Please be patient as the page loads
|
SAP_MOUSE
|
||||||
| SwissProt Accessions | Q61207, Q60861, Q64006, Q64219 | Gene names | Psap, Sgp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sulfated glycoprotein 1 precursor (SGP-1) (Prosaposin). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SAP_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.979211 (rank : 2) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P07602, P07292, P15793, P78538, P78541, P78546, P78547, P78558, Q6IBQ6, Q92739, Q92740, Q92741, Q92742 | Gene names | PSAP | |||
|
Domain Architecture |

|
|||||
| Description | Proactivator polypeptide precursor [Contains: Saposin A (Protein A); Saposin B-Val; Saposin B (Sphingolipid activator protein 1) (SAP-1) (Cerebroside sulfate activator) (CSAct) (Dispersin) (Sulfatide/GM1 activator); Saposin C (Co-beta-glucosidase) (A1 activator) (Glucosylceramidase activator) (Sphingolipid activator protein 2) (SAP-2); Saposin D (Protein C) (Component C)]. | |||||
|
SAP_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q61207, Q60861, Q64006, Q64219 | Gene names | Psap, Sgp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sulfated glycoprotein 1 precursor (SGP-1) (Prosaposin). | |||||
|
PSPB_MOUSE
|
||||||
| θ value | 8.36355e-22 (rank : 3) | NC score | 0.671588 (rank : 3) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P50405 | Gene names | Sftpb, Sftp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pulmonary surfactant-associated protein B precursor (SP-B) (Pulmonary surfactant-associated proteolipid SPL(Phe)). | |||||
|
PSPB_HUMAN
|
||||||
| θ value | 2.35696e-16 (rank : 4) | NC score | 0.632409 (rank : 4) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P07988, Q96R04 | Gene names | SFTPB, SFTP3 | |||
|
Domain Architecture |
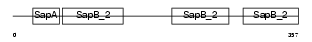
|
|||||
| Description | Pulmonary surfactant-associated protein B precursor (SP-B) (6 kDa protein) (Pulmonary surfactant-associated proteolipid SPL(Phe)) (18 kDa pulmonary-surfactant protein). | |||||
|
HRX_HUMAN
|
||||||
| θ value | 0.365318 (rank : 5) | NC score | 0.031989 (rank : 12) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
TRAIP_HUMAN
|
||||||
| θ value | 0.365318 (rank : 6) | NC score | 0.035405 (rank : 9) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BWF2, O00467 | Gene names | TRAIP, TRIP | |||
|
Domain Architecture |

|
|||||
| Description | TRAF-interacting protein. | |||||
|
ANGL5_HUMAN
|
||||||
| θ value | 0.47712 (rank : 7) | NC score | 0.025159 (rank : 14) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q86XS5, Q86VR9 | Gene names | ANGPTL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiopoietin-related protein 5 precursor (Angiopoietin-like 5). | |||||
|
TRAIP_MOUSE
|
||||||
| θ value | 0.813845 (rank : 8) | NC score | 0.032214 (rank : 11) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 745 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VIG6, O08854, Q922M8, Q9CPP4 | Gene names | Traip, Trip | |||
|
Domain Architecture |

|
|||||
| Description | TRAF-interacting protein. | |||||
|
AOAH_HUMAN
|
||||||
| θ value | 1.38821 (rank : 9) | NC score | 0.101128 (rank : 5) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P28039, Q53F13 | Gene names | AOAH | |||
|
Domain Architecture |

|
|||||
| Description | Acyloxyacyl hydrolase precursor (EC 3.1.1.77) [Contains: Acyloxyacyl hydrolase small subunit; Acyloxyacyl hydrolase large subunit]. | |||||
|
LIGA_HUMAN
|
||||||
| θ value | 2.36792 (rank : 10) | NC score | 0.046973 (rank : 7) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P41214, Q9NR27, Q9NSN0, Q9NV18, Q9NZ21 | Gene names | LGTN, HCA56 | |||
|
Domain Architecture |

