Please be patient as the page loads
|
STAU1_MOUSE
|
||||||
| SwissProt Accessions | Q9Z108 | Gene names | Stau1, Stau | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-binding protein Staufen homolog 1. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
STAU1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.993254 (rank : 2) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O95793, Q6GTM4, Q9H5B4, Q9H5B5, Q9Y3Q2 | Gene names | STAU1, STAU | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-binding protein Staufen homolog 1. | |||||
|
STAU1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9Z108 | Gene names | Stau1, Stau | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-binding protein Staufen homolog 1. | |||||
|
STAU2_MOUSE
|
||||||
| θ value | 1.77149e-80 (rank : 3) | NC score | 0.921731 (rank : 4) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8CJ67, Q8BSY8, Q8CJ66, Q8R175, Q91Z19, Q9D5N7 | Gene names | Stau2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double-stranded RNA-binding protein Staufen homolog 2. | |||||
|
STAU2_HUMAN
|
||||||
| θ value | 2.1655e-78 (rank : 4) | NC score | 0.923561 (rank : 3) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NUL3, Q6AHY7, Q96HM0, Q96HM1, Q9NVI5, Q9UGG6 | Gene names | STAU2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double-stranded RNA-binding protein Staufen homolog 2. | |||||
|
PRKRA_MOUSE
|
||||||
| θ value | 6.00763e-12 (rank : 5) | NC score | 0.570869 (rank : 5) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9WTX2, Q9CZB7 | Gene names | Prkra, Rax | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-inducible double stranded RNA-dependent protein kinase activator A (Protein kinase, interferon-inducible double stranded RNA- dependent activator) (Protein activator of the interferon-induced protein kinase) (PKR-associated protein X) (PKR-associating protein X) (RAX). | |||||
|
PRKRA_HUMAN
|
||||||
| θ value | 7.84624e-12 (rank : 6) | NC score | 0.568228 (rank : 6) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O75569, Q53G24, Q6X7T5, Q8NDK4 | Gene names | PRKRA, PACT, RAX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-inducible double stranded RNA-dependent protein kinase activator A (Protein kinase, interferon-inducible double stranded RNA- dependent activator) (Protein activator of the interferon-induced protein kinase) (PKR-associated protein X) (PKR-associating protein X). | |||||
|
TRBP2_MOUSE
|
||||||
| θ value | 3.41135e-07 (rank : 7) | NC score | 0.480676 (rank : 8) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P97473 | Gene names | Tarbp2, Prbp | |||
|
Domain Architecture |

|
|||||
| Description | TAR RNA-binding protein 2 (Protamine-1 RNA-binding protein) (PRM-1 RNA-binding protein). | |||||
|
TRBP2_HUMAN
|
||||||
| θ value | 5.81887e-07 (rank : 8) | NC score | 0.495989 (rank : 7) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q15633, Q12878 | Gene names | TARBP2, TRBP | |||
|
Domain Architecture |

|
|||||
| Description | TAR RNA-binding protein 2 (Trans-activation-responsive RNA-binding protein). | |||||
|
ILF3_HUMAN
|
||||||
| θ value | 8.40245e-06 (rank : 9) | NC score | 0.349496 (rank : 10) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q12906, O43409, Q6P1X1, Q86XY7, Q99544, Q99545, Q9BZH4, Q9BZH5, Q9NQ95, Q9NQ96, Q9NQ97, Q9NQ98, Q9NQ99, Q9NQA0, Q9NQA1, Q9NQA2, Q9NRN2, Q9NRN3, Q9NRN4, Q9UMZ9, Q9UN00, Q9UN84, Q9UNA2 | Gene names | ILF3, DRBF, MPHOSPH4, NF90 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin enhancer-binding factor 3 (Nuclear factor of activated T- cells 90 kDa) (NF-AT-90) (Double-stranded RNA-binding protein 76) (DRBP76) (Translational control protein 80) (TCP80) (Nuclear factor associated with dsRNA) (NFAR) (M-phase phosphoprotein 4) (MPP4). | |||||
|
ILF3_MOUSE
|
||||||
