Please be patient as the page loads
|
TTC19_HUMAN
|
||||||
| SwissProt Accessions | Q6DKK2, Q7L3U8, Q9NXB2 | Gene names | TTC19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 19 (TPR repeat protein 19). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
TTC19_HUMAN
|
||||||
| θ value | 1.7349e-152 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q6DKK2, Q7L3U8, Q9NXB2 | Gene names | TTC19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 19 (TPR repeat protein 19). | |||||
|
TTC19_MOUSE
|
||||||
| θ value | 1.68038e-147 (rank : 2) | NC score | 0.986970 (rank : 2) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8CC21 | Gene names | Ttc19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 19 (TPR repeat protein 19). | |||||
|
KLC1_HUMAN
|
||||||
| θ value | 1.99992e-07 (rank : 3) | NC score | 0.470492 (rank : 10) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q07866, Q7RTM2, Q7RTM3, Q7RTM5, Q7RTP9, Q7RTQ5, Q86SF5, Q86TF5, Q86V74, Q86V75, Q86V76, Q86V77, Q86V78, Q86V79, Q96H62 | Gene names | KNS2, KLC, KLC1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin light chain 1 (KLC 1). | |||||
|
KLC4_HUMAN
|
||||||
| θ value | 3.41135e-07 (rank : 4) | NC score | 0.482982 (rank : 7) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9NSK0 | Gene names | KLC4, KNSL8 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin light chain 4 (KLC 4) (Kinesin-like protein 8). | |||||
|
KLC3_MOUSE
|
||||||
| θ value | 4.45536e-07 (rank : 5) | NC score | 0.512681 (rank : 3) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q91W40, Q3TZ56 | Gene names | Klc3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin light chain 3. | |||||
|
NPHP3_MOUSE
|
||||||
| θ value | 4.45536e-07 (rank : 6) | NC score | 0.458001 (rank : 11) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q7TNH6, Q69Z39, Q8C798, Q8C7Z3, Q9D6D1 | Gene names | Nphp3, Kiaa2000 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nephrocystin-3. | |||||
|
KLC4_MOUSE
|
||||||
| θ value | 7.59969e-07 (rank : 7) | NC score | 0.481387 (rank : 8) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9DBS5, Q3TCC5 | Gene names | Klc4, Knsl8 | |||
|
Domain Architecture |
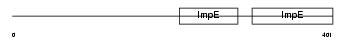
|
|||||
| Description | Kinesin light chain 4 (KLC 4) (Kinesin-like protein 8). | |||||
|
KLC3_HUMAN
|
||||||
| θ value | 9.92553e-07 (rank : 8) | NC score | 0.508613 (rank : 4) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6P597, Q6GMU2, Q8NAL1, Q8WWJ9 | Gene names | KLC3, KLC2, KLC2L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin light chain 3 (kinesin light chain 2) (KLC2-like). | |||||
|
KLC1_MOUSE
|
||||||
| θ value | 2.21117e-06 (rank : 9) | NC score | 0.476414 (rank : 9) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O88447 | Gene names | Kns2, Klc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin light chain 1 (KLC 1). | |||||
|
KLC2_HUMAN
|
||||||
| θ value | 2.21117e-06 (rank : 10) | NC score | 0.491594 (rank : 5) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9H0B6, Q9H9C8, Q9HA20 | Gene names | KLC2 | |||
|
Domain Architecture |
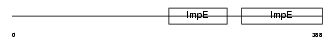
|
|||||
| Description | Kinesin light chain 2 (KLC 2). | |||||
|
KLC2_MOUSE
|
||||||
| θ value | 2.21117e-06 (rank : 11) | NC score | 0.489363 (rank : 6) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O88448 | Gene names | Klc2 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin light chain 2 (KLC 2). | |||||
|
NPHP3_HUMAN
|
||||||
| θ value | 2.88788e-06 (rank : 12) | NC score | 0.453482 (rank : 12) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 304 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q7Z494, Q5JPE3, Q5JPE6, Q68D99, Q6NVH3, Q7Z492, Q7Z493, Q8N9R2, Q8NCM5, Q96N70, Q96NK2 | Gene names | NPHP3, KIAA2000 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nephrocystin-3. | |||||
|
IF3X_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 13) | NC score | 0.340349 (rank : 13) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O75153, Q6AHY2, Q6P3X7, Q6ZUG8, Q8N4U7, Q9BTA3, Q9H979 | Gene names | KIAA0664 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative eukaryotic translation initiation factor 3 subunit (eIF-3). | |||||
