Please be patient as the page loads
|
ACE_MOUSE
|
||||||
| SwissProt Accessions | P09470, Q6GTS2 | Gene names | Ace, Dcp1 | |||
|
Domain Architecture |

|
|||||
| Description | Angiotensin-converting enzyme, somatic isoform precursor (EC 3.4.15.1) (Dipeptidyl carboxypeptidase I) (Kininase II) [Contains: Angiotensin- converting enzyme, somatic isoform, soluble form]. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ACET_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.993175 (rank : 4) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P22966 | Gene names | ACE, DCP, DCP1 | |||
|
Domain Architecture |

|
|||||
| Description | Angiotensin-converting enzyme, testis-specific isoform precursor (EC 3.4.15.1) (EC 3.2.1.-) (ACE-T) (Dipeptidyl carboxypeptidase I) (Kininase II) [Contains: Angiotensin-converting enzyme, testis- specific isoform, soluble form]. | |||||
|
ACET_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.994691 (rank : 3) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P22967 | Gene names | Ace, Dcp1 | |||
|
Domain Architecture |

|
|||||
| Description | Angiotensin-converting enzyme, testis-specific isoform precursor (EC 3.4.15.1) (EC 3.2.1.-) (ACE-T) (Dipeptidyl carboxypeptidase I) (Kininase II) [Contains: Angiotensin-converting enzyme, testis- specific isoform, soluble form]. | |||||
|
ACE_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.997544 (rank : 2) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P12821, Q53YX9, Q59GY8 | Gene names | ACE, DCP, DCP1 | |||
|
Domain Architecture |

|
|||||
| Description | Angiotensin-converting enzyme, somatic isoform precursor (EC 3.4.15.1) (Dipeptidyl carboxypeptidase I) (Kininase II) (CD143 antigen) [Contains: Angiotensin-converting enzyme, somatic isoform, soluble form]. | |||||
|
ACE_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P09470, Q6GTS2 | Gene names | Ace, Dcp1 | |||
|
Domain Architecture |

|
|||||
| Description | Angiotensin-converting enzyme, somatic isoform precursor (EC 3.4.15.1) (Dipeptidyl carboxypeptidase I) (Kininase II) [Contains: Angiotensin- converting enzyme, somatic isoform, soluble form]. | |||||
|
ACE2_MOUSE
|
||||||
| θ value | 1.51984e-148 (rank : 5) | NC score | 0.920704 (rank : 5) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8R0I0, Q99N70, Q99N71 | Gene names | Ace2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiotensin-converting enzyme 2 precursor (EC 3.4.17.-) (ACE-related carboxypeptidase). | |||||
|
ACE2_HUMAN
|
||||||
| θ value | 4.57613e-145 (rank : 6) | NC score | 0.916493 (rank : 6) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BYF1, Q6UWP0, Q86WT0, Q9NRA7, Q9UFZ6 | Gene names | ACE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiotensin-converting enzyme 2 precursor (EC 3.4.17.-) (ACE-related carboxypeptidase) (Angiotensin-converting enzyme homolog) (ACEH). | |||||
|
SIN3B_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 7) | NC score | 0.051972 (rank : 9) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q62141, O54976, Q6A013, Q8VCB8, Q8VDZ5 | Gene names | Sin3b, Kiaa0700 | |||
|
Domain Architecture |
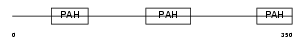
|
|||||
| Description | Paired amphipathic helix protein Sin3b (Transcriptional corepressor Sin3b) (Histone deacetylase complex subunit Sin3b). | |||||
|
SIN3B_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 8) | NC score | 0.049988 (rank : 10) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75182, Q68GC2, Q6P4B8, Q8TB34, Q9BSC8 | Gene names | SIN3B, KIAA0700 | |||
|
Domain Architecture |
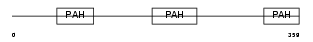
|
|||||
| Description | Paired amphipathic helix protein Sin3b (Transcriptional corepressor Sin3b) (Histone deacetylase complex subunit Sin3b). | |||||
|
CASP_MOUSE
|
||||||
| θ value | 1.06291 (rank : 9) | NC score | 0.015075 (rank : 16) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 1152 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P70403, Q7TQJ4, Q91WN6 | Gene names | Cutl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
CHIT1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 10) | NC score | 0.048960 (rank : 11) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9D7Q1, Q6QJD2 | Gene names | Chit1 | |||
|
Domain Architecture |
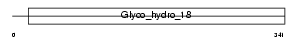
|
|||||
| Description | Chitotriosidase-1 precursor (EC 3.2.1.14) (Chitinase-1). | |||||
|
CUTL1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 11) | NC score | 0.015573 (rank : 15) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P53564, O08994, P70301, Q571L6, Q91ZD2 | Gene names | Cutl1, Cux, Cux1, Kiaa4047 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP) (Homeobox protein Cux). | |||||
|
KLH11_HUMAN
|
||||||
| θ value | 1.06291 (rank : 12) | NC score | 0.011108 (rank : 19) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NVR0 | Gene names | KLHL11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch-like protein 11 precursor. | |||||
|
SETD2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 13) | NC score | 0.017226 (rank : 14) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BYW2, O75397, O75405, Q17RW8, Q5BKS9, Q5QGN2, Q69YI5, Q6IN64, Q6ZN53, Q6ZS25, Q8N3R0, Q8TCN0, Q9C0D1, Q9H696, Q9NZW9 | Gene names | SETD2, HIF1, HYPB, KIAA1732, SET2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36-specific (EC 2.1.1.43) (SET domain-containing protein 2) (hSET2) (Huntingtin- interacting protein HYPB) (Huntingtin-interacting protein 1) (HIF-1) (p231HBP). | |||||
|
CASP_HUMAN
|
||||||
| θ value | 2.36792 (rank : 14) | NC score | 0.014279 (rank : 17) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q13948, Q53GU9, Q8TBS3 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
CHIT1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 15) | NC score | 0.021788 (rank : 12) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13231, Q3ZAR1, Q6ISC2, Q9H3V8 | Gene names | CHIT1 | |||
|
Domain Architecture |

