Please be patient as the page loads
|
TAGL3_HUMAN
|
||||||
| SwissProt Accessions | Q9UI15, Q96A74 | Gene names | TAGLN3, NP25 | |||
|
Domain Architecture |
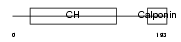
|
|||||
| Description | Transgelin-3 (Neuronal protein NP25) (Neuronal protein 22) (NP22). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
TAGL3_HUMAN
|
||||||
| θ value | 2.44908e-114 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9UI15, Q96A74 | Gene names | TAGLN3, NP25 | |||
|
Domain Architecture |

|
|||||
| Description | Transgelin-3 (Neuronal protein NP25) (Neuronal protein 22) (NP22). | |||||
|
TAGL3_MOUSE
|
||||||
| θ value | 9.30652e-114 (rank : 2) | NC score | 0.999866 (rank : 2) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9R1Q8 | Gene names | Tagln3, Np25 | |||
|
Domain Architecture |
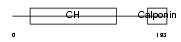
|
|||||
| Description | Transgelin-3 (Neuronal protein NP25). | |||||
|
TAGL2_HUMAN
|
||||||
| θ value | 6.08852e-81 (rank : 3) | NC score | 0.991180 (rank : 3) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P37802, Q5JRQ6, Q5JRQ7, Q9BUH5, Q9H4P0 | Gene names | TAGLN2, KIAA0120 | |||
|
Domain Architecture |
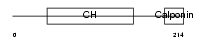
|
|||||
| Description | Transgelin-2 (SM22-alpha homolog). | |||||
|
TAGL2_MOUSE
|
||||||
| θ value | 4.36332e-79 (rank : 4) | NC score | 0.990052 (rank : 6) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9WVA4 | Gene names | Tagln2 | |||
|
Domain Architecture |
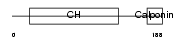
|
|||||
| Description | Transgelin-2. | |||||
|
TAGL_HUMAN
|
||||||
| θ value | 2.92676e-75 (rank : 5) | NC score | 0.991087 (rank : 4) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q01995, O15542 | Gene names | TAGLN, SM22, WS3-10 | |||
|
Domain Architecture |
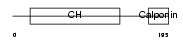
|
|||||
| Description | Transgelin (Smooth muscle protein 22-alpha) (SM22-alpha) (WS3-10) (22 kDa actin-binding protein). | |||||
|
TAGL_MOUSE
|
||||||
| θ value | 4.99229e-75 (rank : 6) | NC score | 0.990955 (rank : 5) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P37804, Q545W0 | Gene names | Tagln, Sm22, Sm22a | |||
|
Domain Architecture |
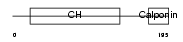
|
|||||
| Description | Transgelin (Smooth muscle protein 22-alpha) (SM22-alpha) (Actin- associated protein p27). | |||||
|
CNN3_HUMAN
|
||||||
| θ value | 8.61488e-35 (rank : 7) | NC score | 0.887993 (rank : 10) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q15417 | Gene names | CNN3 | |||
|
Domain Architecture |
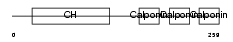
|
|||||
| Description | Calponin-3 (Calponin, acidic isoform). | |||||
|
CNN3_MOUSE
|
||||||
| θ value | 8.61488e-35 (rank : 8) | NC score | 0.888578 (rank : 9) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9DAW9, Q3TW23 | Gene names | Cnn3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calponin-3 (Calponin, acidic isoform). | |||||
|
CNN1_MOUSE
|
||||||
| θ value | 1.46948e-34 (rank : 9) | NC score | 0.893407 (rank : 7) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q08091 | Gene names | Cnn1 | |||
|
Domain Architecture |
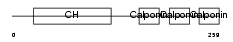
|
|||||
| Description | Calponin-1 (Calponin H1, smooth muscle) (Basic calponin). | |||||
|
CNN1_HUMAN
|
||||||
| θ value | 7.29293e-34 (rank : 10) | NC score | 0.892435 (rank : 8) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P51911, O00638, Q15416, Q8IY93, Q99438 | Gene names | CNN1 | |||
|
Domain Architecture |
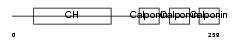
|
|||||
| Description | Calponin-1 (Calponin H1, smooth muscle) (Basic calponin). | |||||
|
CNN2_HUMAN
|
||||||
| θ value | 1.98606e-31 (rank : 11) | NC score | 0.883683 (rank : 11) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q99439, Q92578 | Gene names | CNN2 | |||
|
Domain Architecture |
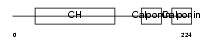
|
|||||
| Description | Calponin-2 (Calponin H2, smooth muscle) (Neutral calponin). | |||||
|
CNN2_MOUSE
|
||||||
| θ value | 5.77852e-31 (rank : 12) | NC score | 0.882021 (rank : 12) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q08093 | Gene names | Cnn2 | |||
|
Domain Architecture |
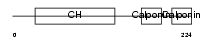
|
|||||
| Description | Calponin-2 (Calponin H2, smooth muscle) (Neutral calponin). | |||||
|
ARHG6_MOUSE
|
||||||
| θ value | 0.00102713 (rank : 13) | NC score | 0.200455 (rank : 16) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8K4I3, Q8C9V4 | Gene names | Arhgef6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 6 (Rac/Cdc42 guanine nucleotide exchange factor 6). | |||||
|
VAV3_MOUSE
|
||||||
