Please be patient as the page loads
|
STAU2_HUMAN
|
||||||
| SwissProt Accessions | Q9NUL3, Q6AHY7, Q96HM0, Q96HM1, Q9NVI5, Q9UGG6 | Gene names | STAU2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double-stranded RNA-binding protein Staufen homolog 2. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
STAU2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9NUL3, Q6AHY7, Q96HM0, Q96HM1, Q9NVI5, Q9UGG6 | Gene names | STAU2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double-stranded RNA-binding protein Staufen homolog 2. | |||||
|
STAU2_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.998388 (rank : 2) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8CJ67, Q8BSY8, Q8CJ66, Q8R175, Q91Z19, Q9D5N7 | Gene names | Stau2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double-stranded RNA-binding protein Staufen homolog 2. | |||||
|
STAU1_HUMAN
|
||||||
| θ value | 7.66637e-92 (rank : 3) | NC score | 0.930971 (rank : 3) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O95793, Q6GTM4, Q9H5B4, Q9H5B5, Q9Y3Q2 | Gene names | STAU1, STAU | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-binding protein Staufen homolog 1. | |||||
|
STAU1_MOUSE
|
||||||
| θ value | 2.1655e-78 (rank : 4) | NC score | 0.923561 (rank : 4) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Z108 | Gene names | Stau1, Stau | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-binding protein Staufen homolog 1. | |||||
|
PRKRA_MOUSE
|
||||||
| θ value | 7.58209e-15 (rank : 5) | NC score | 0.595981 (rank : 5) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9WTX2, Q9CZB7 | Gene names | Prkra, Rax | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-inducible double stranded RNA-dependent protein kinase activator A (Protein kinase, interferon-inducible double stranded RNA- dependent activator) (Protein activator of the interferon-induced protein kinase) (PKR-associated protein X) (PKR-associating protein X) (RAX). | |||||
|
PRKRA_HUMAN
|
||||||
| θ value | 1.68911e-14 (rank : 6) | NC score | 0.592697 (rank : 6) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O75569, Q53G24, Q6X7T5, Q8NDK4 | Gene names | PRKRA, PACT, RAX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-inducible double stranded RNA-dependent protein kinase activator A (Protein kinase, interferon-inducible double stranded RNA- dependent activator) (Protein activator of the interferon-induced protein kinase) (PKR-associated protein X) (PKR-associating protein X). | |||||
|
ILF3_MOUSE
|
||||||
| θ value | 7.34386e-10 (rank : 7) | NC score | 0.436981 (rank : 9) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Z1X4, Q80VD5, Q812A1, Q8BP80, Q8C2H8, Q8K588 | Gene names | Ilf3 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin enhancer-binding factor 3. | |||||
|
RED1_MOUSE
|
||||||
| θ value | 2.79066e-09 (rank : 8) | NC score | 0.430855 (rank : 10) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q91ZS8, Q8K3X1, Q91ZS6, Q91ZS7, Q91ZS9, Q99MU8 | Gene names | Adarb1, Adar2, Red1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double-stranded RNA-specific editase 1 (EC 3.5.-.-) (dsRNA adenosine deaminase) (RNA-editing deaminase 1) (RNA-editing enzyme 1). | |||||
|
RED1_HUMAN
|
||||||
| θ value | 3.64472e-09 (rank : 9) | NC score | 0.430691 (rank : 11) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P78563, O00395, O00465, O00691, O00692, P78555, Q8NFD1 | Gene names | ADARB1, ADAR2, DRADA2, RED1 | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-specific editase 1 (EC 3.5.-.-) (dsRNA adenosine deaminase) (RNA-editing deaminase 1) (RNA-editing enzyme 1). | |||||
|
TRBP2_HUMAN
