Please be patient as the page loads
|
CSR2B_HUMAN
|
||||||
| SwissProt Accessions | Q9H8E8, Q96GW6, Q96IH3, Q9HBF0, Q9UIY5 | Gene names | CSRP2BP | |||
|
Domain Architecture |
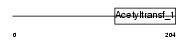
|
|||||
| Description | Cysteine-rich protein 2-binding protein (CSRP2-binding protein) (CRP2BP) (CRP2-binding partner). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CSR2B_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9H8E8, Q96GW6, Q96IH3, Q9HBF0, Q9UIY5 | Gene names | CSRP2BP | |||
|
Domain Architecture |

|
|||||
| Description | Cysteine-rich protein 2-binding protein (CSRP2-binding protein) (CRP2BP) (CRP2-binding partner). | |||||
|
MTF2_MOUSE
|
||||||
| θ value | 8.11959e-09 (rank : 2) | NC score | 0.376340 (rank : 2) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q02395, Q569Z8, Q6PG89, Q8BGP9, Q8BSJ7 | Gene names | Mtf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metal-response element-binding transcription factor 2 (Zinc-regulated factor 1) (ZiRF1) (Metal-response element DNA-binding protein M96) (Metal-regulatory transcription factor 2) (PCL2). | |||||
|
MTF2_HUMAN
|
||||||
| θ value | 4.0297e-08 (rank : 3) | NC score | 0.372821 (rank : 3) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y483, Q9UES9, Q9UP40 | Gene names | MTF2 | |||
|
Domain Architecture |
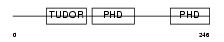
|
|||||
| Description | Metal-response element-binding transcription factor 2 (Metal-response element DNA-binding protein M96) (Metal-regulatory transcription factor 2) (PCL2). | |||||
|
PHF1_HUMAN
|
||||||
| θ value | 3.19293e-05 (rank : 4) | NC score | 0.339481 (rank : 4) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O43189, O60929, Q96KM7 | Gene names | PHF1 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 1 (Protein PHF1). | |||||
|
PHF1_MOUSE
|
||||||
| θ value | 3.19293e-05 (rank : 5) | NC score | 0.338312 (rank : 5) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Z1B8, O54808 | Gene names | Phf1, Tctex-3, Tctex3 | |||
|
Domain Architecture |
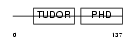
|
|||||
| Description | PHD finger protein 1 (Protein PHF1) (T-complex testis-expressed 3) (Polycomblike 1) (mPCl1). | |||||
|
ASH2L_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 6) | NC score | 0.165476 (rank : 6) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91X20, Q3UIF9, Q9Z2X4 | Gene names | Ash2l | |||
|
Domain Architecture |

|
|||||
| Description | Set1/Ash2 histone methyltransferase complex subunit ASH2 (ASH2-like protein). | |||||
|
ASH2L_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 7) | NC score | 0.161939 (rank : 7) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UBL3, O60659, O60660, Q96B62 | Gene names | ASH2L | |||
|
Domain Architecture |

|
|||||
| Description | Set1/Ash2 histone methyltransferase complex subunit ASH2 (ASH2-like protein). | |||||
|
TP53B_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 8) | NC score | 0.069158 (rank : 11) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
FARP2_HUMAN
|
||||||
| θ value | 0.163984 (rank : 9) | NC score | 0.029484 (rank : 23) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O94887, Q53QM5, Q8WU27, Q9UFE7 | Gene names | FARP2, KIAA0793 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 2 (FERM domain including RhoGEF) (FIR). | |||||
|
RL1D1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 10) | NC score | 0.092115 (rank : 8) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O76021, Q6PL22, Q8IWS7, Q8WUZ1, Q9HDA9, Q9Y3Z9 | Gene names | RSL1D1, CATX11, CSIG, PBK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribosomal L1 domain-containing protein 1 (Cellular senescence- inhibited gene protein) (Protein PBK1) (CATX-11). | |||||
|
SYBU_MOUSE
|
||||||
| θ value | 0.365318 (rank : 11) | NC score | 0.060351 (rank : 14) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BHS8, Q3TQN7, Q401F3, Q401F4, Q571C1, Q6P1J2, Q80XH0, Q8BHS7, Q8BI27 | Gene names | Sybu, Golsyn, Kiaa1472 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Syntabulin (Syntaxin-1-binding protein) (Golgi-localized syntaphilin- related protein) (m-Golsyn). | |||||
|
EVI1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 12) | NC score | 0.007497 (rank : 33) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 924 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q03112, Q16122, Q99917 | Gene names | EVI1 | |||
|
Domain Architecture |

