon protein) (Kindred of IgLON) (Neurotractin). | |||||
|
NFM_HUMAN
|
||||||
| NC score | 0.006790 (rank : 45) | θ value | 3.0926 (rank : 23) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
NEGR1_HUMAN
|
||||||
| NC score | 0.005775 (rank : 46) | θ value | 8.99809 (rank : 46) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7Z3B1, Q5VT21, Q6UY06, Q8NAQ3 | Gene names | NEGR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal growth regulator 1 precursor. | |||||
|
AT10D_HUMAN
|
||||||
| NC score | 0.004614 (rank : 47) | θ value | 8.99809 (rank : 42) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9P241, Q8NC70, Q96SR3 | Gene names | ATP10D, KIAA1487 | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase VD (EC 3.6.3.1) (ATPVD). | |||||
|
SLK_HUMAN
|
||||||
| NC score | 0.004449 (rank : 48) | θ value | 3.0926 (rank : 25) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1775 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9H2G2, O00211, Q6P1Z4, Q86WU7, Q86WW1, Q92603, Q9NQL0, Q9NQL1 | Gene names | SLK, KIAA0204, STK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (hSLK) (Serine/threonine-protein kinase 2) (CTCL tumor antigen se20-9). | |||||
|
ACHB3_HUMAN
|
||||||
| NC score | 0.004300 (rank : 49) | θ value | 6.88961 (rank : 39) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q05901, Q15827 | Gene names | CHRNB3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal acetylcholine receptor protein subunit beta-3 precursor. | |||||