|
||||||
| θ value | 1.80886e-08 (rank : 10) | NC score | 0.514409 (rank : 7) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q15633, Q12878 | Gene names | TARBP2, TRBP | |||
|
Domain Architecture |

|
|||||
| Description | TAR RNA-binding protein 2 (Trans-activation-responsive RNA-binding protein). | |||||
|
DSRAD_MOUSE
|
||||||
| θ value | 1.53129e-07 (rank : 11) | NC score | 0.375865 (rank : 15) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q99MU3, O70375, Q80UZ6, Q8C222, Q99MU2, Q99MU4, Q99MU7 | Gene names | Adar | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-specific adenosine deaminase (EC 3.5.4.-) (DRADA) (RNA adenosine deaminase 1). | |||||
|
ILF3_HUMAN
|
||||||
| θ value | 1.99992e-07 (rank : 12) | NC score | 0.421163 (rank : 12) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q12906, O43409, Q6P1X1, Q86XY7, Q99544, Q99545, Q9BZH4, Q9BZH5, Q9NQ95, Q9NQ96, Q9NQ97, Q9NQ98, Q9NQ99, Q9NQA0, Q9NQA1, Q9NQA2, Q9NRN2, Q9NRN3, Q9NRN4, Q9UMZ9, Q9UN00, Q9UN84, Q9UNA2 | Gene names | ILF3, DRBF, MPHOSPH4, NF90 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin enhancer-binding factor 3 (Nuclear factor of activated T- cells 90 kDa) (NF-AT-90) (Double-stranded RNA-binding protein 76) (DRBP76) (Translational control protein 80) (TCP80) (Nuclear factor associated with dsRNA) (NFAR) (M-phase phosphoprotein 4) (MPP4). | |||||
|
TRBP2_MOUSE
|
||||||
| θ value | 7.59969e-07 (rank : 13) | NC score | 0.498609 (rank : 8) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P97473 | Gene names | Tarbp2, Prbp | |||
|
Domain Architecture |

|
|||||
| Description | TAR RNA-binding protein 2 (Protamine-1 RNA-binding protein) (PRM-1 RNA-binding protein). | |||||
|
RED2_MOUSE
|
||||||
| θ value | 2.21117e-06 (rank : 14) | NC score | 0.381134 (rank : 13) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9JI20, Q3UTP1, Q8CC51, Q9JIE5 | Gene names | Adarb2, Adar3, Red2 | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-specific editase B2 (EC 3.5.-.-) (dsRNA adenosine deaminase B2) (RNA-dependent adenosine deaminase 3) (RNA-editing deaminase 2) (RNA-editing enzyme 2). | |||||
|
DSRAD_HUMAN
|
||||||
| θ value | 0.000158464 (rank : 15) | NC score | 0.380507 (rank : 14) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P55265, O15223, O43859, O43860, Q9BYM3, Q9BYM4 | Gene names | ADAR, ADAR1, DSRAD, IFI4 | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-specific adenosine deaminase (EC 3.5.4.-) (DRADA) (136 kDa double-stranded RNA-binding protein) (P136) (K88DSRBP) (Interferon-inducible protein 4) (IFI-4 protein). | |||||
|
RED2_HUMAN
|
||||||
| θ value | 0.000602161 (rank : 16) | NC score | 0.370259 (rank : 16) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NS39 | Gene names | ADARB2, ADAR3, RED2 | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-specific editase B2 (EC 3.5.-.-) (dsRNA adenosine deaminase B2) (RNA-dependent adenosine deaminase 3) (RNA-editing deaminase 2) (RNA-editing enzyme 2). | |||||
|
E2AK2_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 17) | NC score | 0.036360 (rank : 21) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P19525 | Gene names | EIF2AK2, PKR, PRKR | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced, double-stranded RNA-activated protein kinase (EC 2.7.11.1) (Interferon-inducible RNA-dependent protein kinase) (Protein kinase RNA-activated) (PKR) (p68 kinase) (P1/eIF-2A protein kinase). | |||||
|
E2AK2_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 18) | NC score | 0.028622 (rank : 24) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 864 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q03963, Q61742, Q62026 | Gene names | Eif2ak2, Pkr, Prkr, Tik | |||