|
|||||
| Description | Ligatin (Hepatocellular carcinoma-associated antigen 56). | |||||
|
TF2AA_MOUSE
|
||||||
| θ value | 2.36792 (rank : 11) | NC score | 0.032377 (rank : 10) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99PM3, Q8C812 | Gene names | Gtf2a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor IIA subunit 1 (General transcription factor IIA1) [Contains: Transcription initiation factor IIA alpha chain (TFIIA p35 subunit); Transcription initiation factor IIA beta chain (TFIIA p19 subunit)]. | |||||
|
GDIB_HUMAN
|
||||||
| θ value | 3.0926 (rank : 12) | NC score | 0.027179 (rank : 13) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P50395, O43928, Q9UQM6 | Gene names | GDI2, RABGDIB | |||
|
Domain Architecture |
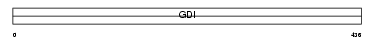
|
|||||
| Description | Rab GDP dissociation inhibitor beta (Rab GDI beta) (Guanosine diphosphate dissociation inhibitor 2) (GDI-2). | |||||
|
LIGA_MOUSE
|
||||||
| θ value | 3.0926 (rank : 13) | NC score | 0.045451 (rank : 8) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61211, Q3TDE0, Q3THV5, Q3TTP4, Q8C491, Q8CBF1, Q8CC17, Q8R1I9, Q8R3M5 | Gene names | Lgtn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ligatin. | |||||
|
PER3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 14) | NC score | 0.016372 (rank : 17) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P56645, Q5H8X4, Q969K6, Q96S77, Q96S78, Q9C0J3, Q9NSP9, Q9UGU8 | Gene names | PER3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (hPER3). | |||||
|
RBBP6_MOUSE
|
||||||
| θ value | 3.0926 (rank : 15) | NC score | 0.017463 (rank : 16) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
CHD9_HUMAN
|
||||||
| θ value | 4.03905 (rank : 16) | NC score | 0.010055 (rank : 19) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q3L8U1, O15025, Q1WF12, Q6DTK9, Q9H9V7 | Gene names | CHD9, KIAA0308, KISH2, PRIC320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Chromatin-related mesenchymal modulator) (CReMM) (Chromatin remodeling factor CHROM1) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha-interacting complex protein 320 kDa) (Kismet homolog 2). | |||||
|
TITIN_HUMAN
|
||||||
| θ value | 4.03905 (rank : 17) | NC score | 0.002740 (rank : 22) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
GDIB_MOUSE
|
||||||
| θ value | 5.27518 (rank : 18) | NC score | 0.024631 (rank : 15) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61598, Q541Z9, Q8C530, Q9D8M9 | Gene names | Gdi2, Gdi3 | |||
|
Domain Architecture |
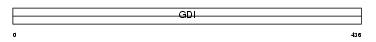
|
|||||
| Description | Rab GDP dissociation inhibitor beta (Rab GDI beta) (Guanosine diphosphate dissociation inhibitor 2) (GDI-2) (GDI-3). | |||||
|
CTR1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 19) | NC score | 0.010185 (rank : 18) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q09143, P30824 | Gene names | Slc7a1, Atrc1, Rec-1 | |||
|
Domain Architecture |
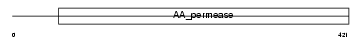
|
|||||
| Description | High-affinity cationic amino acid transporter 1 (CAT-1) (CAT1) (System Y+ basic amino acid transporter) (Ecotropic retroviral leukemia receptor) (ERR) (Ecotropic retrovirus receptor). | |||||
|
KCNV2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 20) | NC score | 0.004990 (rank : 21) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TDN2 | Gene names | KCNV2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily V member 2 (Voltage-gated potassium channel subunit Kv8.2). | |||||
|
NDRG4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 21) | NC score | 0.009133 (rank : 20) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BTG7 | Gene names | Ndrg4, Ndr4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein NDRG4 (Protein Ndr4). | |||||
|
ZN236_HUMAN
|
||||||
| θ value | 8.99809 (rank : 22) | NC score | 0.000082 (rank : 23) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 844 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UL36, Q9UL37 | Gene names | ZNF236 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 236. | |||||
|
AOAH_MOUSE
|
||||||
| θ value | θ > 10 (rank : 23) | NC score | 0.071050 (rank : 6) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O35298 | Gene names | Aoah | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acyloxyacyl hydrolase precursor (EC 3.1.1.77) [Contains: Acyloxyacyl hydrolase small subunit; Acyloxyacyl hydrolase large subunit]. | |||||
|
SAP_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q61207, Q60861, Q64006, Q64219 | Gene names | Psap, Sgp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sulfated glycoprotein 1 precursor (SGP-1) (Prosaposin). | |||||
|
SAP_HUMAN
|
||||||
| NC score | 0.979211 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P07602, P07292, P15793, P78538, P78541, P78546, P78547, P78558, Q6IBQ6, Q92739, Q92740, Q92741, Q92742 | Gene names | PSAP | |||
|
Domain Architecture |

