Please be patient as the page loads
|
TF3C4_HUMAN
|
||||||
| SwissProt Accessions | Q9UKN8 | Gene names | GTF3C4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | General transcription factor 3C polypeptide 4 (EC 2.3.1.48) (Transcription factor IIIC subunit delta) (TF3C-delta) (TFIIIC 90 kDa subunit) (TFIIIC 90). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
TF3C4_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UKN8 | Gene names | GTF3C4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | General transcription factor 3C polypeptide 4 (EC 2.3.1.48) (Transcription factor IIIC subunit delta) (TF3C-delta) (TFIIIC 90 kDa subunit) (TFIIIC 90). | |||||
|
TF3C4_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.995308 (rank : 2) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BMQ2, Q8BKZ4 | Gene names | Gtf3c4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | General transcription factor 3C polypeptide 4 (EC 2.3.1.48) (Transcription factor IIIC subunit delta) (TF3C-delta) (TFIIIC 90 kDa subunit) (TFIIIC 90). | |||||
|
FYB_HUMAN
|
||||||
| θ value | 0.125558 (rank : 3) | NC score | 0.061122 (rank : 6) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15117, O00359 | Gene names | FYB, SLAP130 | |||
|
Domain Architecture |

|
|||||
| Description | FYN-binding protein (FYN-T-binding protein) (FYB-120/130) (p120/p130) (SLP-76-associated phosphoprotein) (SLAP-130). | |||||
|
NFM_MOUSE
|
||||||
| θ value | 0.125558 (rank : 4) | NC score | 0.013616 (rank : 19) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P08553, Q61961 | Gene names | Nef3, Nefm, Nfm | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M). | |||||
|
KCD13_HUMAN
|
||||||
| θ value | 0.47712 (rank : 5) | NC score | 0.066323 (rank : 5) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WZ19, Q96P93, Q96SA1 | Gene names | KCTD13, PDIP1, POLDIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD13 (Polymerase delta-interacting protein 1) (TNFAIP1-like protein). | |||||
|
KCD13_MOUSE
|
||||||
| θ value | 0.47712 (rank : 6) | NC score | 0.066494 (rank : 4) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BGV7, Q6AXE9 | Gene names | Kctd13, Poldip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD13 (Polymerase delta-interacting protein 1). | |||||
|
KU86_HUMAN
|
||||||
| θ value | 1.38821 (rank : 7) | NC score | 0.066726 (rank : 3) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P13010, Q4VBQ5, Q9UCQ0, Q9UCQ1 | Gene names | XRCC5, G22P2 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase 2 subunit 2 (EC 3.6.1.-) (ATP-dependent DNA helicase II 80 kDa subunit) (Lupus Ku autoantigen protein p86) (Ku86) (Ku80) (86 kDa subunit of Ku antigen) (Thyroid-lupus autoantigen) (TLAA) (CTC box-binding factor 85 kDa subunit) (CTCBF) (CTC85) (Nuclear factor IV) (DNA-repair protein XRCC5). | |||||
|
ZN598_HUMAN
|
||||||
| θ value | 1.38821 (rank : 8) | NC score | 0.050890 (rank : 8) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q86UK7, Q8IW49, Q8N3D9, Q96FG3, Q9H7J3 | Gene names | ZNF598 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 598. | |||||
|
BPL1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 9) | NC score | 0.033957 (rank : 11) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P50747, Q99451 | Gene names | HLCS | |||
|
Domain Architecture |