|
Domain Architecture |
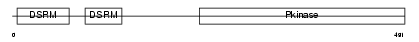
|
|||||
| Description | Interferon-induced, double-stranded RNA-activated protein kinase (EC 2.7.11.1) (Interferon-inducible RNA-dependent protein kinase) (Protein kinase RNA-activated) (PKR) (p68 kinase) (P1/eIF-2A protein kinase) (Serine/threonine-protein kinase TIK). | |||||
|
RNC_HUMAN
|
||||||
| θ value | 0.47712 (rank : 19) | NC score | 0.082844 (rank : 17) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9NRR4, Q7Z5V2, Q86YH0, Q9NW73, Q9Y2V9, Q9Y4Y0 | Gene names | RNASEN, RN3, RNASE3L | |||
|
Domain Architecture |

|
|||||
| Description | Ribonuclease III (EC 3.1.26.3) (RNase III) (Drosha) (p241). | |||||
|
BAT2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 20) | NC score | 0.024481 (rank : 25) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
UB7I1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 21) | NC score | 0.034854 (rank : 22) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P58283, Q3U493, Q3UE56, Q3UGM3, Q68FN0, Q6P1H8, Q6PWY5, Q8BN27, Q8C1U3 | Gene names | Ubce7ip1, Triad3, Uip83, Zin | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin ligase TRIAD3 (EC 6.3.2.-) (Ubiquitin-conjugating enzyme 7-interacting protein 1) (UbcM4-interacting protein 83) (Triad domain- containing protein 3). | |||||
|
HRX_HUMAN
|
||||||
| θ value | 2.36792 (rank : 22) | NC score | 0.020527 (rank : 27) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
AFF2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 23) | NC score | 0.020172 (rank : 28) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O55112 | Gene names | Aff2, Fmr2, Ox19 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 2 (Fragile X mental retardation protein 2 homolog) (Protein FMR-2) (FMR2P) (Protein Ox19). | |||||
|
SON_HUMAN
|
||||||
| θ value | 3.0926 (rank : 24) | NC score | 0.029172 (rank : 23) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
SON_MOUSE
|
||||||
| θ value | 3.0926 (rank : 25) | NC score | 0.042781 (rank : 20) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
UBP42_HUMAN
|
||||||
| θ value | 4.03905 (rank : 26) | NC score | 0.008601 (rank : 35) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H9J4, Q3C166, Q6P9B4 | Gene names | USP42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 42 (EC 3.1.2.15) (Ubiquitin thioesterase 42) (Ubiquitin-specific-processing protease 42) (Deubiquitinating enzyme 42). | |||||
|
CEND3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 27) | NC score | 0.010900 (rank : 32) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R5G7, Q5DTN4, Q6NVF1, Q8R5G6 | Gene names | Centd3, Arap3, Drag1, Kiaa4097 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 3 (Cnt-d3) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 3) (Dual specificity Rho- and Arf-GTPase-activating protein 1). | |||||
|
CEP35_HUMAN
|
||||||
| θ value | 5.27518 (rank : 28) | NC score | 0.009970 (rank : 33) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5VT06, O75068, Q8TDK3, Q8WY20 | Gene names | CEP350, CAP350, GM133 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein 350 (Centrosome-associated protein of 350 kDa). | |||||
|
CO039_HUMAN
|
||||||
| θ value | 5.27518 (rank : 29) | NC score | 0.022575 (rank : 26) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6ZRI6, Q71JB1, Q7L3S0, Q8N3F2, Q96FB6, Q9NTU5 | Gene names | C15orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf39. | |||||
|
PRR12_HUMAN
|
||||||
| θ value | 5.27518 (rank : 30) | NC score | 0.018534 (rank : 29) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