| θ value | 8.40245e-06 (rank : 10) | NC score | 0.359453 (rank : 9) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Z1X4, Q80VD5, Q812A1, Q8BP80, Q8C2H8, Q8K588 | Gene names | Ilf3 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin enhancer-binding factor 3. | |||||
|
RED1_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 11) | NC score | 0.316379 (rank : 11) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P78563, O00395, O00465, O00691, O00692, P78555, Q8NFD1 | Gene names | ADARB1, ADAR2, DRADA2, RED1 | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-specific editase 1 (EC 3.5.-.-) (dsRNA adenosine deaminase) (RNA-editing deaminase 1) (RNA-editing enzyme 1). | |||||
|
RED1_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 12) | NC score | 0.315635 (rank : 12) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q91ZS8, Q8K3X1, Q91ZS6, Q91ZS7, Q91ZS9, Q99MU8 | Gene names | Adarb1, Adar2, Red1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double-stranded RNA-specific editase 1 (EC 3.5.-.-) (dsRNA adenosine deaminase) (RNA-editing deaminase 1) (RNA-editing enzyme 1). | |||||
|
DSRAD_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 13) | NC score | 0.271868 (rank : 13) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P55265, O15223, O43859, O43860, Q9BYM3, Q9BYM4 | Gene names | ADAR, ADAR1, DSRAD, IFI4 | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-specific adenosine deaminase (EC 3.5.4.-) (DRADA) (136 kDa double-stranded RNA-binding protein) (P136) (K88DSRBP) (Interferon-inducible protein 4) (IFI-4 protein). | |||||
|
RED2_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 14) | NC score | 0.264935 (rank : 15) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NS39 | Gene names | ADARB2, ADAR3, RED2 | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-specific editase B2 (EC 3.5.-.-) (dsRNA adenosine deaminase B2) (RNA-dependent adenosine deaminase 3) (RNA-editing deaminase 2) (RNA-editing enzyme 2). | |||||
|
DCP1A_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 15) | NC score | 0.073843 (rank : 18) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NPI6 | Gene names | DCP1A, SMIF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | mRNA decapping enzyme 1A (EC 3.-.-.-) (Transcription factor SMIF) (Smad4-interacting transcriptional co-activator). | |||||
|
RED2_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 16) | NC score | 0.265919 (rank : 14) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9JI20, Q3UTP1, Q8CC51, Q9JIE5 | Gene names | Adarb2, Adar3, Red2 | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-specific editase B2 (EC 3.5.-.-) (dsRNA adenosine deaminase B2) (RNA-dependent adenosine deaminase 3) (RNA-editing deaminase 2) (RNA-editing enzyme 2). | |||||
|
MAP1B_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 17) | NC score | 0.061877 (rank : 20) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
MAP1B_MOUSE
|
||||||
| θ value | 0.125558 (rank : 18) | NC score | 0.066612 (rank : 19) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
E2AK2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 19) | NC score | 0.026436 (rank : 23) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P19525 | Gene names | EIF2AK2, PKR, PRKR | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced, double-stranded RNA-activated protein kinase (EC 2.7.11.1) (Interferon-inducible RNA-dependent protein kinase) (Protein kinase RNA-activated) (PKR) (p68 kinase) (P1/eIF-2A protein kinase). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 0.62314 (rank : 20) | NC score | 0.009228 (rank : 34) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
BAT2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 21) | NC score | 0.026330 (rank : 24) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
DCP1A_MOUSE
|
||||||
| θ value | 0.813845 (rank : 22) | NC score | 0.075559 (rank : 17) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q91YD3, Q6NZE3 | Gene names | Dcp1a, Mitc1, Smif | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | mRNA decapping enzyme 1A (EC 3.-.-.-) (Transcription factor SMIF) (MAD homolog 4-interacting transcription coactivator 1) (Smad4-interacting transcriptional co-activator). | |||||