|
|||||
| Description | Ecotropic virus integration 1 site protein. | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 13) | NC score | 0.037073 (rank : 20) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
NIP30_MOUSE
|
||||||
| θ value | 1.38821 (rank : 14) | NC score | 0.074539 (rank : 9) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91WE2, Q8R0C9 | Gene names | Nip30 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NEFA-interacting nuclear protein NIP30. | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 15) | NC score | 0.037150 (rank : 19) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 1.81305 (rank : 16) | NC score | 0.031413 (rank : 21) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
SMG1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 17) | NC score | 0.029784 (rank : 22) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96Q15, O43305, Q13284, Q8NFX2, Q96QV0, Q96RW3 | Gene names | SMG1, ATX, KIAA0421, LIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SMG1 (EC 2.7.11.1) (SMG-1) (hSMG-1) (Lambda/iota protein kinase C-interacting protein) (Lambda-interacting protein) (61E3.4). | |||||
|
SF3B1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 18) | NC score | 0.052602 (rank : 15) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75533 | Gene names | SF3B1, SAP155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
SF3B1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 19) | NC score | 0.052550 (rank : 16) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99NB9, Q9CSK5 | Gene names | Sf3b1, Sap155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
NIP30_HUMAN
|
||||||
| θ value | 3.0926 (rank : 20) | NC score | 0.064804 (rank : 12) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9GZU8 | Gene names | NIP30 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NEFA-interacting nuclear protein NIP30. | |||||
|
ZN435_HUMAN
|
||||||
| θ value | 3.0926 (rank : 21) | NC score | 0.004894 (rank : 34) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 774 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H4T2, Q9H6K2 | Gene names | ZNF435, ZSCAN16 | |||
|
Domain Architecture |
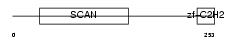
|
|||||
| Description | Zinc finger protein 435 (Zinc finger and SCAN domain-containing protein 16). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 4.03905 (rank : 22) | NC score | 0.028482 (rank : 25) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
CATA_MOUSE
|
||||||
| θ value | 5.27518 (rank : 23) | NC score | 0.028831 (rank : 24) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P24270 | Gene names | Cat, Cas-1, Cas1 | |||
|
Domain Architecture |