DEPD5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 31) | NC score | 0.018356 (rank : 30) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75140, Q5K3V5, Q5THY9, Q5THZ0, Q5THZ1, Q5THZ3, Q68DR1, Q6MZX3, Q6PEZ1, Q9UGV8, Q9UH13 | Gene names | DEPDC5, KIAA0645 | |||
|
Domain Architecture |

|
|||||
| Description | DEP domain-containing protein 5. | |||||
|
MPP8_HUMAN
|
||||||
| θ value | 8.99809 (rank : 32) | NC score | 0.006023 (rank : 36) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 948 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99549, Q86TK3, Q9BTP1 | Gene names | MPHOSPH8, MPP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | M-phase phosphoprotein 8. | |||||
|
WHRN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 33) | NC score | 0.008618 (rank : 34) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9P202, Q96MZ9, Q9H9F4, Q9UFZ3 | Gene names | WHRN, DFNB31, KIAA1526 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Whirlin (Autosomal recessive deafness type 31 protein). | |||||
|
ZN533_HUMAN
|
||||||
| θ value | 8.99809 (rank : 34) | NC score | 0.014265 (rank : 31) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q569K4, Q49A04, Q6ZMZ7, Q8IY01, Q8N8H2, Q96DK4 | Gene names | ZNF533 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 533. | |||||
|
ILF2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 35) | NC score | 0.057420 (rank : 18) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q12905, Q5SR10, Q5SR11, Q7L7R3, Q9BWD4, Q9P1N0 | Gene names | ILF2, NF45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin enhancer-binding factor 2 (Nuclear factor of activated T- cells 45 kDa). | |||||
|
ILF2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 36) | NC score | 0.057420 (rank : 19) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CXY6, Q3U083, Q5RKG0, Q8CCY9, Q99KS3 | Gene names | Ilf2, Nf45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin enhancer-binding factor 2 (Nuclear factor of activated T- cells 45 kDa). | |||||
|
STAU2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9NUL3, Q6AHY7, Q96HM0, Q96HM1, Q9NVI5, Q9UGG6 | Gene names | STAU2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double-stranded RNA-binding protein Staufen homolog 2. | |||||
|
STAU2_MOUSE
|
||||||
| NC score | 0.998388 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8CJ67, Q8BSY8, Q8CJ66, Q8R175, Q91Z19, Q9D5N7 | Gene names | Stau2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double-stranded RNA-binding protein Staufen homolog 2. | |||||
|
STAU1_HUMAN
|
||||||
| NC score | 0.930971 (rank : 3) | θ value | 7.66637e-92 (rank : 3) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O95793, Q6GTM4, Q9H5B4, Q9H5B5, Q9Y3Q2 | Gene names | STAU1, STAU | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-binding protein Staufen homolog 1. | |||||
|
STAU1_MOUSE
|
||||||
| NC score | 0.923561 (rank : 4) | θ value | 2.1655e-78 (rank : 4) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Z108 | Gene names | Stau1, Stau | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-binding protein Staufen homolog 1. | |||||
|
PRKRA_MOUSE
|
||||||
| NC score | 0.595981 (rank : 5) | θ value | 7.58209e-15 (rank : 5) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9WTX2, Q9CZB7 | Gene names | Prkra, Rax | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-inducible double stranded RNA-dependent protein kinase activator A (Protein kinase, interferon-inducible double stranded RNA- dependent activator) (Protein activator of the interferon-induced protein kinase) (PKR-associated protein X) (PKR-associating protein X) (RAX). | |||||