|
TTC28_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 14) | NC score | 0.165346 (rank : 17) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96AY4, O95928, O95929, Q9NTE4, Q9UG31, Q9UGG5, Q9UPV8, Q9Y3S5 | Gene names | TTC28, KIAA1043 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 28 (TPR repeat protein 28). | |||||
|
TTC28_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 15) | NC score | 0.167178 (rank : 16) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q80XJ3, Q6P9J6, Q80TL5, Q8BV10, Q8C0F2 | Gene names | Ttc28, Kiaa1043 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 28 (TPR repeat protein 28). | |||||
|
SYNE2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 16) | NC score | 0.026805 (rank : 39) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
STIP1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 17) | NC score | 0.117966 (rank : 19) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q60864, Q3TT16, Q8BPH3, Q99L66 | Gene names | Stip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stress-induced-phosphoprotein 1 (STI1) (Hsc70/Hsp90-organizing protein) (Hop) (mSTI1). | |||||
|
APBP2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 18) | NC score | 0.251293 (rank : 15) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q92624, O95095, Q8WVC9 | Gene names | APPBP2, KIAA0228, PAT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid protein-binding protein 2 (Amyloid beta precursor protein- binding protein 2) (APP-BP2) (Protein interacting with APP tail 1). | |||||
|
APBP2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 19) | NC score | 0.251298 (rank : 14) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9DAX9, Q80U61 | Gene names | Appbp2, Kiaa0228 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid protein-binding protein 2 (Amyloid beta precursor protein- binding protein 2) (APP-BP2). | |||||
|
RAD50_HUMAN
|
||||||
| θ value | 1.81305 (rank : 20) | NC score | 0.031297 (rank : 38) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q92878, O43254, Q6GMT7, Q6P5X3, Q9UP86 | Gene names | RAD50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (hRAD50). | |||||
|
STIP1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 21) | NC score | 0.114214 (rank : 20) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P31948 | Gene names | STIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Stress-induced-phosphoprotein 1 (STI1) (Hsc70/Hsp90-organizing protein) (Hop) (Transformation-sensitive protein IEF SSP 3521) (NY- REN-11 antigen). | |||||
|
TPM3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 22) | NC score | 0.059943 (rank : 27) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 689 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P06753, P12324, Q969Q2, Q9NQH8 | Gene names | TPM3 | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin alpha-3 chain (Tropomyosin-3) (Tropomyosin gamma) (hTM5). | |||||
|
TPM3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 23) | NC score | 0.060149 (rank : 26) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P21107, Q09021, Q60606, Q80SW6, Q9EPW3 | Gene names | Tpm3, Tpm-5, Tpm5 | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin alpha-3 chain (Tropomyosin-3) (Tropomyosin gamma). | |||||
|
TPM2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 24) | NC score | 0.053038 (rank : 34) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 677 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P07951, P06468, Q13894, Q53FM4, Q5TCU4, Q5TCU7, Q9UH67 | Gene names | TPM2, TMSB | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin beta chain (Tropomyosin 2) (Beta-tropomyosin). | |||||
|
TPM2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 25) | NC score | 0.053080 (rank : 33) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P58774, P02560, P46901 | Gene names | Tpm2, Tpm-2 | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin beta chain (Tropomyosin 2) (Beta-tropomyosin). | |||||
|
TPR_HUMAN
|
||||||
| θ value | 2.36792 (rank : 26) | NC score | 0.021340 (rank : 42) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