|
|||||
| Description | Biotin--protein ligase (EC 6.3.4.-) (Biotin apo-protein ligase) [Includes: Biotin--[methylmalonyl-CoA-carboxytransferase] ligase (EC 6.3.4.9); Biotin--[propionyl-CoA-carboxylase [ATP-hydrolyzing]] ligase (EC 6.3.4.10) (Holocarboxylase synthetase) (HCS); Biotin-- [methylcrotonoyl-CoA-carboxylase] ligase (EC 6.3.4.11); Biotin-- [acetyl-CoA-carboxylase] ligase (EC 6.3.4.15)]. | |||||
|
KU86_MOUSE
|
||||||
| θ value | 2.36792 (rank : 10) | NC score | 0.055160 (rank : 7) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P27641 | Gene names | Xrcc5, G22p2 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase 2 subunit 2 (EC 3.6.1.-) (ATP-dependent DNA helicase II 80 kDa subunit) (Ku autoantigen protein p86 homolog) (Ku80) (CTC box-binding factor 85 kDa subunit) (CTCBF) (CTC85) (Nuclear factor IV) (DNA-repair protein XRCC5). | |||||
|
SET_HUMAN
|
||||||
| θ value | 2.36792 (rank : 11) | NC score | 0.022443 (rank : 14) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q01105, Q15541, Q5VXV1 | Gene names | SET | |||
|
Domain Architecture |
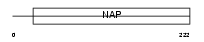
|
|||||
| Description | Protein SET (Phosphatase 2A inhibitor I2PP2A) (I-2PP2A) (Template- activating factor I) (TAF-I) (HLA-DR-associated protein II) (PHAPII) (Inhibitor of granzyme A-activated DNase) (IGAAD). | |||||
|
MYH10_MOUSE
|
||||||
| θ value | 3.0926 (rank : 12) | NC score | 0.006664 (rank : 22) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 1612 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61879, Q5SV63 | Gene names | Myh10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
RPO3G_HUMAN
|
||||||
| θ value | 5.27518 (rank : 13) | NC score | 0.029063 (rank : 13) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15318 | Gene names | POLR3G, RPC32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA-directed RNA polymerase III subunit G (EC 2.7.7.6) (DNA-directed RNA polymerase III 32 kDa polypeptide) (RNA polymerase III C32 subunit). | |||||
|
CMGA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 14) | NC score | 0.019123 (rank : 16) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P10645, Q96E84, Q96GL7, Q9BQB5 | Gene names | CHGA | |||
|
Domain Architecture |

|
|||||
| Description | Chromogranin A precursor (CgA) (Pituitary secretory protein I) (SP-I) [Contains: Vasostatin-1 (Vasostatin I); Vasostatin-2 (Vasostatin II); EA-92; ES-43; Pancreastatin; SS-18; WA-8; WE-14; LF-19; AL-11; GV-19; GR-44; ER-37]. | |||||
|
FYB_MOUSE
|
||||||
| θ value | 6.88961 (rank : 15) | NC score | 0.043989 (rank : 9) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35601, Q9Z2H3 | Gene names | Fyb | |||
|
Domain Architecture |

