Please be patient as the page loads
|
PGRP2_HUMAN
|
||||||
| SwissProt Accessions | Q96PD5, Q96N74, Q9UC60 | Gene names | PGLYRP2, PGLYRPL, PGRPL | |||
|
Domain Architecture |

|
|||||
| Description | N-acetylmuramoyl-L-alanine amidase precursor (EC 3.5.1.28) (Peptidoglycan recognition protein long) (PGRP-L) (Peptidoglycan recognition protein 2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PGRP2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q96PD5, Q96N74, Q9UC60 | Gene names | PGLYRP2, PGLYRPL, PGRPL | |||
|
Domain Architecture |

|
|||||
| Description | N-acetylmuramoyl-L-alanine amidase precursor (EC 3.5.1.28) (Peptidoglycan recognition protein long) (PGRP-L) (Peptidoglycan recognition protein 2). | |||||
|
PGRP2_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.990723 (rank : 2) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VCS0, Q8K4I8, Q9QXZ1, Q9QXZ2 | Gene names | Pglyrp2, Pglyrpl, Pgrpl | |||
|
Domain Architecture |

|
|||||
| Description | N-acetylmuramoyl-L-alanine amidase precursor (EC 3.5.1.28) (Peptidoglycan recognition protein long) (PGRP-L) (Peptidoglycan recognition protein 2) (TagL). | |||||
|
PGRP_HUMAN
|
||||||
| θ value | 1.16434e-31 (rank : 3) | NC score | 0.841724 (rank : 3) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75594 | Gene names | PGLYRP1, PGLYRP, PGRP | |||
|
Domain Architecture |

|
|||||
| Description | Peptidoglycan recognition protein precursor (PGRP-S). | |||||
|
PGRP3_HUMAN
|
||||||
| θ value | 4.42448e-31 (rank : 4) | NC score | 0.806161 (rank : 5) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96LB9 | Gene names | PGLYRP3, PGRPIA | |||
|
Domain Architecture |
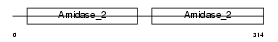
|
|||||
| Description | Peptidoglycan recognition protein I-alpha precursor (Peptidoglycan recognition protein intermediate alpha) (PGRP-I-alpha) (PGLYRPIalpha) (Peptidoglycan recognition protein 3). | |||||
|
PGRP4_HUMAN
|
||||||
| θ value | 1.92365e-26 (rank : 5) | NC score | 0.785922 (rank : 6) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96LB8, Q9HD75 | Gene names | PGLYRP4, PGRPIB | |||
|
Domain Architecture |
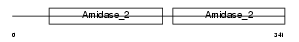
|
|||||
| Description | Peptidoglycan recognition protein I-beta precursor (Peptidoglycan recognition protein intermediate beta) (PGRP-I-beta) (PGLYRPIbeta) (Peptidoglycan recognition protein 4). | |||||
|
PGRP_MOUSE
|
||||||
| θ value | 3.28125e-26 (rank : 6) | NC score | 0.828460 (rank : 4) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O88593, Q62185 | Gene names | Pglyrp1, Pglyrp, Pgrp, Pgrps, Tag7 | |||
|
Domain Architecture |

|
|||||
| Description | Peptidoglycan recognition protein precursor (Peptidoglycan recognition protein short) (PGRP-S) (Cytokine tag7). | |||||
|
DOT1L_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 7) | NC score | 0.066392 (rank : 7) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8TEK3, O60379, Q96JL1 | Gene names | DOT1L, KIAA1814 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-79 specific (EC 2.1.1.43) (Histone H3-K79 methyltransferase) (H3-K79-HMTase) (DOT1-like protein). | |||||
|
URP2_MOUSE
|
||||||
| θ value | 0.62314 (rank : 8) | NC score | 0.028984 (rank : 11) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K1B8 | Gene names | Urp2, Kind3 | |||
|
Domain Architecture |

|
|||||
| Description | Unc-112-related protein 2 (Kindlin-3). | |||||
|
CRSP2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 9) | NC score | 0.038639 (rank : 9) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60244, Q9UNB3 | Gene names | CRSP2, ARC150, DRIP150, EXLM1, TRAP170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CRSP complex subunit 2 (Cofactor required for Sp1 transcriptional activation subunit 2) (Transcriptional coactivator CRSP150) (Vitamin D3 receptor-interacting protein complex 150 kDa component) (DRIP150) (Thyroid hormone receptor-associated protein complex 170 kDa component) (Trap170) (Activator-recruited cofactor 150 kDa component) (ARC150). | |||||
|
AATK_HUMAN
|
||||||
| θ value | 1.38821 (rank : 10) | NC score | 0.006110 (rank : 17) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 1243 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6ZMQ8, O75136, Q6ZN31, Q86X28 | Gene names | AATK, KIAA0641 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis-associated tyrosine-protein kinase (EC 2.7.10.2) (AATYK) (Brain apoptosis-associated tyrosine kinase) (CDK5-binding protein) (p35-binding protein) (p35BP). | |||||
|
DAG1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 11) | NC score | 0.042590 (rank : 8) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14118 | Gene names | DAG1 | |||
|
Domain Architecture |