|
|||||
| Description | Chitotriosidase-1 precursor (EC 3.2.1.14) (Chitinase-1). | |||||
|
CUTL1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 16) | NC score | 0.014142 (rank : 18) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
UACA_MOUSE
|
||||||
| θ value | 4.03905 (rank : 17) | NC score | 0.005403 (rank : 26) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8CGB3, Q69ZG3, Q7TN77, Q8BJC8 | Gene names | Uaca, Kiaa1561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uveal autoantigen with coiled-coil domains and ankyrin repeats protein (Nucling) (Nuclear membrane-binding protein). | |||||
|
NK3R_MOUSE
|
||||||
| θ value | 5.27518 (rank : 18) | NC score | 0.001733 (rank : 30) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 902 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P47937, Q61968, Q8BL44, Q9JKN0 | Gene names | Tacr3, Tac3r | |||
|
Domain Architecture |

|
|||||
| Description | Neuromedin K receptor (NKR) (Neurokinin B receptor) (NK-3 receptor) (NK-3R) (Tachykinin receptor 3). | |||||
|
PTN4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 19) | NC score | 0.004205 (rank : 29) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P29074 | Gene names | PTPN4 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 4 (EC 3.1.3.48) (Protein-tyrosine phosphatase MEG1) (PTPase-MEG1) (MEG). | |||||
|
SRBP1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 20) | NC score | 0.009579 (rank : 20) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P36956, Q16062 | Gene names | SREBF1, SREBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
DYH11_HUMAN
|
||||||
| θ value | 6.88961 (rank : 21) | NC score | 0.006464 (rank : 23) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96DT5, Q9UJ82 | Gene names | DNAH11 | |||
|
Domain Architecture |

|
|||||
| Description | Ciliary dynein heavy chain 11 (Axonemal beta dynein heavy chain 11). | |||||
|
GAN_HUMAN
|
||||||
| θ value | 6.88961 (rank : 22) | NC score | 0.004654 (rank : 28) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H2C0 | Gene names | GAN, GAN1, KLHL16 | |||
|
Domain Architecture |

|
|||||
| Description | Gigaxonin (Kelch-like protein 16). | |||||
|
GOGA2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 23) | NC score | 0.006460 (rank : 24) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q08379, Q9BRB0, Q9NYF9 | Gene names | GOLGA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgin subfamily A member 2 (Cis-Golgi matrix protein GM130) (Gm130 autoantigen) (Golgin-95). | |||||
|
CENPJ_HUMAN
|
||||||
| θ value | 8.99809 (rank : 24) | NC score | 0.020926 (rank : 13) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 721 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9HC77, Q5T6R5, Q96KS5, Q9C067 | Gene names | CENPJ, CPAP, LAP, LIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein J (CENP-J) (Centrosomal P4.1-associated protein) (LAG-3-associated protein) (LYST-interacting protein 1). | |||||
|
FRAP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 25) | NC score | 0.008798 (rank : 21) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P42345, Q6LE87, Q96QG3, Q9Y4I3 | Gene names | FRAP1, FRAP, FRAP2 | |||
|
Domain Architecture |