|
|||||
| Description | FYN-binding protein (FYN-T-binding protein) (FYB-120/130) (p120/p130) (SLP-76-associated phosphoprotein) (SLAP-130). | |||||
|
GNL1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 16) | NC score | 0.017524 (rank : 17) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P36915 | Gene names | GNL1, HSR1 | |||
|
Domain Architecture |
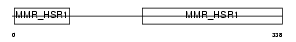
|
|||||
| Description | Guanine nucleotide-binding protein-like 1 (GTP-binding protein HSR1). | |||||
|
KLC3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 17) | NC score | 0.009680 (rank : 21) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6P597, Q6GMU2, Q8NAL1, Q8WWJ9 | Gene names | KLC3, KLC2, KLC2L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin light chain 3 (kinesin light chain 2) (KLC2-like). | |||||
|
MYH10_HUMAN
|
||||||
| θ value | 6.88961 (rank : 18) | NC score | 0.005761 (rank : 24) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P35580, Q16087 | Gene names | MYH10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
PTMA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 19) | NC score | 0.032598 (rank : 12) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P06454, Q15249, Q15592 | Gene names | PTMA, TMSA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha-1]. | |||||
|
AKT1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 20) | NC score | 0.000181 (rank : 25) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 885 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P31749, Q9BWB6 | Gene names | AKT1, PKB, RAC | |||
|
Domain Architecture |
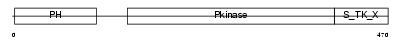
|
|||||
| Description | RAC-alpha serine/threonine-protein kinase (EC 2.7.11.1) (RAC-PK-alpha) (Protein kinase B) (PKB) (C-AKT). | |||||
|
ATS20_MOUSE
|
||||||
| θ value | 8.99809 (rank : 21) | NC score | 0.006298 (rank : 23) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P59511 | Gene names | Adamts20 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-20 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 20) (ADAM-TS 20) (ADAM-TS20). | |||||
|
C8AP2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 22) | NC score | 0.021440 (rank : 15) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 548 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UKL3, Q6PH76, Q7LCQ7, Q86YD9, Q9NUQ4, Q9NZV9, Q9P2N1, Q9Y563 | Gene names | CASP8AP2, FLASH, KIAA1315, RIP25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CASP8-associated protein 2 (FLICE-associated huge protein). | |||||
|
CLUL1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 23) | NC score | 0.017205 (rank : 18) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15846 | Gene names | CLUL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Clusterin-like protein 1 precursor (Retinal-specific clusterin-like protein). | |||||
|
PDXL2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 24) | NC score | 0.013521 (rank : 20) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CAE9, Q8CFW3 | Gene names | Podxl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Podocalyxin-like protein 2 precursor (Endoglycan). | |||||
|
TNAP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 25) | NC score | 0.038939 (rank : 10) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13829 | Gene names | TNFAIP1, EDP1 | |||
|
Domain Architecture |
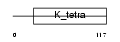
|
|||||
| Description | BTB/POZ domain-containing protein TNFAIP1 (Tumor necrosis factor, alpha-induced protein 1, endothelial) (Protein B12). | |||||
|
TF3C4_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UKN8 | Gene names | GTF3C4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | General transcription factor 3C polypeptide 4 (EC 2.3.1.48) (Transcription factor IIIC subunit delta) (TF3C-delta) (TFIIIC 90 kDa subunit) (TFIIIC 90). | |||||
|
TF3C4_MOUSE
|
||||||
| NC score | 0.995308 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BMQ2, Q8BKZ4 | Gene names | Gtf3c4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | General transcription factor 3C polypeptide 4 (EC 2.3.1.48) (Transcription factor IIIC subunit delta) (TF3C-delta) (TFIIIC 90 kDa subunit) (TFIIIC 90). | |||||
|
KU86_HUMAN
|
||||||
| NC score | 0.066726 (rank : 3) | θ value | 1.38821 (rank : 7) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P13010, Q4VBQ5, Q9UCQ0, Q9UCQ1 | Gene names | XRCC5, G22P2 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase 2 subunit 2 (EC 3.6.1.-) (ATP-dependent DNA helicase II 80 kDa subunit) (Lupus Ku autoantigen protein p86) (Ku86) (Ku80) (86 kDa subunit of Ku antigen) (Thyroid-lupus autoantigen) (TLAA) (CTC box-binding factor 85 kDa subunit) (CTCBF) (CTC85) (Nuclear factor IV) (DNA-repair protein XRCC5). | |||||
|
KCD13_MOUSE
|
||||||
| NC score | 0.066494 (rank : 4) | θ value | 0.47712 (rank : 6) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BGV7, Q6AXE9 | Gene names | Kctd13, Poldip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD13 (Polymerase delta-interacting protein 1). | |||||
|
KCD13_HUMAN
|
||||||
| NC score | 0.066323 (rank : 5) | θ value | 0.47712 (rank : 5) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WZ19, Q96P93, Q96SA1 | Gene names | KCTD13, PDIP1, POLDIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD13 (Polymerase delta-interacting protein 1) (TNFAIP1-like protein). | |||||
|
FYB_HUMAN
|
||||||
| NC score | 0.061122 (rank : 6) | θ value | 0.125558 (rank : 3) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15117, O00359 | Gene names | FYB, SLAP130 | |||
|
Domain Architecture |