|
|||||
| Description | Proactivator polypeptide precursor [Contains: Saposin A (Protein A); Saposin B-Val; Saposin B (Sphingolipid activator protein 1) (SAP-1) (Cerebroside sulfate activator) (CSAct) (Dispersin) (Sulfatide/GM1 activator); Saposin C (Co-beta-glucosidase) (A1 activator) (Glucosylceramidase activator) (Sphingolipid activator protein 2) (SAP-2); Saposin D (Protein C) (Component C)]. | |||||
|
PSPB_MOUSE
|
||||||
| NC score | 0.671588 (rank : 3) | θ value | 8.36355e-22 (rank : 3) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P50405 | Gene names | Sftpb, Sftp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pulmonary surfactant-associated protein B precursor (SP-B) (Pulmonary surfactant-associated proteolipid SPL(Phe)). | |||||
|
PSPB_HUMAN
|
||||||
| NC score | 0.632409 (rank : 4) | θ value | 2.35696e-16 (rank : 4) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P07988, Q96R04 | Gene names | SFTPB, SFTP3 | |||
|
Domain Architecture |

|
|||||
| Description | Pulmonary surfactant-associated protein B precursor (SP-B) (6 kDa protein) (Pulmonary surfactant-associated proteolipid SPL(Phe)) (18 kDa pulmonary-surfactant protein). | |||||
|
AOAH_HUMAN
|
||||||
| NC score | 0.101128 (rank : 5) | θ value | 1.38821 (rank : 9) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P28039, Q53F13 | Gene names | AOAH | |||
|
Domain Architecture |

|
|||||
| Description | Acyloxyacyl hydrolase precursor (EC 3.1.1.77) [Contains: Acyloxyacyl hydrolase small subunit; Acyloxyacyl hydrolase large subunit]. | |||||
|
AOAH_MOUSE
|
||||||
| NC score | 0.071050 (rank : 6) | θ value | θ > 10 (rank : 23) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O35298 | Gene names | Aoah | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acyloxyacyl hydrolase precursor (EC 3.1.1.77) [Contains: Acyloxyacyl hydrolase small subunit; Acyloxyacyl hydrolase large subunit]. | |||||
|
LIGA_HUMAN
|
||||||
| NC score | 0.046973 (rank : 7) | θ value | 2.36792 (rank : 10) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P41214, Q9NR27, Q9NSN0, Q9NV18, Q9NZ21 | Gene names | LGTN, HCA56 | |||
|
Domain Architecture |

|
|||||
| Description | Ligatin (Hepatocellular carcinoma-associated antigen 56). | |||||
|
LIGA_MOUSE
|
||||||
| NC score | 0.045451 (rank : 8) | θ value | 3.0926 (rank : 13) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61211, Q3TDE0, Q3THV5, Q3TTP4, Q8C491, Q8CBF1, Q8CC17, Q8R1I9, Q8R3M5 | Gene names | Lgtn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ligatin. | |||||
|
TRAIP_HUMAN
|
||||||
| NC score | 0.035405 (rank : 9) | θ value | 0.365318 (rank : 6) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BWF2, O00467 | Gene names | TRAIP, TRIP | |||
|
Domain Architecture |