| θ value | 0.00102713 (rank : 14) | NC score | 0.203054 (rank : 14) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 404 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9R0C8, Q7TS85 | Gene names | Vav3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein vav-3. | |||||
|
LRCH1_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 15) | NC score | 0.176473 (rank : 23) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P62046 | Gene names | Lrch1, Chdc1, Kiaa1016 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-rich repeats and calponin homology domain-containing protein 1 (Calponin homology domain-containing protein 1). | |||||
|
VAV_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 16) | NC score | 0.201764 (rank : 15) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P27870, Q8BTV7 | Gene names | Vav1, Vav | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene vav (p95vav). | |||||
|
LMO7_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 17) | NC score | 0.338868 (rank : 13) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8WWI1, O15462, O95346, Q9UKC1, Q9UQM5, Q9Y6A7 | Gene names | LMO7, FBX20, FBXO20, KIAA0858 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain only protein 7 (LOMP) (F-box only protein 20). | |||||
|
LRCH1_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 18) | NC score | 0.179558 (rank : 22) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9Y2L9, Q7Z5F6, Q7Z5F7 | Gene names | LRCH1, CHDC1, KIAA1016 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-rich repeats and calponin homology domain-containing protein 1 (Calponin homology domain-containing protein 1) (Neuronal protein 81) (NP81). | |||||
|
VAV_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 19) | NC score | 0.194548 (rank : 17) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P15498, Q15860 | Gene names | VAV1, VAV | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene vav. | |||||
|
VAV3_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 20) | NC score | 0.189593 (rank : 18) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 398 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UKW4, O95230, Q9Y5X8 | Gene names | VAV3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein vav-3. | |||||
|
ARHG6_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 21) | NC score | 0.179750 (rank : 21) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q15052, Q15396, Q5JQ66, Q86XH0 | Gene names | ARHGEF6, COOL2, KIAA0006, PIXA | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 6 (Rac/Cdc42 guanine nucleotide exchange factor 6) (PAK-interacting exchange factor alpha) (Alpha-Pix) (COOL-2). | |||||
|
VAV2_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 22) | NC score | 0.185969 (rank : 20) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q60992 | Gene names | Vav2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein vav-2. | |||||
|
VAV2_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 23) | NC score | 0.186666 (rank : 19) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P52735 | Gene names | VAV2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein vav-2. | |||||
|
LRCH2_HUMAN
|
||||||
| θ value | 0.125558 (rank : 24) | NC score | 0.143932 (rank : 25) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q5VUJ6, Q9HA88, Q9P233 | Gene names | LRCH2, KIAA1495 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeats and calponin homology domain-containing protein 2. | |||||
|
LRCH2_MOUSE
|
||||||
| θ value | 0.125558 (rank : 25) | NC score | 0.141997 (rank : 26) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q3UMG5, Q7TNP9, Q80TC9 | Gene names | Lrch2, Kiaa1495 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeats and calponin homology domain-containing protein 2. | |||||
|
LRCH3_HUMAN
|
||||||
| θ value | 0.163984 (rank : 26) | NC score | 0.150261 (rank : 24) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 270 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96II8, Q96FP9, Q9NT52 | Gene names | LRCH3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeats and calponin homology domain-containing protein 3 precursor. | |||||
|
LRCH3_MOUSE
|
||||||
| θ value | 0.365318 (rank : 27) | NC score | 0.139563 (rank : 27) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BVU0, Q3U222, Q3UZ74 | Gene names | Lrch3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeats and calponin homology domain-containing protein 3 precursor. | |||||
|
PLSI_HUMAN
|
||||||
| θ value | 1.38821 (rank : 28) | NC score | 0.036777 (rank : 36) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q14651 | Gene names | PLS1 | |||
|
Domain Architecture |