|
|||||
| Description | FYN-binding protein (FYN-T-binding protein) (FYB-120/130) (p120/p130) (SLP-76-associated phosphoprotein) (SLAP-130). | |||||
|
KU86_MOUSE
|
||||||
| NC score | 0.055160 (rank : 7) | θ value | 2.36792 (rank : 10) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P27641 | Gene names | Xrcc5, G22p2 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase 2 subunit 2 (EC 3.6.1.-) (ATP-dependent DNA helicase II 80 kDa subunit) (Ku autoantigen protein p86 homolog) (Ku80) (CTC box-binding factor 85 kDa subunit) (CTCBF) (CTC85) (Nuclear factor IV) (DNA-repair protein XRCC5). | |||||
|
ZN598_HUMAN
|
||||||
| NC score | 0.050890 (rank : 8) | θ value | 1.38821 (rank : 8) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q86UK7, Q8IW49, Q8N3D9, Q96FG3, Q9H7J3 | Gene names | ZNF598 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 598. | |||||
|
FYB_MOUSE
|
||||||
| NC score | 0.043989 (rank : 9) | θ value | 6.88961 (rank : 15) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35601, Q9Z2H3 | Gene names | Fyb | |||
|
Domain Architecture |

|
|||||
| Description | FYN-binding protein (FYN-T-binding protein) (FYB-120/130) (p120/p130) (SLP-76-associated phosphoprotein) (SLAP-130). | |||||
|
TNAP1_HUMAN
|
||||||
| NC score | 0.038939 (rank : 10) | θ value | 8.99809 (rank : 25) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13829 | Gene names | TNFAIP1, EDP1 | |||
|
Domain Architecture |

|
|||||
| Description | BTB/POZ domain-containing protein TNFAIP1 (Tumor necrosis factor, alpha-induced protein 1, endothelial) (Protein B12). | |||||
|
BPL1_HUMAN
|
||||||
| NC score | 0.033957 (rank : 11) | θ value | 1.81305 (rank : 9) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P50747, Q99451 | Gene names | HLCS | |||
|
Domain Architecture |

|
|||||
| Description | Biotin--protein ligase (EC 6.3.4.-) (Biotin apo-protein ligase) [Includes: Biotin--[methylmalonyl-CoA-carboxytransferase] ligase (EC 6.3.4.9); Biotin--[propionyl-CoA-carboxylase [ATP-hydrolyzing]] ligase (EC 6.3.4.10) (Holocarboxylase synthetase) (HCS); Biotin-- [methylcrotonoyl-CoA-carboxylase] ligase (EC 6.3.4.11); Biotin-- [acetyl-CoA-carboxylase] ligase (EC 6.3.4.15)]. | |||||
|
PTMA_HUMAN
|
||||||
| NC score | 0.032598 (rank : 12) | θ value | 6.88961 (rank : 19) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P06454, Q15249, Q15592 | Gene names | PTMA, TMSA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha-1]. | |||||
|
RPO3G_HUMAN
|
||||||
| NC score | 0.029063 (rank : 13) | θ value | 5.27518 (rank : 13) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15318 | Gene names | POLR3G, RPC32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA-directed RNA polymerase III subunit G (EC 2.7.7.6) (DNA-directed RNA polymerase III 32 kDa polypeptide) (RNA polymerase III C32 subunit). | |||||
|
SET_HUMAN
|
||||||
| NC score | 0.022443 (rank : 14) | θ value | 2.36792 (rank : 11) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q01105, Q15541, Q5VXV1 | Gene names | SET | |||
|
Domain Architecture |