|
|||||
| Description | FKBP12-rapamycin complex-associated protein (FK506-binding protein 12- rapamycin complex-associated protein 1) (Rapamycin target protein) (RAPT1) (Mammalian target of rapamycin) (mTOR). | |||||
|
FRAP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 26) | NC score | 0.008795 (rank : 22) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JLN9, Q811J5, Q9CST1 | Gene names | Frap1, Frap | |||
|
Domain Architecture |

|
|||||
| Description | FKBP12-rapamycin complex-associated protein (FK506-binding protein 12- rapamycin complex-associated protein 1) (Rapamycin target protein) (RAPT1) (Mammalian target of rapamycin) (mTOR). | |||||
|
JADE3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 27) | NC score | 0.006405 (rank : 25) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6IE82, Q6ZQG2 | Gene names | Phf16, Jade3, Kiaa0215 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-3 (PHD finger protein 16). | |||||
|
UTRO_HUMAN
|
||||||
| θ value | 8.99809 (rank : 28) | NC score | 0.005378 (rank : 27) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P46939 | Gene names | UTRN, DMDL | |||
|
Domain Architecture |

|
|||||
| Description | Utrophin (Dystrophin-related protein 1) (DRP1) (DRP). | |||||
|
TMM27_HUMAN
|
||||||
| θ value | θ > 10 (rank : 29) | NC score | 0.205077 (rank : 8) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9HBJ8, Q6UW07 | Gene names | TMEM27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collectrin precursor (Transmembrane protein 27). | |||||
|
TMM27_MOUSE
|
||||||
| θ value | θ > 10 (rank : 30) | NC score | 0.229463 (rank : 7) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9ESG4, Q3UFF6 | Gene names | Tmem27, Nx17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collectrin precursor (Transmembrane protein 27). | |||||
|
ACE_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P09470, Q6GTS2 | Gene names | Ace, Dcp1 | |||
|
Domain Architecture |

|
|||||
| Description | Angiotensin-converting enzyme, somatic isoform precursor (EC 3.4.15.1) (Dipeptidyl carboxypeptidase I) (Kininase II) [Contains: Angiotensin- converting enzyme, somatic isoform, soluble form]. | |||||
|
ACE_HUMAN
|
||||||
| NC score | 0.997544 (rank : 2) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P12821, Q53YX9, Q59GY8 | Gene names | ACE, DCP, DCP1 | |||
|
Domain Architecture |

|
|||||
| Description | Angiotensin-converting enzyme, somatic isoform precursor (EC 3.4.15.1) (Dipeptidyl carboxypeptidase I) (Kininase II) (CD143 antigen) [Contains: Angiotensin-converting enzyme, somatic isoform, soluble form]. | |||||
|
ACET_MOUSE
|
||||||
| NC score | 0.994691 (rank : 3) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P22967 | Gene names | Ace, Dcp1 | |||
|
Domain Architecture |

|
|||||
| Description | Angiotensin-converting enzyme, testis-specific isoform precursor (EC 3.4.15.1) (EC 3.2.1.-) (ACE-T) (Dipeptidyl carboxypeptidase I) (Kininase II) [Contains: Angiotensin-converting enzyme, testis- specific isoform, soluble form]. | |||||
|
ACET_HUMAN
|
||||||
| NC score | 0.993175 (rank : 4) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P22966 | Gene names | ACE, DCP, DCP1 | |||
|
Domain Architecture |