|
|||||
| Description | Plastin-1 (I-plastin) (Intestine-specific plastin). | |||||
|
IQGA3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 29) | NC score | 0.058685 (rank : 31) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q86VI3 | Gene names | IQGAP3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating-like protein IQGAP3. | |||||
|
PLSL_HUMAN
|
||||||
| θ value | 6.88961 (rank : 30) | NC score | 0.035550 (rank : 37) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P13796 | Gene names | LCP1, PLS2 | |||
|
Domain Architecture |

|
|||||
| Description | Plastin-2 (L-plastin) (Lymphocyte cytosolic protein 1) (LCP-1) (LC64P). | |||||
|
PLSL_MOUSE
|
||||||
| θ value | 6.88961 (rank : 31) | NC score | 0.036980 (rank : 35) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q61233, Q3UE24, Q8R1X3, Q9CV77 | Gene names | Lcp1, Pls2 | |||
|
Domain Architecture |

|
|||||
| Description | Plastin-2 (L-plastin) (Lymphocyte cytosolic protein 1) (LCP-1) (65 kDa macrophage protein) (pp65). | |||||
|
PLST_HUMAN
|
||||||
| θ value | 8.99809 (rank : 32) | NC score | 0.030530 (rank : 39) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P13797, Q86YI6 | Gene names | PLS3 | |||
|
Domain Architecture |

|
|||||
| Description | Plastin-3 (T-plastin). | |||||
|
PLST_MOUSE
|
||||||
| θ value | 8.99809 (rank : 33) | NC score | 0.030725 (rank : 38) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q99K51 | Gene names | Pls3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plastin-3 (T-plastin). | |||||
|
ARHG7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 34) | NC score | 0.099330 (rank : 30) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q14155, Q6P9G3, Q6PII2, Q86W63, Q8N3M1 | Gene names | ARHGEF7, COOL1, KIAA0142, P85SPR, PAK3BP, PIXB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 7 (PAK-interacting exchange factor beta) (Beta-Pix) (COOL-1) (p85). | |||||
|
GA2L2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 35) | NC score | 0.052800 (rank : 34) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8NHY3, Q8NHY4 | Gene names | GAS2L2, GAR17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 2 (Growth arrest-specific 2-like 2) (GAS2-related protein on chromosome 17) (GAR17 protein). | |||||
|
IQGA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 36) | NC score | 0.053195 (rank : 33) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P46940 | Gene names | IQGAP1, KIAA0051 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating-like protein IQGAP1 (p195). | |||||
|
IQGA1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 37) | NC score | 0.053963 (rank : 32) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9JKF1 | Gene names | Iqgap1 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating-like protein IQGAP1. | |||||
|
LRCH4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 38) | NC score | 0.113289 (rank : 29) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O75427, Q8WV85, Q96ID0 | Gene names | LRCH4, LRN, LRRN4 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-rich repeats and calponin homology domain-containing protein 4 (Leucine-rich neuronal protein). | |||||
|
LRCH4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 39) | NC score | 0.127244 (rank : 28) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q921G6 | Gene names | Lrch4 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-rich repeats and calponin homology domain-containing protein 4. | |||||
|
TAGL3_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 2.44908e-114 (rank : 1) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9UI15, Q96A74 | Gene names | TAGLN3, NP25 | |||
|
Domain Architecture |