|
BMP2K_HUMAN
|
||||||
| θ value | 1.38821 (rank : 23) | NC score | 0.014607 (rank : 31) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 867 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NSY1, O94791, Q8IYF2, Q8N2G7, Q8NHG9, Q9NTG8 | Gene names | BMP2K, BIKE | |||
|
Domain Architecture |
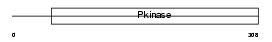
|
|||||
| Description | BMP-2-inducible protein kinase (EC 2.7.11.1) (BIKe). | |||||
|
DSRAD_MOUSE
|
||||||
| θ value | 1.38821 (rank : 24) | NC score | 0.248059 (rank : 16) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q99MU3, O70375, Q80UZ6, Q8C222, Q99MU2, Q99MU4, Q99MU7 | Gene names | Adar | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-specific adenosine deaminase (EC 3.5.4.-) (DRADA) (RNA adenosine deaminase 1). | |||||
|
PAF49_HUMAN
|
||||||
| θ value | 3.0926 (rank : 25) | NC score | 0.029408 (rank : 22) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O15446, Q32N11, Q7Z5U2, Q9UPF6 | Gene names | CD3EAP, ASE1, CAST, PAF49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase I-associated factor PAF49 (Anti-sense to ERCC-1 protein) (ASE-1) (CD3-epsilon-associated protein) (CD3E-associated protein) (CAST). | |||||
|
SYTL5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 26) | NC score | 0.009551 (rank : 33) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8TDW5 | Gene names | SYTL5, SLP5 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 5. | |||||
|
THAP4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 27) | NC score | 0.020935 (rank : 28) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WY91, Q53NU7, Q6GRN0, Q6IPJ3, Q9NW26, Q9Y325 | Gene names | THAP4 | |||
|
Domain Architecture |
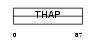
|
|||||
| Description | THAP domain-containing protein 4. | |||||
|
SMAP1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 28) | NC score | 0.020849 (rank : 30) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q91VZ6, Q497I6, Q68EF3 | Gene names | Smap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1. | |||||
|
CCD12_MOUSE
|
||||||
| θ value | 5.27518 (rank : 29) | NC score | 0.025939 (rank : 25) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8R344, Q9CZH5 | Gene names | Ccdc12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 12. | |||||
|
SON_HUMAN
|
||||||
| θ value | 5.27518 (rank : 30) | NC score | 0.025586 (rank : 26) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
SON_MOUSE
|
||||||
| θ value | 5.27518 (rank : 31) | NC score | 0.038557 (rank : 21) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
UBN1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 32) | NC score | 0.020923 (rank : 29) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NPG3, Q13079, Q9P1P7 | Gene names | UBN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubinuclein (Ubiquitously expressed nuclear protein) (VT4). | |||||
|
DHX9_HUMAN
|
||||||
| θ value | 6.88961 (rank : 33) | NC score | 0.013490 (rank : 32) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q08211, Q5VY62, Q6PD69, Q99556 | Gene names | DHX9, DDX9, NDH2 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase A (EC 3.6.1.-) (Nuclear DNA helicase II) (NDH II) (DEAH box protein 9). | |||||
|
TCOF_MOUSE
|
||||||
| θ value | 6.88961 (rank : 34) | NC score | 0.023460 (rank : 27) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
UBP44_HUMAN
|
||||||
| θ value | 6.88961 (rank : 35) | NC score | 0.006698 (rank : 36) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H0E7 | Gene names | USP44 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 44 (EC 3.1.2.15) (Ubiquitin thioesterase 44) (Ubiquitin-specific-processing protease 44) (Deubiquitinating enzyme 44). | |||||
|