P85A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 27) | NC score | 0.022314 (rank : 41) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P27986 | Gene names | PIK3R1, GRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit alpha (PI3-kinase p85-subunit alpha) (PtdIns-3-kinase p85-alpha) (PI3K). | |||||
|
TPM1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 28) | NC score | 0.053886 (rank : 32) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 716 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P09493, P09494, P10469, Q86W64, Q96IK2, Q9UCY9 | Gene names | TPM1, TMSA | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin 1 alpha chain (Alpha-tropomyosin). | |||||
|
TPM1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 29) | NC score | 0.052307 (rank : 35) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 729 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P58771, P02558, P19354, P46902, P99034 | Gene names | Tpm1, Tpm-1, Tpma | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin 1 alpha chain (Alpha-tropomyosin). | |||||
|
E41L1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 30) | NC score | 0.007620 (rank : 46) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H4G0, O15046, Q4VXN4 | Gene names | EPB41L1, KIAA0338 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 1 (Neuronal protein 4.1) (4.1N). | |||||
|
FES_HUMAN
|
||||||
| θ value | 5.27518 (rank : 31) | NC score | 0.000587 (rank : 47) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 978 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P07332 | Gene names | FES, FPS | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase Fes/Fps (EC 2.7.10.2) (C-Fes). | |||||
|
GPSM2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 32) | NC score | 0.062487 (rank : 23) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P81274, Q8N0Z5 | Gene names | GPSM2, LGN | |||
|
Domain Architecture |

|
|||||
| Description | G-protein signaling modulator 2 (Mosaic protein LGN). | |||||
|
GPSM2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 33) | NC score | 0.062229 (rank : 24) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8VDU0, Q8BLX3 | Gene names | Gpsm2, Lgn, Pins | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein signaling modulator 2 (Pins homolog). | |||||
|
P85A_MOUSE
|
||||||
| θ value | 5.27518 (rank : 34) | NC score | 0.017205 (rank : 44) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P26450 | Gene names | Pik3r1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit alpha (PI3-kinase p85-subunit alpha) (PtdIns-3-kinase p85-alpha) (PI3K). | |||||
|
ARI1A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 35) | NC score | 0.010342 (rank : 45) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
OGT1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 36) | NC score | 0.163348 (rank : 18) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O15294, Q7Z3K0, Q8WWM8, Q96CC1, Q9UG57 | Gene names | OGT | |||
|
Domain Architecture |

|
|||||
| Description | UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit (EC 2.4.1.-) (O-GlcNAc transferase p110 subunit). | |||||
|
SRP72_HUMAN
|
||||||
| θ value | 6.88961 (rank : 37) | NC score | 0.064505 (rank : 21) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O76094 | Gene names | SRP72 | |||
|
Domain Architecture |

|
|||||
| Description | Signal recognition particle 72 kDa protein (SRP72). | |||||
|
CCD21_HUMAN
|
||||||
| θ value | 8.99809 (rank : 38) | NC score | 0.018093 (rank : 43) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1228 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6P2H3, Q5VY68, Q5VY70, Q9H6Q1, Q9H828, Q9UF52 | Gene names | CCDC21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
NUMA1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 39) | NC score | 0.025845 (rank : 40) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
TPM4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 40) | NC score | 0.039985 (rank : 37) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 525 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P67936, P07226, Q15659, Q9BU85, Q9H8Q3 | Gene names | TPM4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tropomyosin alpha-4 chain (Tropomyosin-4) (TM30p1). | |||||
|