|
|||||
| Description | Transgelin-3 (Neuronal protein NP25) (Neuronal protein 22) (NP22). | |||||
|
TAGL3_MOUSE
|
||||||
| NC score | 0.999866 (rank : 2) | θ value | 9.30652e-114 (rank : 2) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9R1Q8 | Gene names | Tagln3, Np25 | |||
|
Domain Architecture |

|
|||||
| Description | Transgelin-3 (Neuronal protein NP25). | |||||
|
TAGL2_HUMAN
|
||||||
| NC score | 0.991180 (rank : 3) | θ value | 6.08852e-81 (rank : 3) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P37802, Q5JRQ6, Q5JRQ7, Q9BUH5, Q9H4P0 | Gene names | TAGLN2, KIAA0120 | |||
|
Domain Architecture |

|
|||||
| Description | Transgelin-2 (SM22-alpha homolog). | |||||
|
TAGL_HUMAN
|
||||||
| NC score | 0.991087 (rank : 4) | θ value | 2.92676e-75 (rank : 5) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q01995, O15542 | Gene names | TAGLN, SM22, WS3-10 | |||
|
Domain Architecture |

|
|||||
| Description | Transgelin (Smooth muscle protein 22-alpha) (SM22-alpha) (WS3-10) (22 kDa actin-binding protein). | |||||
|
TAGL_MOUSE
|
||||||
| NC score | 0.990955 (rank : 5) | θ value | 4.99229e-75 (rank : 6) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P37804, Q545W0 | Gene names | Tagln, Sm22, Sm22a | |||
|
Domain Architecture |

|
|||||
| Description | Transgelin (Smooth muscle protein 22-alpha) (SM22-alpha) (Actin- associated protein p27). | |||||
|
TAGL2_MOUSE
|
||||||
| NC score | 0.990052 (rank : 6) | θ value | 4.36332e-79 (rank : 4) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9WVA4 | Gene names | Tagln2 | |||
|
Domain Architecture |

|
|||||
| Description | Transgelin-2. | |||||
|
CNN1_MOUSE
|
||||||
| NC score | 0.893407 (rank : 7) | θ value | 1.46948e-34 (rank : 9) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q08091 | Gene names | Cnn1 | |||
|
Domain Architecture |

|
|||||
| Description | Calponin-1 (Calponin H1, smooth muscle) (Basic calponin). | |||||
|
CNN1_HUMAN
|
||||||
| NC score | 0.892435 (rank : 8) | θ value | 7.29293e-34 (rank : 10) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P51911, O00638, Q15416, Q8IY93, Q99438 | Gene names | CNN1 | |||
|
Domain Architecture |

|
|||||
| Description | Calponin-1 (Calponin H1, smooth muscle) (Basic calponin). | |||||
|
CNN3_MOUSE
|
||||||
| NC score | 0.888578 (rank : 9) | θ value | 8.61488e-35 (rank : 8) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9DAW9, Q3TW23 | Gene names | Cnn3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calponin-3 (Calponin, acidic isoform). | |||||
|
CNN3_HUMAN
|
||||||
| NC score | 0.887993 (rank : 10) | θ value | 8.61488e-35 (rank : 7) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q15417 | Gene names | CNN3 | |||
|
Domain Architecture |

|
|||||
| Description | Calponin-3 (Calponin, acidic isoform). | |||||
|
CNN2_HUMAN
|
||||||
| NC score | 0.883683 (rank : 11) | θ value | 1.98606e-31 (rank : 11) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q99439, Q92578 | Gene names | CNN2 | |||
|
Domain Architecture |

|
|||||
| Description | Calponin-2 (Calponin H2, smooth muscle) (Neutral calponin). | |||||
|
CNN2_MOUSE
|
||||||
| NC score | 0.882021 (rank : 12) | θ value | 5.77852e-31 (rank : 12) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q08093 | Gene names | Cnn2 | |||
|
Domain Architecture |