|
|||||
| Description | Angiotensin-converting enzyme, testis-specific isoform precursor (EC 3.4.15.1) (EC 3.2.1.-) (ACE-T) (Dipeptidyl carboxypeptidase I) (Kininase II) [Contains: Angiotensin-converting enzyme, testis- specific isoform, soluble form]. | |||||
|
ACE2_MOUSE
|
||||||
| NC score | 0.920704 (rank : 5) | θ value | 1.51984e-148 (rank : 5) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8R0I0, Q99N70, Q99N71 | Gene names | Ace2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiotensin-converting enzyme 2 precursor (EC 3.4.17.-) (ACE-related carboxypeptidase). | |||||
|
ACE2_HUMAN
|
||||||
| NC score | 0.916493 (rank : 6) | θ value | 4.57613e-145 (rank : 6) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BYF1, Q6UWP0, Q86WT0, Q9NRA7, Q9UFZ6 | Gene names | ACE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiotensin-converting enzyme 2 precursor (EC 3.4.17.-) (ACE-related carboxypeptidase) (Angiotensin-converting enzyme homolog) (ACEH). | |||||
|
TMM27_MOUSE
|
||||||
| NC score | 0.229463 (rank : 7) | θ value | θ > 10 (rank : 30) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9ESG4, Q3UFF6 | Gene names | Tmem27, Nx17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collectrin precursor (Transmembrane protein 27). | |||||
|
TMM27_HUMAN
|
||||||
| NC score | 0.205077 (rank : 8) | θ value | θ > 10 (rank : 29) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9HBJ8, Q6UW07 | Gene names | TMEM27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collectrin precursor (Transmembrane protein 27). | |||||
|
SIN3B_MOUSE
|
||||||
| NC score | 0.051972 (rank : 9) | θ value | 0.0736092 (rank : 7) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q62141, O54976, Q6A013, Q8VCB8, Q8VDZ5 | Gene names | Sin3b, Kiaa0700 | |||
|
Domain Architecture |

|
|||||
| Description | Paired amphipathic helix protein Sin3b (Transcriptional corepressor Sin3b) (Histone deacetylase complex subunit Sin3b). | |||||
|
SIN3B_HUMAN
|
||||||
| NC score | 0.049988 (rank : 10) | θ value | 0.0961366 (rank : 8) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75182, Q68GC2, Q6P4B8, Q8TB34, Q9BSC8 | Gene names | SIN3B, KIAA0700 | |||
|
Domain Architecture |

|
|||||
| Description | Paired amphipathic helix protein Sin3b (Transcriptional corepressor Sin3b) (Histone deacetylase complex subunit Sin3b). | |||||
|
CHIT1_MOUSE
|
||||||
| NC score | 0.048960 (rank : 11) | θ value | 1.06291 (rank : 10) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9D7Q1, Q6QJD2 | Gene names | Chit1 | |||
|
Domain Architecture |
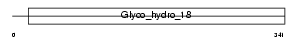
|
|||||
| Description | Chitotriosidase-1 precursor (EC 3.2.1.14) (Chitinase-1). | |||||
|
CHIT1_HUMAN
|
||||||
| NC score | 0.021788 (rank : 12) | θ value | 2.36792 (rank : 15) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13231, Q3ZAR1, Q6ISC2, Q9H3V8 | Gene names | CHIT1 | |||
|
Domain Architecture |

|
|||||
| Description | Chitotriosidase-1 precursor (EC 3.2.1.14) (Chitinase-1). | |||||
|
CENPJ_HUMAN
|
||||||
| NC score | 0.020926 (rank : 13) | θ value | 8.99809 (rank : 24) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 721 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9HC77, Q5T6R5, Q96KS5, Q9C067 | Gene names | CENPJ, CPAP, LAP, LIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein J (CENP-J) (Centrosomal P4.1-associated protein) (LAG-3-associated protein) (LYST-interacting protein 1). | |||||
|
SETD2_HUMAN
|
||||||
| NC score | 0.017226 (rank : 14) | θ value | 1.38821 (rank : 13) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BYW2, O75397, O75405, Q17RW8, Q5BKS9, Q5QGN2, Q69YI5, Q6IN64, Q6ZN53, Q6ZS25, Q8N3R0, Q8TCN0, Q9C0D1, Q9H696, Q9NZW9 | Gene names | SETD2, HIF1, HYPB, KIAA1732, SET2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36-specific (EC 2.1.1.43) (SET domain-containing protein 2) (hSET2) (Huntingtin- interacting protein HYPB) (Huntingtin-interacting protein 1) (HIF-1) (p231HBP). | |||||
|
CUTL1_MOUSE
|
||||||
| NC score | 0.015573 (rank : 15) | θ value | 1.06291 (rank : 11) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P53564, O08994, P70301, Q571L6, Q91ZD2 | Gene names | Cutl1, Cux, Cux1, Kiaa4047 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP) (Homeobox protein Cux). | |||||
|
CASP_MOUSE
|
||||||
| NC score | 0.015075 (rank : 16) | θ value | 1.06291 (rank : 9) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 1152 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P70403, Q7TQJ4, Q91WN6 | Gene names | Cutl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
CASP_HUMAN
|
||||||
| NC score | 0.014279 (rank : 17) | θ value | 2.36792 (rank : 14) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q13948, Q53GU9, Q8TBS3 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
CUTL1_HUMAN
|
||||||
| NC score | 0.014142 (rank : 18) | θ value | 2.36792 (rank : 16) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
KLH11_HUMAN
|
||||||
| NC score | 0.011108 (rank : 19) | θ value | 1.06291 (rank : 12) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NVR0 | Gene names | KLHL11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch-like protein 11 precursor. | |||||
|
SRBP1_HUMAN
|
||||||
| NC score | 0.009579 (rank : 20) | θ value | 5.27518 (rank : 20) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P36956, Q16062 | Gene names | SREBF1, SREBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
FRAP_HUMAN
|
||||||
| NC score | 0.008798 (rank : 21) | θ value | 8.99809 (rank : 25) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P42345, Q6LE87, Q96QG3, Q9Y4I3 | Gene names | FRAP1, FRAP, FRAP2 | |||
|
Domain Architecture |