CTR9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 41) | NC score | 0.062997 (rank : 22) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6PD62, Q15015 | Gene names | CTR9, KIAA0155, SH2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1). | |||||
|
CTR9_MOUSE
|
||||||
| θ value | θ > 10 (rank : 42) | NC score | 0.061951 (rank : 25) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q62018, Q3UFF5, Q3UY40, Q66JX4, Q7TPS6, Q8BND9, Q8BRD1, Q8C9W7, Q8C9Y3, Q8CHI1 | Gene names | Ctr9, Kiaa0155, Sh2bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1) (Tetratricopeptide repeat-containing, SH2-binding phosphoprotein of 150 kDa) (TPR-containing, SH2-binding phosphoprotein of 150 kDa) (p150TSP). | |||||
|
GPSM1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 43) | NC score | 0.054792 (rank : 30) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q86YR5, Q86SR5, Q969T1, Q9UFS8 | Gene names | GPSM1, AGS3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein signaling modulator 1 (Activator of G-protein signaling 3). | |||||
|
GPSM1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 44) | NC score | 0.056682 (rank : 29) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6IR34, Q61366, Q8BUK4, Q8BX78, Q8R0R9 | Gene names | Gpsm1, Ags3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein signaling modulator 1 (Activator of G-protein signaling 3). | |||||
|
K0372_HUMAN
|
||||||
| θ value | θ > 10 (rank : 45) | NC score | 0.051788 (rank : 36) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6PGP7, O15077, Q6PJI3 | Gene names | KIAA0372 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein KIAA0372 (TPR repeat protein KIAA0372). | |||||
|
SGTA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 46) | NC score | 0.054298 (rank : 31) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O43765, Q9BTZ9 | Gene names | SGTA, SGT | |||
|
Domain Architecture |
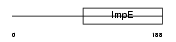
|
|||||
| Description | Small glutamine-rich tetratricopeptide repeat-containing protein A (Vpu-binding protein) (UBP). | |||||
|
SGTA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 47) | NC score | 0.059410 (rank : 28) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BJU0, Q8BGA6, Q99L52 | Gene names | Sgta, Sgt | |||
|
Domain Architecture |
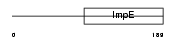
|
|||||
| Description | Small glutamine-rich tetratricopeptide repeat-containing protein A. | |||||
|
TTC19_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 1.7349e-152 (rank : 1) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q6DKK2, Q7L3U8, Q9NXB2 | Gene names | TTC19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 19 (TPR repeat protein 19). | |||||
|
TTC19_MOUSE
|
||||||
| NC score | 0.986970 (rank : 2) | θ value | 1.68038e-147 (rank : 2) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8CC21 | Gene names | Ttc19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 19 (TPR repeat protein 19). | |||||
|
KLC3_MOUSE
|
||||||
| NC score | 0.512681 (rank : 3) | θ value | 4.45536e-07 (rank : 5) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q91W40, Q3TZ56 | Gene names | Klc3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin light chain 3. | |||||
|
KLC3_HUMAN
|
||||||
| NC score | 0.508613 (rank : 4) | θ value | 9.92553e-07 (rank : 8) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6P597, Q6GMU2, Q8NAL1, Q8WWJ9 | Gene names | KLC3, KLC2, KLC2L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin light chain 3 (kinesin light chain 2) (KLC2-like). | |||||
|
KLC2_HUMAN
|
||||||
| NC score | 0.491594 (rank : 5) | θ value | 2.21117e-06 (rank : 10) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9H0B6, Q9H9C8, Q9HA20 | Gene names | KLC2 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin light chain 2 (KLC 2). | |||||
|
KLC2_MOUSE
|
||||||
| NC score | 0.489363 (rank : 6) | θ value | 2.21117e-06 (rank : 11) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O88448 | Gene names | Klc2 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin light chain 2 (KLC 2). | |||||
|
KLC4_HUMAN
|
||||||