|
PRKRA_HUMAN
|
||||||
| NC score | 0.592697 (rank : 6) | θ value | 1.68911e-14 (rank : 6) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O75569, Q53G24, Q6X7T5, Q8NDK4 | Gene names | PRKRA, PACT, RAX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-inducible double stranded RNA-dependent protein kinase activator A (Protein kinase, interferon-inducible double stranded RNA- dependent activator) (Protein activator of the interferon-induced protein kinase) (PKR-associated protein X) (PKR-associating protein X). | |||||
|
TRBP2_HUMAN
|
||||||
| NC score | 0.514409 (rank : 7) | θ value | 1.80886e-08 (rank : 10) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q15633, Q12878 | Gene names | TARBP2, TRBP | |||
|
Domain Architecture |

|
|||||
| Description | TAR RNA-binding protein 2 (Trans-activation-responsive RNA-binding protein). | |||||
|
TRBP2_MOUSE
|
||||||
| NC score | 0.498609 (rank : 8) | θ value | 7.59969e-07 (rank : 13) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P97473 | Gene names | Tarbp2, Prbp | |||
|
Domain Architecture |

|
|||||
| Description | TAR RNA-binding protein 2 (Protamine-1 RNA-binding protein) (PRM-1 RNA-binding protein). | |||||
|
ILF3_MOUSE
|
||||||
| NC score | 0.436981 (rank : 9) | θ value | 7.34386e-10 (rank : 7) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Z1X4, Q80VD5, Q812A1, Q8BP80, Q8C2H8, Q8K588 | Gene names | Ilf3 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin enhancer-binding factor 3. | |||||
|
RED1_MOUSE
|
||||||
| NC score | 0.430855 (rank : 10) | θ value | 2.79066e-09 (rank : 8) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q91ZS8, Q8K3X1, Q91ZS6, Q91ZS7, Q91ZS9, Q99MU8 | Gene names | Adarb1, Adar2, Red1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double-stranded RNA-specific editase 1 (EC 3.5.-.-) (dsRNA adenosine deaminase) (RNA-editing deaminase 1) (RNA-editing enzyme 1). | |||||
|
RED1_HUMAN
|
||||||
| NC score | 0.430691 (rank : 11) | θ value | 3.64472e-09 (rank : 9) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P78563, O00395, O00465, O00691, O00692, P78555, Q8NFD1 | Gene names | ADARB1, ADAR2, DRADA2, RED1 | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-specific editase 1 (EC 3.5.-.-) (dsRNA adenosine deaminase) (RNA-editing deaminase 1) (RNA-editing enzyme 1). | |||||
|
ILF3_HUMAN
|
||||||
| NC score | 0.421163 (rank : 12) | θ value | 1.99992e-07 (rank : 12) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q12906, O43409, Q6P1X1, Q86XY7, Q99544, Q99545, Q9BZH4, Q9BZH5, Q9NQ95, Q9NQ96, Q9NQ97, Q9NQ98, Q9NQ99, Q9NQA0, Q9NQA1, Q9NQA2, Q9NRN2, Q9NRN3, Q9NRN4, Q9UMZ9, Q9UN00, Q9UN84, Q9UNA2 | Gene names | ILF3, DRBF, MPHOSPH4, NF90 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin enhancer-binding factor 3 (Nuclear factor of activated T- cells 90 kDa) (NF-AT-90) (Double-stranded RNA-binding protein 76) (DRBP76) (Translational control protein 80) (TCP80) (Nuclear factor associated with dsRNA) (NFAR) (M-phase phosphoprotein 4) (MPP4). | |||||
|
RED2_MOUSE
|
||||||
| NC score | 0.381134 (rank : 13) | θ value | 2.21117e-06 (rank : 14) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9JI20, Q3UTP1, Q8CC51, Q9JIE5 | Gene names | Adarb2, Adar3, Red2 | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-specific editase B2 (EC 3.5.-.-) (dsRNA adenosine deaminase B2) (RNA-dependent adenosine deaminase 3) (RNA-editing deaminase 2) (RNA-editing enzyme 2). | |||||
|
DSRAD_HUMAN
|
||||||