|
|||||
| Description | Dystroglycan precursor (Dystrophin-associated glycoprotein 1) [Contains: Alpha-dystroglycan (Alpha-DG); Beta-dystroglycan (Beta- DG)]. | |||||
|
TRAK2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 12) | NC score | 0.017943 (rank : 14) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 476 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60296, Q8WVH7, Q96NS2, Q9C0K5, Q9C0K6 | Gene names | TRAK2, ALS2CR3, KIAA0549 | |||
|
Domain Architecture |
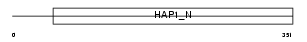
|
|||||
| Description | Trafficking kinesin-binding protein 2 (Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 3 protein). | |||||
|
TRM1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 13) | NC score | 0.028811 (rank : 12) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NXH9, O76103, Q548Y5, Q8WVA6 | Gene names | TRMT1 | |||
|
Domain Architecture |

|
|||||
| Description | N(2),N(2)-dimethylguanosine tRNA methyltransferase (EC 2.1.1.32) (tRNA(guanine-26,N(2)-N(2)) methyltransferase) (tRNA 2,2- dimethylguanosine-26 methyltransferase) (tRNA(m(2,2)G26)dimethyltransferase). | |||||
|
LAP4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 14) | NC score | 0.008478 (rank : 15) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 590 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80U72, Q6P5H7, Q7TPH8, Q80VB1, Q8CI48, Q8VII1, Q922S3 | Gene names | Scrib, Kiaa0147, Lap4, Scrib1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP4 (Protein scribble homolog). | |||||
|
DAG1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 15) | NC score | 0.037310 (rank : 10) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q62165, Q61094, Q61141, Q61497 | Gene names | Dag1, Dag-1 | |||
|
Domain Architecture |

|
|||||
| Description | Dystroglycan precursor (Dystrophin-associated glycoprotein 1) [Contains: Alpha-dystroglycan (Alpha-DG); Beta-dystroglycan (Beta- DG)]. | |||||
|
LAT_HUMAN
|
||||||
| θ value | 6.88961 (rank : 16) | NC score | 0.022834 (rank : 13) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O43561, O43919 | Gene names | LAT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Linker for activation of T-cells family member 1 (36 kDa phospho- tyrosine adapter protein) (pp36) (p36-38). | |||||
|
PLXA1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 17) | NC score | 0.005884 (rank : 18) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UIW2 | Gene names | PLXNA1, NOV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-A1 precursor (Semaphorin receptor NOV). | |||||
|
LAP4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 18) | NC score | 0.006643 (rank : 16) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 608 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14160, Q6P496, Q7Z5D1, Q8WWV8, Q96C69, Q96GG1 | Gene names | SCRIB, CRIB1, KIAA0147, LAP4, SCRB1, VARTUL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP4 (Protein scribble homolog) (hScrib). | |||||
|
RIPK3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 19) | NC score | 0.001224 (rank : 19) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y572 | Gene names | RIPK3, RIP3 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-interacting serine/threonine-protein kinase 3 (EC 2.7.11.1) (RIP-like protein kinase 3) (Receptor-interacting protein 3) (RIP-3). | |||||
|
PGRP2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q96PD5, Q96N74, Q9UC60 | Gene names | PGLYRP2, PGLYRPL, PGRPL | |||
|
Domain Architecture |

|
|||||
| Description | N-acetylmuramoyl-L-alanine amidase precursor (EC 3.5.1.28) (Peptidoglycan recognition protein long) (PGRP-L) (Peptidoglycan recognition protein 2). | |||||
|
PGRP2_MOUSE
|
||||||
| NC score | 0.990723 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VCS0, Q8K4I8, Q9QXZ1, Q9QXZ2 | Gene names | Pglyrp2, Pglyrpl, Pgrpl | |||
|
Domain Architecture |

|
|||||
| Description | N-acetylmuramoyl-L-alanine amidase precursor (EC 3.5.1.28) (Peptidoglycan recognition protein long) (PGRP-L) (Peptidoglycan recognition protein 2) (TagL). | |||||
|
PGRP_HUMAN
|
||||||
| NC score | 0.841724 (rank : 3) | θ value | 1.16434e-31 (rank : 3) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75594 | Gene names | PGLYRP1, PGLYRP, PGRP | |||
|
Domain Architecture |