| NC score | 0.482982 (rank : 7) | θ value | 3.41135e-07 (rank : 4) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9NSK0 | Gene names | KLC4, KNSL8 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin light chain 4 (KLC 4) (Kinesin-like protein 8). | |||||
|
KLC4_MOUSE
|
||||||
| NC score | 0.481387 (rank : 8) | θ value | 7.59969e-07 (rank : 7) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9DBS5, Q3TCC5 | Gene names | Klc4, Knsl8 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin light chain 4 (KLC 4) (Kinesin-like protein 8). | |||||
|
KLC1_MOUSE
|
||||||
| NC score | 0.476414 (rank : 9) | θ value | 2.21117e-06 (rank : 9) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O88447 | Gene names | Kns2, Klc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin light chain 1 (KLC 1). | |||||
|
KLC1_HUMAN
|
||||||
| NC score | 0.470492 (rank : 10) | θ value | 1.99992e-07 (rank : 3) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q07866, Q7RTM2, Q7RTM3, Q7RTM5, Q7RTP9, Q7RTQ5, Q86SF5, Q86TF5, Q86V74, Q86V75, Q86V76, Q86V77, Q86V78, Q86V79, Q96H62 | Gene names | KNS2, KLC, KLC1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin light chain 1 (KLC 1). | |||||
|
NPHP3_MOUSE
|
||||||
| NC score | 0.458001 (rank : 11) | θ value | 4.45536e-07 (rank : 6) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q7TNH6, Q69Z39, Q8C798, Q8C7Z3, Q9D6D1 | Gene names | Nphp3, Kiaa2000 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nephrocystin-3. | |||||
|
NPHP3_HUMAN
|
||||||
| NC score | 0.453482 (rank : 12) | θ value | 2.88788e-06 (rank : 12) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 304 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q7Z494, Q5JPE3, Q5JPE6, Q68D99, Q6NVH3, Q7Z492, Q7Z493, Q8N9R2, Q8NCM5, Q96N70, Q96NK2 | Gene names | NPHP3, KIAA2000 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nephrocystin-3. | |||||
|
IF3X_HUMAN
|
||||||
| NC score | 0.340349 (rank : 13) | θ value | 0.00869519 (rank : 13) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O75153, Q6AHY2, Q6P3X7, Q6ZUG8, Q8N4U7, Q9BTA3, Q9H979 | Gene names | KIAA0664 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative eukaryotic translation initiation factor 3 subunit (eIF-3). | |||||
|
APBP2_MOUSE
|
||||||
| NC score | 0.251298 (rank : 14) | θ value | 1.81305 (rank : 19) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9DAX9, Q80U61 | Gene names | Appbp2, Kiaa0228 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid protein-binding protein 2 (Amyloid beta precursor protein- binding protein 2) (APP-BP2). | |||||
|
APBP2_HUMAN
|
||||||
| NC score | 0.251293 (rank : 15) | θ value | 1.81305 (rank : 18) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q92624, O95095, Q8WVC9 | Gene names | APPBP2, KIAA0228, PAT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid protein-binding protein 2 (Amyloid beta precursor protein- binding protein 2) (APP-BP2) (Protein interacting with APP tail 1). | |||||
|
TTC28_MOUSE
|
||||||
| NC score | 0.167178 (rank : 16) | θ value | 0.0431538 (rank : 15) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q80XJ3, Q6P9J6, Q80TL5, Q8BV10, Q8C0F2 | Gene names | Ttc28, Kiaa1043 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 28 (TPR repeat protein 28). | |||||
|
TTC28_HUMAN
|
||||||
| NC score | 0.165346 (rank : 17) | θ value | 0.0330416 (rank : 14) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96AY4, O95928, O95929, Q9NTE4, Q9UG31, Q9UGG5, Q9UPV8, Q9Y3S5 | Gene names | TTC28, KIAA1043 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 28 (TPR repeat protein 28). | |||||
|
OGT1_HUMAN
|
||||||
| NC score | 0.163348 (rank : 18) | θ value | 6.88961 (rank : 36) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O15294, Q7Z3K0, Q8WWM8, Q96CC1, Q9UG57 | Gene names | OGT | |||
|
Domain Architecture |

|
|||||
| Description | UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit (EC 2.4.1.-) (O-GlcNAc transferase p110 subunit). | |||||
|
STIP1_MOUSE
|
||||||
| NC score | 0.117966 (rank : 19) | θ value | 1.06291 (rank : 17) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q60864, Q3TT16, Q8BPH3, Q99L66 | Gene names | Stip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stress-induced-phosphoprotein 1 (STI1) (Hsc70/Hsp90-organizing protein) (Hop) (mSTI1). | |||||
|
STIP1_HUMAN
|