|
|||||
| Description | Calponin-2 (Calponin H2, smooth muscle) (Neutral calponin). | |||||
|
LMO7_HUMAN
|
||||||
| NC score | 0.338868 (rank : 13) | θ value | 0.00228821 (rank : 17) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8WWI1, O15462, O95346, Q9UKC1, Q9UQM5, Q9Y6A7 | Gene names | LMO7, FBX20, FBXO20, KIAA0858 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain only protein 7 (LOMP) (F-box only protein 20). | |||||
|
VAV3_MOUSE
|
||||||
| NC score | 0.203054 (rank : 14) | θ value | 0.00102713 (rank : 14) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 404 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9R0C8, Q7TS85 | Gene names | Vav3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein vav-3. | |||||
|
VAV_MOUSE
|
||||||
| NC score | 0.201764 (rank : 15) | θ value | 0.00175202 (rank : 16) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P27870, Q8BTV7 | Gene names | Vav1, Vav | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene vav (p95vav). | |||||
|
ARHG6_MOUSE
|
||||||
| NC score | 0.200455 (rank : 16) | θ value | 0.00102713 (rank : 13) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8K4I3, Q8C9V4 | Gene names | Arhgef6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 6 (Rac/Cdc42 guanine nucleotide exchange factor 6). | |||||
|
VAV_HUMAN
|
||||||
| NC score | 0.194548 (rank : 17) | θ value | 0.00298849 (rank : 19) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P15498, Q15860 | Gene names | VAV1, VAV | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene vav. | |||||
|
VAV3_HUMAN
|
||||||
| NC score | 0.189593 (rank : 18) | θ value | 0.00390308 (rank : 20) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 398 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UKW4, O95230, Q9Y5X8 | Gene names | VAV3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein vav-3. | |||||
|
VAV2_HUMAN
|
||||||
| NC score | 0.186666 (rank : 19) | θ value | 0.0252991 (rank : 23) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P52735 | Gene names | VAV2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein vav-2. | |||||
|
VAV2_MOUSE
|
||||||
| NC score | 0.185969 (rank : 20) | θ value | 0.0193708 (rank : 22) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q60992 | Gene names | Vav2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein vav-2. | |||||
|
ARHG6_HUMAN
|
||||||
| NC score | 0.179750 (rank : 21) | θ value | 0.0193708 (rank : 21) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q15052, Q15396, Q5JQ66, Q86XH0 | Gene names | ARHGEF6, COOL2, KIAA0006, PIXA | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 6 (Rac/Cdc42 guanine nucleotide exchange factor 6) (PAK-interacting exchange factor alpha) (Alpha-Pix) (COOL-2). | |||||
|
LRCH1_HUMAN
|
||||||
| NC score | 0.179558 (rank : 22) | θ value | 0.00298849 (rank : 18) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9Y2L9, Q7Z5F6, Q7Z5F7 | Gene names | LRCH1, CHDC1, KIAA1016 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-rich repeats and calponin homology domain-containing protein 1 (Calponin homology domain-containing protein 1) (Neuronal protein 81) (NP81). | |||||
|
LRCH1_MOUSE
|
||||||
| NC score | 0.176473 (rank : 23) | θ value | 0.00175202 (rank : 15) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P62046 | Gene names | Lrch1, Chdc1, Kiaa1016 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-rich repeats and calponin homology domain-containing protein 1 (Calponin homology domain-containing protein 1). | |||||
|
LRCH3_HUMAN
|
||||||
| NC score | 0.150261 (rank : 24) | θ value | 0.163984 (rank : 26) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 270 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96II8, Q96FP9, Q9NT52 | Gene names | LRCH3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeats and calponin homology domain-containing protein 3 precursor. | |||||
|
LRCH2_HUMAN
|
||||||
| NC score | 0.143932 (rank : 25) | θ value | 0.125558 (rank : 24) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q5VUJ6, Q9HA88, Q9P233 | Gene names | LRCH2, KIAA1495 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeats and calponin homology domain-containing protein 2. | |||||
|
LRCH2_MOUSE
|
||||||
| NC score | 0.141997 (rank : 26) | θ value | 0.125558 (rank : 25) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q3UMG5, Q7TNP9, Q80TC9 | Gene names | Lrch2, Kiaa1495 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeats and calponin homology domain-containing protein 2. | |||||
|
LRCH3_MOUSE
|
||||||
| NC score | 0.139563 (rank : 27) | θ value | 0.365318 (rank : 27) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BVU0, Q3U222, Q3UZ74 | Gene names | Lrch3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeats and calponin homology domain-containing protein 3 precursor. | |||||
|
LRCH4_MOUSE
|
||||||
| NC score | 0.127244 (rank : 28) | θ value | θ > 10 (rank : 39) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q921G6 | Gene names | Lrch4 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-rich repeats and calponin homology domain-containing protein 4. | |||||
|
LRCH4_HUMAN
|
||||||
| NC score | 0.113289 (rank : 29) | θ value | θ > 10 (rank : 38) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O75427, Q8WV85, Q96ID0 | Gene names | LRCH4, LRN, LRRN4 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-rich repeats and calponin homology domain-containing protein 4 (Leucine-rich neuronal protein). | |||||
|
ARHG7_HUMAN
|
||||||
| NC score | 0.099330 (rank : 30) | θ value | θ > 10 (rank : 34) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q14155, Q6P9G3, Q6PII2, Q86W63, Q8N3M1 | Gene names | ARHGEF7, COOL1, KIAA0142, P85SPR, PAK3BP, PIXB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 7 (PAK-interacting exchange factor beta) (Beta-Pix) (COOL-1) (p85). | |||||
|
IQGA3_HUMAN
|
||||||
| NC score | 0.058685 (rank : 31) | θ value | 3.0926 (rank : 29) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q86VI3 | Gene names | IQGAP3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating-like protein IQGAP3. | |||||
|
IQGA1_MOUSE
|
||||||
| NC score | 0.053963 (rank : 32) | θ value | θ > 10 (rank : 37) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9JKF1 | Gene names | Iqgap1 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating-like protein IQGAP1. | |||||
|
IQGA1_HUMAN
|
||||||
| NC score | 0.053195 (rank : 33) | θ value | θ > 10 (rank : 36) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P46940 | Gene names | IQGAP1, KIAA0051 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating-like protein IQGAP1 (p195). | |||||
|
GA2L2_HUMAN
|
||||||
| NC score | 0.052800 (rank : 34) | θ value | θ > 10 (rank : 35) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8NHY3, Q8NHY4 | Gene names | GAS2L2, GAR17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 2 (Growth arrest-specific 2-like 2) (GAS2-related protein on chromosome 17) (GAR17 protein). | |||||
|
PLSL_MOUSE
|
||||||
| NC score | 0.036980 (rank : 35) | θ value | 6.88961 (rank : 31) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q61233, Q3UE24, Q8R1X3, Q9CV77 | Gene names | Lcp1, Pls2 | |||
|
Domain Architecture |