|
|||||
| Description | FKBP12-rapamycin complex-associated protein (FK506-binding protein 12- rapamycin complex-associated protein 1) (Rapamycin target protein) (RAPT1) (Mammalian target of rapamycin) (mTOR). | |||||
|
FRAP_MOUSE
|
||||||
| NC score | 0.008795 (rank : 22) | θ value | 8.99809 (rank : 26) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JLN9, Q811J5, Q9CST1 | Gene names | Frap1, Frap | |||
|
Domain Architecture |

|
|||||
| Description | FKBP12-rapamycin complex-associated protein (FK506-binding protein 12- rapamycin complex-associated protein 1) (Rapamycin target protein) (RAPT1) (Mammalian target of rapamycin) (mTOR). | |||||
|
DYH11_HUMAN
|
||||||
| NC score | 0.006464 (rank : 23) | θ value | 6.88961 (rank : 21) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96DT5, Q9UJ82 | Gene names | DNAH11 | |||
|
Domain Architecture |

|
|||||
| Description | Ciliary dynein heavy chain 11 (Axonemal beta dynein heavy chain 11). | |||||
|
GOGA2_HUMAN
|
||||||
| NC score | 0.006460 (rank : 24) | θ value | 6.88961 (rank : 23) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q08379, Q9BRB0, Q9NYF9 | Gene names | GOLGA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgin subfamily A member 2 (Cis-Golgi matrix protein GM130) (Gm130 autoantigen) (Golgin-95). | |||||
|
JADE3_MOUSE
|
||||||
| NC score | 0.006405 (rank : 25) | θ value | 8.99809 (rank : 27) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6IE82, Q6ZQG2 | Gene names | Phf16, Jade3, Kiaa0215 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-3 (PHD finger protein 16). | |||||
|
UACA_MOUSE
|
||||||
| NC score | 0.005403 (rank : 26) | θ value | 4.03905 (rank : 17) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8CGB3, Q69ZG3, Q7TN77, Q8BJC8 | Gene names | Uaca, Kiaa1561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uveal autoantigen with coiled-coil domains and ankyrin repeats protein (Nucling) (Nuclear membrane-binding protein). | |||||
|
UTRO_HUMAN
|
||||||
| NC score | 0.005378 (rank : 27) | θ value | 8.99809 (rank : 28) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P46939 | Gene names | UTRN, DMDL | |||
|
Domain Architecture |

|
|||||
| Description | Utrophin (Dystrophin-related protein 1) (DRP1) (DRP). | |||||
|
GAN_HUMAN
|
||||||
| NC score | 0.004654 (rank : 28) | θ value | 6.88961 (rank : 22) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H2C0 | Gene names | GAN, GAN1, KLHL16 | |||
|
Domain Architecture |

|
|||||
| Description | Gigaxonin (Kelch-like protein 16). | |||||
|
PTN4_HUMAN
|
||||||
| NC score | 0.004205 (rank : 29) | θ value | 5.27518 (rank : 19) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P29074 | Gene names | PTPN4 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 4 (EC 3.1.3.48) (Protein-tyrosine phosphatase MEG1) (PTPase-MEG1) (MEG). | |||||
|
NK3R_MOUSE
|
||||||
| NC score | 0.001733 (rank : 30) | θ value | 5.27518 (rank : 18) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 902 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P47937, Q61968, Q8BL44, Q9JKN0 | Gene names | Tacr3, Tac3r | |||
|
Domain Architecture |

|
|||||
| Description | Neuromedin K receptor (NKR) (Neurokinin B receptor) (NK-3 receptor) (NK-3R) (Tachykinin receptor 3). | |||||