||||||
| NC score | 0.114214 (rank : 20) | θ value | 1.81305 (rank : 21) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P31948 | Gene names | STIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Stress-induced-phosphoprotein 1 (STI1) (Hsc70/Hsp90-organizing protein) (Hop) (Transformation-sensitive protein IEF SSP 3521) (NY- REN-11 antigen). | |||||
|
SRP72_HUMAN
|
||||||
| NC score | 0.064505 (rank : 21) | θ value | 6.88961 (rank : 37) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O76094 | Gene names | SRP72 | |||
|
Domain Architecture |

|
|||||
| Description | Signal recognition particle 72 kDa protein (SRP72). | |||||
|
CTR9_HUMAN
|
||||||
| NC score | 0.062997 (rank : 22) | θ value | θ > 10 (rank : 41) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6PD62, Q15015 | Gene names | CTR9, KIAA0155, SH2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1). | |||||
|
GPSM2_HUMAN
|
||||||
| NC score | 0.062487 (rank : 23) | θ value | 5.27518 (rank : 32) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P81274, Q8N0Z5 | Gene names | GPSM2, LGN | |||
|
Domain Architecture |

|
|||||
| Description | G-protein signaling modulator 2 (Mosaic protein LGN). | |||||
|
GPSM2_MOUSE
|
||||||
| NC score | 0.062229 (rank : 24) | θ value | 5.27518 (rank : 33) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8VDU0, Q8BLX3 | Gene names | Gpsm2, Lgn, Pins | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein signaling modulator 2 (Pins homolog). | |||||
|
CTR9_MOUSE
|
||||||
| NC score | 0.061951 (rank : 25) | θ value | θ > 10 (rank : 42) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q62018, Q3UFF5, Q3UY40, Q66JX4, Q7TPS6, Q8BND9, Q8BRD1, Q8C9W7, Q8C9Y3, Q8CHI1 | Gene names | Ctr9, Kiaa0155, Sh2bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1) (Tetratricopeptide repeat-containing, SH2-binding phosphoprotein of 150 kDa) (TPR-containing, SH2-binding phosphoprotein of 150 kDa) (p150TSP). | |||||
|
TPM3_MOUSE
|
||||||
| NC score | 0.060149 (rank : 26) | θ value | 1.81305 (rank : 23) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P21107, Q09021, Q60606, Q80SW6, Q9EPW3 | Gene names | Tpm3, Tpm-5, Tpm5 | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin alpha-3 chain (Tropomyosin-3) (Tropomyosin gamma). | |||||
|
TPM3_HUMAN
|
||||||
| NC score | 0.059943 (rank : 27) | θ value | 1.81305 (rank : 22) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 689 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P06753, P12324, Q969Q2, Q9NQH8 | Gene names | TPM3 | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin alpha-3 chain (Tropomyosin-3) (Tropomyosin gamma) (hTM5). | |||||
|
SGTA_MOUSE
|
||||||
| NC score | 0.059410 (rank : 28) | θ value | θ > 10 (rank : 47) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BJU0, Q8BGA6, Q99L52 | Gene names | Sgta, Sgt | |||
|
Domain Architecture |

|
|||||
| Description | Small glutamine-rich tetratricopeptide repeat-containing protein A. | |||||
|
GPSM1_MOUSE
|
||||||
| NC score | 0.056682 (rank : 29) | θ value | θ > 10 (rank : 44) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6IR34, Q61366, Q8BUK4, Q8BX78, Q8R0R9 | Gene names | Gpsm1, Ags3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein signaling modulator 1 (Activator of G-protein signaling 3). | |||||
|
GPSM1_HUMAN
|
||||||
| NC score | 0.054792 (rank : 30) | θ value | θ > 10 (rank : 43) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q86YR5, Q86SR5, Q969T1, Q9UFS8 | Gene names | GPSM1, AGS3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein signaling modulator 1 (Activator of G-protein signaling 3). | |||||
|
SGTA_HUMAN
|
||||||
| NC score | 0.054298 (rank : 31) | θ value | θ > 10 (rank : 46) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O43765, Q9BTZ9 | Gene names | SGTA, SGT | |||
|
Domain Architecture |

|
|||||
| Description | Small glutamine-rich tetratricopeptide repeat-containing protein A (Vpu-binding protein) (UBP). | |||||
|
TPM1_HUMAN
|
||||||
| NC score | 0.053886 (rank : 32) | θ value | 4.03905 (rank : 28) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 716 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P09493, P09494, P10469, Q86W64, Q96IK2, Q9UCY9 | Gene names | TPM1, TMSA | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin 1 alpha chain (Alpha-tropomyosin). | |||||