| NC score | 0.380507 (rank : 14) | θ value | 0.000158464 (rank : 15) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P55265, O15223, O43859, O43860, Q9BYM3, Q9BYM4 | Gene names | ADAR, ADAR1, DSRAD, IFI4 | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-specific adenosine deaminase (EC 3.5.4.-) (DRADA) (136 kDa double-stranded RNA-binding protein) (P136) (K88DSRBP) (Interferon-inducible protein 4) (IFI-4 protein). | |||||
|
DSRAD_MOUSE
|
||||||
| NC score | 0.375865 (rank : 15) | θ value | 1.53129e-07 (rank : 11) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q99MU3, O70375, Q80UZ6, Q8C222, Q99MU2, Q99MU4, Q99MU7 | Gene names | Adar | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-specific adenosine deaminase (EC 3.5.4.-) (DRADA) (RNA adenosine deaminase 1). | |||||
|
RED2_HUMAN
|
||||||
| NC score | 0.370259 (rank : 16) | θ value | 0.000602161 (rank : 16) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NS39 | Gene names | ADARB2, ADAR3, RED2 | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-specific editase B2 (EC 3.5.-.-) (dsRNA adenosine deaminase B2) (RNA-dependent adenosine deaminase 3) (RNA-editing deaminase 2) (RNA-editing enzyme 2). | |||||
|
RNC_HUMAN
|
||||||
| NC score | 0.082844 (rank : 17) | θ value | 0.47712 (rank : 19) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9NRR4, Q7Z5V2, Q86YH0, Q9NW73, Q9Y2V9, Q9Y4Y0 | Gene names | RNASEN, RN3, RNASE3L | |||
|
Domain Architecture |

|
|||||
| Description | Ribonuclease III (EC 3.1.26.3) (RNase III) (Drosha) (p241). | |||||
|
ILF2_HUMAN
|
||||||
| NC score | 0.057420 (rank : 18) | θ value | θ > 10 (rank : 35) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q12905, Q5SR10, Q5SR11, Q7L7R3, Q9BWD4, Q9P1N0 | Gene names | ILF2, NF45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin enhancer-binding factor 2 (Nuclear factor of activated T- cells 45 kDa). | |||||
|
ILF2_MOUSE
|
||||||
| NC score | 0.057420 (rank : 19) | θ value | θ > 10 (rank : 36) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CXY6, Q3U083, Q5RKG0, Q8CCY9, Q99KS3 | Gene names | Ilf2, Nf45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin enhancer-binding factor 2 (Nuclear factor of activated T- cells 45 kDa). | |||||
|
SON_MOUSE
|
||||||
| NC score | 0.042781 (rank : 20) | θ value | 3.0926 (rank : 25) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
E2AK2_HUMAN
|
||||||
| NC score | 0.036360 (rank : 21) | θ value | 0.0431538 (rank : 17) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P19525 | Gene names | EIF2AK2, PKR, PRKR | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced, double-stranded RNA-activated protein kinase (EC 2.7.11.1) (Interferon-inducible RNA-dependent protein kinase) (Protein kinase RNA-activated) (PKR) (p68 kinase) (P1/eIF-2A protein kinase). | |||||
|
UB7I1_MOUSE
|
||||||
| NC score | 0.034854 (rank : 22) | θ value | 1.38821 (rank : 21) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P58283, Q3U493, Q3UE56, Q3UGM3, Q68FN0, Q6P1H8, Q6PWY5, Q8BN27, Q8C1U3 | Gene names | Ubce7ip1, Triad3, Uip83, Zin | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin ligase TRIAD3 (EC 6.3.2.-) (Ubiquitin-conjugating enzyme 7-interacting protein 1) (UbcM4-interacting protein 83) (Triad domain- containing protein 3). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.029172 (rank : 23) | θ value | 3.0926 (rank : 24) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
E2AK2_MOUSE
|
||||||
| NC score | 0.028622 (rank : 24) | θ value | 0.0431538 (rank : 18) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 864 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q03963, Q61742, Q62026 | Gene names | Eif2ak2, Pkr, Prkr, Tik | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced, double-stranded RNA-activated protein kinase (EC 2.7.11.1) (Interferon-inducible RNA-dependent protein kinase) (Protein kinase RNA-activated) (PKR) (p68 kinase) (P1/eIF-2A protein kinase) (Serine/threonine-protein kinase TIK). | |||||