|
|||||
| Description | Plastin-2 (L-plastin) (Lymphocyte cytosolic protein 1) (LCP-1) (65 kDa macrophage protein) (pp65). | |||||
|
PLSI_HUMAN
|
||||||
| NC score | 0.036777 (rank : 36) | θ value | 1.38821 (rank : 28) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q14651 | Gene names | PLS1 | |||
|
Domain Architecture |

|
|||||
| Description | Plastin-1 (I-plastin) (Intestine-specific plastin). | |||||
|
PLSL_HUMAN
|
||||||
| NC score | 0.035550 (rank : 37) | θ value | 6.88961 (rank : 30) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P13796 | Gene names | LCP1, PLS2 | |||
|
Domain Architecture |

|
|||||
| Description | Plastin-2 (L-plastin) (Lymphocyte cytosolic protein 1) (LCP-1) (LC64P). | |||||
|
PLST_MOUSE
|
||||||
| NC score | 0.030725 (rank : 38) | θ value | 8.99809 (rank : 33) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q99K51 | Gene names | Pls3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plastin-3 (T-plastin). | |||||
|
PLST_HUMAN
|
||||||
| NC score | 0.030530 (rank : 39) | θ value | 8.99809 (rank : 32) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P13797, Q86YI6 | Gene names | PLS3 | |||
|
Domain Architecture |

|
|||||
| Description | Plastin-3 (T-plastin). | |||||