MKL1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 36) | NC score | 0.008693 (rank : 35) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8K4J6 | Gene names | Mkl1, Bsac | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein homolog) (Basic SAP coiled-coil transcription activator). | |||||
|
STAU1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9Z108 | Gene names | Stau1, Stau | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-binding protein Staufen homolog 1. | |||||
|
STAU1_HUMAN
|
||||||
| NC score | 0.993254 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O95793, Q6GTM4, Q9H5B4, Q9H5B5, Q9Y3Q2 | Gene names | STAU1, STAU | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-binding protein Staufen homolog 1. | |||||
|
STAU2_HUMAN
|
||||||
| NC score | 0.923561 (rank : 3) | θ value | 2.1655e-78 (rank : 4) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NUL3, Q6AHY7, Q96HM0, Q96HM1, Q9NVI5, Q9UGG6 | Gene names | STAU2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double-stranded RNA-binding protein Staufen homolog 2. | |||||
|
STAU2_MOUSE
|
||||||
| NC score | 0.921731 (rank : 4) | θ value | 1.77149e-80 (rank : 3) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8CJ67, Q8BSY8, Q8CJ66, Q8R175, Q91Z19, Q9D5N7 | Gene names | Stau2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double-stranded RNA-binding protein Staufen homolog 2. | |||||
|
PRKRA_MOUSE
|
||||||
| NC score | 0.570869 (rank : 5) | θ value | 6.00763e-12 (rank : 5) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9WTX2, Q9CZB7 | Gene names | Prkra, Rax | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-inducible double stranded RNA-dependent protein kinase activator A (Protein kinase, interferon-inducible double stranded RNA- dependent activator) (Protein activator of the interferon-induced protein kinase) (PKR-associated protein X) (PKR-associating protein X) (RAX). | |||||
|
PRKRA_HUMAN
|
||||||
| NC score | 0.568228 (rank : 6) | θ value | 7.84624e-12 (rank : 6) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O75569, Q53G24, Q6X7T5, Q8NDK4 | Gene names | PRKRA, PACT, RAX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-inducible double stranded RNA-dependent protein kinase activator A (Protein kinase, interferon-inducible double stranded RNA- dependent activator) (Protein activator of the interferon-induced protein kinase) (PKR-associated protein X) (PKR-associating protein X). | |||||
|
TRBP2_HUMAN
|
||||||
| NC score | 0.495989 (rank : 7) | θ value | 5.81887e-07 (rank : 8) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q15633, Q12878 | Gene names | TARBP2, TRBP | |||
|
Domain Architecture |

|
|||||
| Description | TAR RNA-binding protein 2 (Trans-activation-responsive RNA-binding protein). | |||||
|
TRBP2_MOUSE
|
||||||
| NC score | 0.480676 (rank : 8) | θ value | 3.41135e-07 (rank : 7) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P97473 | Gene names | Tarbp2, Prbp | |||
|
Domain Architecture |

|
|||||
| Description | TAR RNA-binding protein 2 (Protamine-1 RNA-binding protein) (PRM-1 RNA-binding protein). | |||||
|
ILF3_MOUSE
|
||||||
| NC score | 0.359453 (rank : 9) | θ value | 8.40245e-06 (rank : 10) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Z1X4, Q80VD5, Q812A1, Q8BP80, Q8C2H8, Q8K588 | Gene names | Ilf3 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin enhancer-binding factor 3. | |||||
|
ILF3_HUMAN
|
||||||
| NC score | 0.349496 (rank : 10) | θ value | 8.40245e-06 (rank : 9) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q12906, O43409, Q6P1X1, Q86XY7, Q99544, Q99545, Q9BZH4, Q9BZH5, Q9NQ95, Q9NQ96, Q9NQ97, Q9NQ98, Q9NQ99, Q9NQA0, Q9NQA1, Q9NQA2, Q9NRN2, Q9NRN3, Q9NRN4, Q9UMZ9, Q9UN00, Q9UN84, Q9UNA2 | Gene names | ILF3, DRBF, MPHOSPH4, NF90 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin enhancer-binding factor 3 (Nuclear factor of activated T- cells 90 kDa) (NF-AT-90) (Double-stranded RNA-binding protein 76) (DRBP76) (Translational control protein 80) (TCP80) (Nuclear factor associated with dsRNA) (NFAR) (M-phase phosphoprotein 4) (MPP4). | |||||