|
|||||
| Description | TRAF-interacting protein. | |||||
|
TF2AA_MOUSE
|
||||||
| NC score | 0.032377 (rank : 10) | θ value | 2.36792 (rank : 11) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99PM3, Q8C812 | Gene names | Gtf2a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor IIA subunit 1 (General transcription factor IIA1) [Contains: Transcription initiation factor IIA alpha chain (TFIIA p35 subunit); Transcription initiation factor IIA beta chain (TFIIA p19 subunit)]. | |||||
|
TRAIP_MOUSE
|
||||||
| NC score | 0.032214 (rank : 11) | θ value | 0.813845 (rank : 8) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 745 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VIG6, O08854, Q922M8, Q9CPP4 | Gene names | Traip, Trip | |||
|
Domain Architecture |

|
|||||
| Description | TRAF-interacting protein. | |||||
|
HRX_HUMAN
|
||||||
| NC score | 0.031989 (rank : 12) | θ value | 0.365318 (rank : 5) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
GDIB_HUMAN
|
||||||
| NC score | 0.027179 (rank : 13) | θ value | 3.0926 (rank : 12) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P50395, O43928, Q9UQM6 | Gene names | GDI2, RABGDIB | |||
|
Domain Architecture |

|
|||||
| Description | Rab GDP dissociation inhibitor beta (Rab GDI beta) (Guanosine diphosphate dissociation inhibitor 2) (GDI-2). | |||||
|
ANGL5_HUMAN
|
||||||
| NC score | 0.025159 (rank : 14) | θ value | 0.47712 (rank : 7) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q86XS5, Q86VR9 | Gene names | ANGPTL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiopoietin-related protein 5 precursor (Angiopoietin-like 5). | |||||
|
GDIB_MOUSE
|
||||||
| NC score | 0.024631 (rank : 15) | θ value | 5.27518 (rank : 18) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61598, Q541Z9, Q8C530, Q9D8M9 | Gene names | Gdi2, Gdi3 | |||
|
Domain Architecture |

|
|||||
| Description | Rab GDP dissociation inhibitor beta (Rab GDI beta) (Guanosine diphosphate dissociation inhibitor 2) (GDI-2) (GDI-3). | |||||
|
RBBP6_MOUSE
|
||||||
| NC score | 0.017463 (rank : 16) | θ value | 3.0926 (rank : 15) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
PER3_HUMAN
|
||||||
| NC score | 0.016372 (rank : 17) | θ value | 3.0926 (rank : 14) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P56645, Q5H8X4, Q969K6, Q96S77, Q96S78, Q9C0J3, Q9NSP9, Q9UGU8 | Gene names | PER3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (hPER3). | |||||
|
CTR1_MOUSE
|
||||||
| NC score | 0.010185 (rank : 18) | θ value | 6.88961 (rank : 19) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q09143, P30824 | Gene names | Slc7a1, Atrc1, Rec-1 | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cationic amino acid transporter 1 (CAT-1) (CAT1) (System Y+ basic amino acid transporter) (Ecotropic retroviral leukemia receptor) (ERR) (Ecotropic retrovirus receptor). | |||||
|
CHD9_HUMAN
|
||||||
| NC score | 0.010055 (rank : 19) | θ value | 4.03905 (rank : 16) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q3L8U1, O15025, Q1WF12, Q6DTK9, Q9H9V7 | Gene names | CHD9, KIAA0308, KISH2, PRIC320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Chromatin-related mesenchymal modulator) (CReMM) (Chromatin remodeling factor CHROM1) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha-interacting complex protein 320 kDa) (Kismet homolog 2). | |||||
|
NDRG4_MOUSE
|
||||||
| NC score | 0.009133 (rank : 20) | θ value | 8.99809 (rank : 21) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BTG7 | Gene names | Ndrg4, Ndr4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein NDRG4 (Protein Ndr4). | |||||
|
KCNV2_HUMAN
|
||||||
| NC score | 0.004990 (rank : 21) | θ value | 8.99809 (rank : 20) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TDN2 | Gene names | KCNV2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily V member 2 (Voltage-gated potassium channel subunit Kv8.2). | |||||
|
TITIN_HUMAN
|
||||||
| NC score | 0.002740 (rank : 22) | θ value | 4.03905 (rank : 17) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
ZN236_HUMAN
|
||||||
| NC score | 0.000082 (rank : 23) | θ value | 8.99809 (rank : 22) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 844 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UL36, Q9UL37 | Gene names | ZNF236 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 236. | |||||