|
TPM2_MOUSE
|
||||||
| NC score | 0.053080 (rank : 33) | θ value | 2.36792 (rank : 25) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P58774, P02560, P46901 | Gene names | Tpm2, Tpm-2 | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin beta chain (Tropomyosin 2) (Beta-tropomyosin). | |||||
|
TPM2_HUMAN
|
||||||
| NC score | 0.053038 (rank : 34) | θ value | 2.36792 (rank : 24) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 677 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P07951, P06468, Q13894, Q53FM4, Q5TCU4, Q5TCU7, Q9UH67 | Gene names | TPM2, TMSB | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin beta chain (Tropomyosin 2) (Beta-tropomyosin). | |||||
|
TPM1_MOUSE
|
||||||
| NC score | 0.052307 (rank : 35) | θ value | 4.03905 (rank : 29) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 729 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P58771, P02558, P19354, P46902, P99034 | Gene names | Tpm1, Tpm-1, Tpma | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin 1 alpha chain (Alpha-tropomyosin). | |||||
|
K0372_HUMAN
|
||||||
| NC score | 0.051788 (rank : 36) | θ value | θ > 10 (rank : 45) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6PGP7, O15077, Q6PJI3 | Gene names | KIAA0372 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein KIAA0372 (TPR repeat protein KIAA0372). | |||||
|
TPM4_HUMAN
|
||||||
| NC score | 0.039985 (rank : 37) | θ value | 8.99809 (rank : 40) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 525 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P67936, P07226, Q15659, Q9BU85, Q9H8Q3 | Gene names | TPM4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tropomyosin alpha-4 chain (Tropomyosin-4) (TM30p1). | |||||
|
RAD50_HUMAN
|
||||||
| NC score | 0.031297 (rank : 38) | θ value | 1.81305 (rank : 20) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q92878, O43254, Q6GMT7, Q6P5X3, Q9UP86 | Gene names | RAD50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (hRAD50). | |||||
|
SYNE2_HUMAN
|
||||||
| NC score | 0.026805 (rank : 39) | θ value | 0.365318 (rank : 16) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
NUMA1_HUMAN
|
||||||
| NC score | 0.025845 (rank : 40) | θ value | 8.99809 (rank : 39) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
P85A_HUMAN
|
||||||
| NC score | 0.022314 (rank : 41) | θ value | 3.0926 (rank : 27) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P27986 | Gene names | PIK3R1, GRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit alpha (PI3-kinase p85-subunit alpha) (PtdIns-3-kinase p85-alpha) (PI3K). | |||||
|
TPR_HUMAN
|
||||||
| NC score | 0.021340 (rank : 42) | θ value | 2.36792 (rank : 26) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
CCD21_HUMAN
|
||||||
| NC score | 0.018093 (rank : 43) | θ value | 8.99809 (rank : 38) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1228 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6P2H3, Q5VY68, Q5VY70, Q9H6Q1, Q9H828, Q9UF52 | Gene names | CCDC21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
P85A_MOUSE
|
||||||
| NC score | 0.017205 (rank : 44) | θ value | 5.27518 (rank : 34) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P26450 | Gene names | Pik3r1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit alpha (PI3-kinase p85-subunit alpha) (PtdIns-3-kinase p85-alpha) (PI3K). | |||||
|
ARI1A_HUMAN
|
||||||
| NC score | 0.010342 (rank : 45) | θ value | 6.88961 (rank : 35) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
E41L1_HUMAN
|
||||||
| NC score | 0.007620 (rank : 46) | θ value | 5.27518 (rank : 30) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H4G0, O15046, Q4VXN4 | Gene names | EPB41L1, KIAA0338 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 1 (Neuronal protein 4.1) (4.1N). | |||||
|
FES_HUMAN
|
||||||
| NC score | 0.000587 (rank : 47) | θ value | 5.27518 (rank : 31) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 978 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P07332 | Gene names | FES, FPS | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase Fes/Fps (EC 2.7.10.2) (C-Fes). | |||||