|
|||||
| Description | Catalase (EC 1.11.1.6). | |||||
|
KTN1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 24) | NC score | 0.008749 (rank : 31) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q86UP2, Q13999, Q14707, Q15387, Q86W57 | Gene names | KTN1, CG1, KIAA0004 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin (Kinesin receptor) (CG-1 antigen). | |||||
|
NAT5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 25) | NC score | 0.037813 (rank : 17) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P61599, Q5TFT7, Q9D7H8, Q9H0Y4, Q9NQH6, Q9Y6D2 | Gene names | NAT5 | |||
|
Domain Architecture |
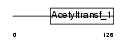
|
|||||
| Description | N-acetyltransferase 5 (EC 2.3.1.-). | |||||
|
NAT5_MOUSE
|
||||||
| θ value | 6.88961 (rank : 26) | NC score | 0.037813 (rank : 18) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P61600, Q14B28, Q4VAC3, Q9D7H8, Q9H0Y4, Q9NQH6, Q9Y6D2 | Gene names | Nat5 | |||
|
Domain Architecture |
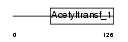
|
|||||
| Description | N-acetyltransferase 5 (EC 2.3.1.-). | |||||
|
NUP53_HUMAN
|
||||||
| θ value | 6.88961 (rank : 27) | NC score | 0.024808 (rank : 26) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NFH5, Q4ZFZ9, Q53S95, Q8TDJ1 | Gene names | NUP35, MP44, NUP53 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin NUP53 (Nuclear pore complex protein Nup53) (Nucleoporin Nup35) (35 kDa nucleoporin) (Mitotic phosphoprotein 44) (MP-44). | |||||
|
BCL9_HUMAN
|
||||||
| θ value | 8.99809 (rank : 28) | NC score | 0.012980 (rank : 30) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
MMGL_MOUSE
|
||||||
| θ value | 8.99809 (rank : 29) | NC score | 0.007653 (rank : 32) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 270 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P49300 | Gene names | Mgl1, Mgl | |||
|
Domain Architecture |
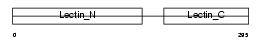
|
|||||
| Description | Macrophage asialoglycoprotein-binding protein 1 (M-ASGP-BP) (Macrophage galactose/N-acetylgalactosamine-specific lectin) (MMGL). | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | 8.99809 (rank : 30) | NC score | 0.019604 (rank : 28) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
NPBL_MOUSE
|
||||||
| θ value | 8.99809 (rank : 31) | NC score | 0.020986 (rank : 27) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
UBXD7_MOUSE
|
||||||
| θ value | 8.99809 (rank : 32) | NC score | 0.019460 (rank : 29) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6P5G6, Q6ZQ44 | Gene names | Ubxd7, Kiaa0794 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UBX domain-containing protein 7. | |||||
|
ZN213_HUMAN
|
||||||
| θ value | 8.99809 (rank : 33) | NC score | 0.002920 (rank : 36) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 755 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O14771, Q96IS1 | Gene names | ZNF213 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 213 (Putative transcription factor CR53). | |||||
|
ZN335_HUMAN
|
||||||
| θ value | 8.99809 (rank : 34) | NC score | 0.004371 (rank : 35) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 973 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H4Z2, Q9H684 | Gene names | ZNF335 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 335. | |||||
|
PHF22_HUMAN
|
||||||
| θ value | θ > 10 (rank : 35) | NC score | 0.063491 (rank : 13) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96CB8, Q9HD71 | Gene names | INTS12, PHF22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Integrator complex subunit 12 (PHD finger protein 22). | |||||
|
PHF22_MOUSE
|
||||||
| θ value | θ > 10 (rank : 36) | NC score | 0.072620 (rank : 10) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D168, Q3U2Q5, Q921U2 | Gene names | Ints12, Phf22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Integrator complex subunit 12 (PHD finger protein 22). | |||||
|
CSR2B_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9H8E8, Q96GW6, Q96IH3, Q9HBF0, Q9UIY5 | Gene names | CSRP2BP | |||
|
Domain Architecture |

|
|||||
| Description | Cysteine-rich protein 2-binding protein (CSRP2-binding protein) (CRP2BP) (CRP2-binding partner). | |||||
|
MTF2_MOUSE
|
||||||
| NC score | 0.376340 (rank : 2) | θ value | 8.11959e-09 (rank : 2) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q02395, Q569Z8, Q6PG89, Q8BGP9, Q8BSJ7 | Gene names | Mtf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metal-response element-binding transcription factor 2 (Zinc-regulated factor 1) (ZiRF1) (Metal-response element DNA-binding protein M96) (Metal-regulatory transcription factor 2) (PCL2). | |||||
|
MTF2_HUMAN
|
||||||
| NC score | 0.372821 (rank : 3) | θ value | 4.0297e-08 (rank : 3) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y483, Q9UES9, Q9UP40 | Gene names | MTF2 | |||
|
Domain Architecture |

|
|||||
| Description | Metal-response element-binding transcription factor 2 (Metal-response element DNA-binding protein M96) (Metal-regulatory transcription factor 2) (PCL2). | |||||
|
PHF1_HUMAN
|
||||||
| NC score | 0.339481 (rank : 4) | θ value | 3.19293e-05 (rank : 4) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O43189, O60929, Q96KM7 | Gene names | PHF1 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 1 (Protein PHF1). | |||||
|
PHF1_MOUSE
|
||||||
| NC score | 0.338312 (rank : 5) | θ value | 3.19293e-05 (rank : 5) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Z1B8, O54808 | Gene names | Phf1, Tctex-3, Tctex3 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 1 (Protein PHF1) (T-complex testis-expressed 3) (Polycomblike 1) (mPCl1). | |||||
|
ASH2L_MOUSE
|
||||||
| NC score | 0.165476 (rank : 6) | θ value | 0.0113563 (rank : 6) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91X20, Q3UIF9, Q9Z2X4 | Gene names | Ash2l | |||
|
Domain Architecture |