|
BAT2_MOUSE
|
||||||
| NC score | 0.024481 (rank : 25) | θ value | 1.06291 (rank : 20) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
CO039_HUMAN
|
||||||
| NC score | 0.022575 (rank : 26) | θ value | 5.27518 (rank : 29) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6ZRI6, Q71JB1, Q7L3S0, Q8N3F2, Q96FB6, Q9NTU5 | Gene names | C15orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf39. | |||||
|
HRX_HUMAN
|
||||||
| NC score | 0.020527 (rank : 27) | θ value | 2.36792 (rank : 22) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
AFF2_MOUSE
|
||||||
| NC score | 0.020172 (rank : 28) | θ value | 3.0926 (rank : 23) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O55112 | Gene names | Aff2, Fmr2, Ox19 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 2 (Fragile X mental retardation protein 2 homolog) (Protein FMR-2) (FMR2P) (Protein Ox19). | |||||
|
PRR12_HUMAN
|
||||||
| NC score | 0.018534 (rank : 29) | θ value | 5.27518 (rank : 30) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
DEPD5_HUMAN
|
||||||
| NC score | 0.018356 (rank : 30) | θ value | 6.88961 (rank : 31) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75140, Q5K3V5, Q5THY9, Q5THZ0, Q5THZ1, Q5THZ3, Q68DR1, Q6MZX3, Q6PEZ1, Q9UGV8, Q9UH13 | Gene names | DEPDC5, KIAA0645 | |||
|
Domain Architecture |

|
|||||
| Description | DEP domain-containing protein 5. | |||||
|
ZN533_HUMAN
|
||||||
| NC score | 0.014265 (rank : 31) | θ value | 8.99809 (rank : 34) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q569K4, Q49A04, Q6ZMZ7, Q8IY01, Q8N8H2, Q96DK4 | Gene names | ZNF533 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 533. | |||||
|
CEND3_MOUSE
|
||||||
| NC score | 0.010900 (rank : 32) | θ value | 5.27518 (rank : 27) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R5G7, Q5DTN4, Q6NVF1, Q8R5G6 | Gene names | Centd3, Arap3, Drag1, Kiaa4097 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 3 (Cnt-d3) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 3) (Dual specificity Rho- and Arf-GTPase-activating protein 1). | |||||
|
CEP35_HUMAN
|
||||||
| NC score | 0.009970 (rank : 33) | θ value | 5.27518 (rank : 28) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5VT06, O75068, Q8TDK3, Q8WY20 | Gene names | CEP350, CAP350, GM133 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein 350 (Centrosome-associated protein of 350 kDa). | |||||
|
WHRN_HUMAN
|
||||||
| NC score | 0.008618 (rank : 34) | θ value | 8.99809 (rank : 33) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9P202, Q96MZ9, Q9H9F4, Q9UFZ3 | Gene names | WHRN, DFNB31, KIAA1526 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Whirlin (Autosomal recessive deafness type 31 protein). | |||||
|
UBP42_HUMAN
|
||||||
| NC score | 0.008601 (rank : 35) | θ value | 4.03905 (rank : 26) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H9J4, Q3C166, Q6P9B4 | Gene names | USP42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 42 (EC 3.1.2.15) (Ubiquitin thioesterase 42) (Ubiquitin-specific-processing protease 42) (Deubiquitinating enzyme 42). | |||||
|
MPP8_HUMAN
|
||||||
| NC score | 0.006023 (rank : 36) | θ value | 8.99809 (rank : 32) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 948 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99549, Q86TK3, Q9BTP1 | Gene names | MPHOSPH8, MPP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | M-phase phosphoprotein 8. | |||||