|
RED1_HUMAN
|
||||||
| NC score | 0.316379 (rank : 11) | θ value | 0.00134147 (rank : 11) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P78563, O00395, O00465, O00691, O00692, P78555, Q8NFD1 | Gene names | ADARB1, ADAR2, DRADA2, RED1 | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-specific editase 1 (EC 3.5.-.-) (dsRNA adenosine deaminase) (RNA-editing deaminase 1) (RNA-editing enzyme 1). | |||||
|
RED1_MOUSE
|
||||||
| NC score | 0.315635 (rank : 12) | θ value | 0.00134147 (rank : 12) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q91ZS8, Q8K3X1, Q91ZS6, Q91ZS7, Q91ZS9, Q99MU8 | Gene names | Adarb1, Adar2, Red1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double-stranded RNA-specific editase 1 (EC 3.5.-.-) (dsRNA adenosine deaminase) (RNA-editing deaminase 1) (RNA-editing enzyme 1). | |||||
|
DSRAD_HUMAN
|
||||||
| NC score | 0.271868 (rank : 13) | θ value | 0.00509761 (rank : 13) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P55265, O15223, O43859, O43860, Q9BYM3, Q9BYM4 | Gene names | ADAR, ADAR1, DSRAD, IFI4 | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-specific adenosine deaminase (EC 3.5.4.-) (DRADA) (136 kDa double-stranded RNA-binding protein) (P136) (K88DSRBP) (Interferon-inducible protein 4) (IFI-4 protein). | |||||
|
RED2_MOUSE
|
||||||
| NC score | 0.265919 (rank : 14) | θ value | 0.0736092 (rank : 16) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9JI20, Q3UTP1, Q8CC51, Q9JIE5 | Gene names | Adarb2, Adar3, Red2 | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-specific editase B2 (EC 3.5.-.-) (dsRNA adenosine deaminase B2) (RNA-dependent adenosine deaminase 3) (RNA-editing deaminase 2) (RNA-editing enzyme 2). | |||||
|
RED2_HUMAN
|
||||||
| NC score | 0.264935 (rank : 15) | θ value | 0.0113563 (rank : 14) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NS39 | Gene names | ADARB2, ADAR3, RED2 | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-specific editase B2 (EC 3.5.-.-) (dsRNA adenosine deaminase B2) (RNA-dependent adenosine deaminase 3) (RNA-editing deaminase 2) (RNA-editing enzyme 2). | |||||
|
DSRAD_MOUSE
|
||||||
| NC score | 0.248059 (rank : 16) | θ value | 1.38821 (rank : 24) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q99MU3, O70375, Q80UZ6, Q8C222, Q99MU2, Q99MU4, Q99MU7 | Gene names | Adar | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-specific adenosine deaminase (EC 3.5.4.-) (DRADA) (RNA adenosine deaminase 1). | |||||
|
DCP1A_MOUSE
|
||||||
| NC score | 0.075559 (rank : 17) | θ value | 0.813845 (rank : 22) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q91YD3, Q6NZE3 | Gene names | Dcp1a, Mitc1, Smif | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | mRNA decapping enzyme 1A (EC 3.-.-.-) (Transcription factor SMIF) (MAD homolog 4-interacting transcription coactivator 1) (Smad4-interacting transcriptional co-activator). | |||||
|
DCP1A_HUMAN
|
||||||
| NC score | 0.073843 (rank : 18) | θ value | 0.0736092 (rank : 15) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NPI6 | Gene names | DCP1A, SMIF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | mRNA decapping enzyme 1A (EC 3.-.-.-) (Transcription factor SMIF) (Smad4-interacting transcriptional co-activator). | |||||
|
MAP1B_MOUSE
|
||||||
| NC score | 0.066612 (rank : 19) | θ value | 0.125558 (rank : 18) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
MAP1B_HUMAN
|
||||||
| NC score | 0.061877 (rank : 20) | θ value | 0.0961366 (rank : 17) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
SON_MOUSE
|
||||||
| NC score | 0.038557 (rank : 21) | θ value | 5.27518 (rank : 31) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
PAF49_HUMAN
|
||||||