|
|||||
| Description | Set1/Ash2 histone methyltransferase complex subunit ASH2 (ASH2-like protein). | |||||
|
ASH2L_HUMAN
|
||||||
| NC score | 0.161939 (rank : 7) | θ value | 0.0193708 (rank : 7) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UBL3, O60659, O60660, Q96B62 | Gene names | ASH2L | |||
|
Domain Architecture |

|
|||||
| Description | Set1/Ash2 histone methyltransferase complex subunit ASH2 (ASH2-like protein). | |||||
|
RL1D1_HUMAN
|
||||||
| NC score | 0.092115 (rank : 8) | θ value | 0.279714 (rank : 10) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O76021, Q6PL22, Q8IWS7, Q8WUZ1, Q9HDA9, Q9Y3Z9 | Gene names | RSL1D1, CATX11, CSIG, PBK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribosomal L1 domain-containing protein 1 (Cellular senescence- inhibited gene protein) (Protein PBK1) (CATX-11). | |||||
|
NIP30_MOUSE
|
||||||
| NC score | 0.074539 (rank : 9) | θ value | 1.38821 (rank : 14) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91WE2, Q8R0C9 | Gene names | Nip30 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NEFA-interacting nuclear protein NIP30. | |||||
|
PHF22_MOUSE
|
||||||
| NC score | 0.072620 (rank : 10) | θ value | θ > 10 (rank : 36) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D168, Q3U2Q5, Q921U2 | Gene names | Ints12, Phf22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Integrator complex subunit 12 (PHD finger protein 22). | |||||
|
TP53B_MOUSE
|
||||||
| NC score | 0.069158 (rank : 11) | θ value | 0.0736092 (rank : 8) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
NIP30_HUMAN
|
||||||
| NC score | 0.064804 (rank : 12) | θ value | 3.0926 (rank : 20) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9GZU8 | Gene names | NIP30 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NEFA-interacting nuclear protein NIP30. | |||||
|
PHF22_HUMAN
|
||||||
| NC score | 0.063491 (rank : 13) | θ value | θ > 10 (rank : 35) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96CB8, Q9HD71 | Gene names | INTS12, PHF22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Integrator complex subunit 12 (PHD finger protein 22). | |||||
|
SYBU_MOUSE
|
||||||
| NC score | 0.060351 (rank : 14) | θ value | 0.365318 (rank : 11) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BHS8, Q3TQN7, Q401F3, Q401F4, Q571C1, Q6P1J2, Q80XH0, Q8BHS7, Q8BI27 | Gene names | Sybu, Golsyn, Kiaa1472 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Syntabulin (Syntaxin-1-binding protein) (Golgi-localized syntaphilin- related protein) (m-Golsyn). | |||||
|
SF3B1_HUMAN
|
||||||
| NC score | 0.052602 (rank : 15) | θ value | 2.36792 (rank : 18) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75533 | Gene names | SF3B1, SAP155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
SF3B1_MOUSE
|
||||||
| NC score | 0.052550 (rank : 16) | θ value | 2.36792 (rank : 19) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99NB9, Q9CSK5 | Gene names | Sf3b1, Sap155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
NAT5_HUMAN
|
||||||
| NC score | 0.037813 (rank : 17) | θ value | 6.88961 (rank : 25) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P61599, Q5TFT7, Q9D7H8, Q9H0Y4, Q9NQH6, Q9Y6D2 | Gene names | NAT5 | |||
|
Domain Architecture |

|
|||||
| Description | N-acetyltransferase 5 (EC 2.3.1.-). | |||||
|
NAT5_MOUSE
|
||||||
| NC score | 0.037813 (rank : 18) | θ value | 6.88961 (rank : 26) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P61600, Q14B28, Q4VAC3, Q9D7H8, Q9H0Y4, Q9NQH6, Q9Y6D2 | Gene names | Nat5 | |||
|
Domain Architecture |