|
|||||
| Description | Protein SET (Phosphatase 2A inhibitor I2PP2A) (I-2PP2A) (Template- activating factor I) (TAF-I) (HLA-DR-associated protein II) (PHAPII) (Inhibitor of granzyme A-activated DNase) (IGAAD). | |||||
|
C8AP2_HUMAN
|
||||||
| NC score | 0.021440 (rank : 15) | θ value | 8.99809 (rank : 22) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 548 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UKL3, Q6PH76, Q7LCQ7, Q86YD9, Q9NUQ4, Q9NZV9, Q9P2N1, Q9Y563 | Gene names | CASP8AP2, FLASH, KIAA1315, RIP25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CASP8-associated protein 2 (FLICE-associated huge protein). | |||||
|
CMGA_HUMAN
|
||||||
| NC score | 0.019123 (rank : 16) | θ value | 6.88961 (rank : 14) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P10645, Q96E84, Q96GL7, Q9BQB5 | Gene names | CHGA | |||
|
Domain Architecture |

|
|||||
| Description | Chromogranin A precursor (CgA) (Pituitary secretory protein I) (SP-I) [Contains: Vasostatin-1 (Vasostatin I); Vasostatin-2 (Vasostatin II); EA-92; ES-43; Pancreastatin; SS-18; WA-8; WE-14; LF-19; AL-11; GV-19; GR-44; ER-37]. | |||||
|
GNL1_HUMAN
|
||||||
| NC score | 0.017524 (rank : 17) | θ value | 6.88961 (rank : 16) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P36915 | Gene names | GNL1, HSR1 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-binding protein-like 1 (GTP-binding protein HSR1). | |||||
|
CLUL1_HUMAN
|
||||||
| NC score | 0.017205 (rank : 18) | θ value | 8.99809 (rank : 23) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15846 | Gene names | CLUL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Clusterin-like protein 1 precursor (Retinal-specific clusterin-like protein). | |||||
|
NFM_MOUSE
|
||||||
| NC score | 0.013616 (rank : 19) | θ value | 0.125558 (rank : 4) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P08553, Q61961 | Gene names | Nef3, Nefm, Nfm | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M). | |||||
|
PDXL2_MOUSE
|
||||||
| NC score | 0.013521 (rank : 20) | θ value | 8.99809 (rank : 24) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CAE9, Q8CFW3 | Gene names | Podxl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Podocalyxin-like protein 2 precursor (Endoglycan). | |||||
|
KLC3_HUMAN
|
||||||
| NC score | 0.009680 (rank : 21) | θ value | 6.88961 (rank : 17) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6P597, Q6GMU2, Q8NAL1, Q8WWJ9 | Gene names | KLC3, KLC2, KLC2L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin light chain 3 (kinesin light chain 2) (KLC2-like). | |||||
|
MYH10_MOUSE
|
||||||
| NC score | 0.006664 (rank : 22) | θ value | 3.0926 (rank : 12) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 1612 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61879, Q5SV63 | Gene names | Myh10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
ATS20_MOUSE
|
||||||
| NC score | 0.006298 (rank : 23) | θ value | 8.99809 (rank : 21) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P59511 | Gene names | Adamts20 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-20 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 20) (ADAM-TS 20) (ADAM-TS20). | |||||
|
MYH10_HUMAN
|
||||||
| NC score | 0.005761 (rank : 24) | θ value | 6.88961 (rank : 18) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P35580, Q16087 | Gene names | MYH10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
AKT1_HUMAN
|
||||||
| NC score | 0.000181 (rank : 25) | θ value | 8.99809 (rank : 20) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 885 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P31749, Q9BWB6 | Gene names | AKT1, PKB, RAC | |||
|
Domain Architecture |

|
|||||
| Description | RAC-alpha serine/threonine-protein kinase (EC 2.7.11.1) (RAC-PK-alpha) (Protein kinase B) (PKB) (C-AKT). | |||||