| NC score | 0.029408 (rank : 22) | θ value | 3.0926 (rank : 25) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O15446, Q32N11, Q7Z5U2, Q9UPF6 | Gene names | CD3EAP, ASE1, CAST, PAF49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase I-associated factor PAF49 (Anti-sense to ERCC-1 protein) (ASE-1) (CD3-epsilon-associated protein) (CD3E-associated protein) (CAST). | |||||
|
E2AK2_HUMAN
|
||||||
| NC score | 0.026436 (rank : 23) | θ value | 0.47712 (rank : 19) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P19525 | Gene names | EIF2AK2, PKR, PRKR | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced, double-stranded RNA-activated protein kinase (EC 2.7.11.1) (Interferon-inducible RNA-dependent protein kinase) (Protein kinase RNA-activated) (PKR) (p68 kinase) (P1/eIF-2A protein kinase). | |||||
|
BAT2_HUMAN
|
||||||
| NC score | 0.026330 (rank : 24) | θ value | 0.813845 (rank : 21) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
CCD12_MOUSE
|
||||||
| NC score | 0.025939 (rank : 25) | θ value | 5.27518 (rank : 29) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8R344, Q9CZH5 | Gene names | Ccdc12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 12. | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.025586 (rank : 26) | θ value | 5.27518 (rank : 30) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
TCOF_MOUSE
|
||||||
| NC score | 0.023460 (rank : 27) | θ value | 6.88961 (rank : 34) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
THAP4_HUMAN
|
||||||
| NC score | 0.020935 (rank : 28) | θ value | 3.0926 (rank : 27) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WY91, Q53NU7, Q6GRN0, Q6IPJ3, Q9NW26, Q9Y325 | Gene names | THAP4 | |||
|
Domain Architecture |

|
|||||
| Description | THAP domain-containing protein 4. | |||||
|
UBN1_HUMAN
|
||||||
| NC score | 0.020923 (rank : 29) | θ value | 5.27518 (rank : 32) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NPG3, Q13079, Q9P1P7 | Gene names | UBN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubinuclein (Ubiquitously expressed nuclear protein) (VT4). | |||||
|
SMAP1_MOUSE
|
||||||
| NC score | 0.020849 (rank : 30) | θ value | 4.03905 (rank : 28) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q91VZ6, Q497I6, Q68EF3 | Gene names | Smap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1. | |||||
|
BMP2K_HUMAN
|
||||||
| NC score | 0.014607 (rank : 31) | θ value | 1.38821 (rank : 23) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 867 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NSY1, O94791, Q8IYF2, Q8N2G7, Q8NHG9, Q9NTG8 | Gene names | BMP2K, BIKE | |||
|
Domain Architecture |

|
|||||
| Description | BMP-2-inducible protein kinase (EC 2.7.11.1) (BIKe). | |||||
|
DHX9_HUMAN
|
||||||
| NC score | 0.013490 (rank : 32) | θ value | 6.88961 (rank : 33) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q08211, Q5VY62, Q6PD69, Q99556 | Gene names | DHX9, DDX9, NDH2 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase A (EC 3.6.1.-) (Nuclear DNA helicase II) (NDH II) (DEAH box protein 9). | |||||
|
SYTL5_HUMAN
|
||||||
| NC score | 0.009551 (rank : 33) | θ value | 3.0926 (rank : 26) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8TDW5 | Gene names | SYTL5, SLP5 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 5. | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.009228 (rank : 34) | θ value | 0.62314 (rank : 20) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
MKL1_MOUSE
|
||||||
| NC score | 0.008693 (rank : 35) | θ value | 8.99809 (rank : 36) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8K4J6 | Gene names | Mkl1, Bsac | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein homolog) (Basic SAP coiled-coil transcription activator). | |||||
|
UBP44_HUMAN
|
||||||
| NC score | 0.006698 (rank : 36) | θ value | 6.88961 (rank : 35) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H0E7 | Gene names | USP44 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 44 (EC 3.1.2.15) (Ubiquitin thioesterase 44) (Ubiquitin-specific-processing protease 44) (Deubiquitinating enzyme 44). | |||||