|
|||||
| Description | N-acetyltransferase 5 (EC 2.3.1.-). | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.037150 (rank : 19) | θ value | 1.81305 (rank : 15) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.037073 (rank : 20) | θ value | 1.38821 (rank : 13) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.031413 (rank : 21) | θ value | 1.81305 (rank : 16) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
SMG1_HUMAN
|
||||||
| NC score | 0.029784 (rank : 22) | θ value | 1.81305 (rank : 17) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96Q15, O43305, Q13284, Q8NFX2, Q96QV0, Q96RW3 | Gene names | SMG1, ATX, KIAA0421, LIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SMG1 (EC 2.7.11.1) (SMG-1) (hSMG-1) (Lambda/iota protein kinase C-interacting protein) (Lambda-interacting protein) (61E3.4). | |||||
|
FARP2_HUMAN
|
||||||
| NC score | 0.029484 (rank : 23) | θ value | 0.163984 (rank : 9) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O94887, Q53QM5, Q8WU27, Q9UFE7 | Gene names | FARP2, KIAA0793 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 2 (FERM domain including RhoGEF) (FIR). | |||||
|
CATA_MOUSE
|
||||||
| NC score | 0.028831 (rank : 24) | θ value | 5.27518 (rank : 23) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P24270 | Gene names | Cat, Cas-1, Cas1 | |||
|
Domain Architecture |

|
|||||
| Description | Catalase (EC 1.11.1.6). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.028482 (rank : 25) | θ value | 4.03905 (rank : 22) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
NUP53_HUMAN
|
||||||
| NC score | 0.024808 (rank : 26) | θ value | 6.88961 (rank : 27) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NFH5, Q4ZFZ9, Q53S95, Q8TDJ1 | Gene names | NUP35, MP44, NUP53 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin NUP53 (Nuclear pore complex protein Nup53) (Nucleoporin Nup35) (35 kDa nucleoporin) (Mitotic phosphoprotein 44) (MP-44). | |||||
|
NPBL_MOUSE
|
||||||
| NC score | 0.020986 (rank : 27) | θ value | 8.99809 (rank : 31) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.019604 (rank : 28) | θ value | 8.99809 (rank : 30) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
UBXD7_MOUSE
|
||||||
| NC score | 0.019460 (rank : 29) | θ value | 8.99809 (rank : 32) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6P5G6, Q6ZQ44 | Gene names | Ubxd7, Kiaa0794 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UBX domain-containing protein 7. | |||||
|
BCL9_HUMAN
|
||||||
| NC score | 0.012980 (rank : 30) | θ value | 8.99809 (rank : 28) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
KTN1_HUMAN
|
||||||
| NC score | 0.008749 (rank : 31) | θ value | 5.27518 (rank : 24) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q86UP2, Q13999, Q14707, Q15387, Q86W57 | Gene names | KTN1, CG1, KIAA0004 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin (Kinesin receptor) (CG-1 antigen). | |||||
|
MMGL_MOUSE
|
||||||
| NC score | 0.007653 (rank : 32) | θ value | 8.99809 (rank : 29) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 270 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P49300 | Gene names | Mgl1, Mgl | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage asialoglycoprotein-binding protein 1 (M-ASGP-BP) (Macrophage galactose/N-acetylgalactosamine-specific lectin) (MMGL). | |||||
|
EVI1_HUMAN
|
||||||
| NC score | 0.007497 (rank : 33) | θ value | 1.38821 (rank : 12) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 924 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q03112, Q16122, Q99917 | Gene names | EVI1 | |||
|
Domain Architecture |

|
|||||
| Description | Ecotropic virus integration 1 site protein. | |||||
|
ZN435_HUMAN
|
||||||
| NC score | 0.004894 (rank : 34) | θ value | 3.0926 (rank : 21) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 774 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H4T2, Q9H6K2 | Gene names | ZNF435, ZSCAN16 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 435 (Zinc finger and SCAN domain-containing protein 16). | |||||
|
ZN335_HUMAN
|
||||||
| NC score | 0.004371 (rank : 35) | θ value | 8.99809 (rank : 34) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 973 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H4Z2, Q9H684 | Gene names | ZNF335 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 335. | |||||
|
ZN213_HUMAN
|
||||||
| NC score | 0.002920 (rank : 36) | θ value | 8.99809 (rank : 33) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 755 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O14771, Q96IS1 | Gene names | ZNF213 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 213 (Putative transcription factor CR53). | |||||