|
|||||
| Description | Peptidoglycan recognition protein precursor (PGRP-S). | |||||
|
PGRP_MOUSE
|
||||||
| NC score | 0.828460 (rank : 4) | θ value | 3.28125e-26 (rank : 6) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O88593, Q62185 | Gene names | Pglyrp1, Pglyrp, Pgrp, Pgrps, Tag7 | |||
|
Domain Architecture |

|
|||||
| Description | Peptidoglycan recognition protein precursor (Peptidoglycan recognition protein short) (PGRP-S) (Cytokine tag7). | |||||
|
PGRP3_HUMAN
|
||||||
| NC score | 0.806161 (rank : 5) | θ value | 4.42448e-31 (rank : 4) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96LB9 | Gene names | PGLYRP3, PGRPIA | |||
|
Domain Architecture |

|
|||||
| Description | Peptidoglycan recognition protein I-alpha precursor (Peptidoglycan recognition protein intermediate alpha) (PGRP-I-alpha) (PGLYRPIalpha) (Peptidoglycan recognition protein 3). | |||||
|
PGRP4_HUMAN
|
||||||
| NC score | 0.785922 (rank : 6) | θ value | 1.92365e-26 (rank : 5) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96LB8, Q9HD75 | Gene names | PGLYRP4, PGRPIB | |||
|
Domain Architecture |

|
|||||
| Description | Peptidoglycan recognition protein I-beta precursor (Peptidoglycan recognition protein intermediate beta) (PGRP-I-beta) (PGLYRPIbeta) (Peptidoglycan recognition protein 4). | |||||
|
DOT1L_HUMAN
|
||||||
| NC score | 0.066392 (rank : 7) | θ value | 0.0252991 (rank : 7) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8TEK3, O60379, Q96JL1 | Gene names | DOT1L, KIAA1814 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-79 specific (EC 2.1.1.43) (Histone H3-K79 methyltransferase) (H3-K79-HMTase) (DOT1-like protein). | |||||
|
DAG1_HUMAN
|
||||||
| NC score | 0.042590 (rank : 8) | θ value | 1.38821 (rank : 11) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14118 | Gene names | DAG1 | |||
|
Domain Architecture |

|
|||||
| Description | Dystroglycan precursor (Dystrophin-associated glycoprotein 1) [Contains: Alpha-dystroglycan (Alpha-DG); Beta-dystroglycan (Beta- DG)]. | |||||
|
CRSP2_HUMAN
|
||||||
| NC score | 0.038639 (rank : 9) | θ value | 1.06291 (rank : 9) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60244, Q9UNB3 | Gene names | CRSP2, ARC150, DRIP150, EXLM1, TRAP170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CRSP complex subunit 2 (Cofactor required for Sp1 transcriptional activation subunit 2) (Transcriptional coactivator CRSP150) (Vitamin D3 receptor-interacting protein complex 150 kDa component) (DRIP150) (Thyroid hormone receptor-associated protein complex 170 kDa component) (Trap170) (Activator-recruited cofactor 150 kDa component) (ARC150). | |||||
|
DAG1_MOUSE
|
||||||
| NC score | 0.037310 (rank : 10) | θ value | 4.03905 (rank : 15) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q62165, Q61094, Q61141, Q61497 | Gene names | Dag1, Dag-1 | |||
|
Domain Architecture |

|
|||||
| Description | Dystroglycan precursor (Dystrophin-associated glycoprotein 1) [Contains: Alpha-dystroglycan (Alpha-DG); Beta-dystroglycan (Beta- DG)]. | |||||
|
URP2_MOUSE
|
||||||
| NC score | 0.028984 (rank : 11) | θ value | 0.62314 (rank : 8) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K1B8 | Gene names | Urp2, Kind3 | |||
|
Domain Architecture |

|
|||||
| Description | Unc-112-related protein 2 (Kindlin-3). | |||||
|
TRM1_HUMAN
|
||||||
| NC score | 0.028811 (rank : 12) | θ value | 2.36792 (rank : 13) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NXH9, O76103, Q548Y5, Q8WVA6 | Gene names | TRMT1 | |||
|
Domain Architecture |

|
|||||
| Description | N(2),N(2)-dimethylguanosine tRNA methyltransferase (EC 2.1.1.32) (tRNA(guanine-26,N(2)-N(2)) methyltransferase) (tRNA 2,2- dimethylguanosine-26 methyltransferase) (tRNA(m(2,2)G26)dimethyltransferase). | |||||
|
LAT_HUMAN
|
||||||
| NC score | 0.022834 (rank : 13) | θ value | 6.88961 (rank : 16) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O43561, O43919 | Gene names | LAT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Linker for activation of T-cells family member 1 (36 kDa phospho- tyrosine adapter protein) (pp36) (p36-38). | |||||
|
TRAK2_HUMAN
|
||||||
| NC score | 0.017943 (rank : 14) | θ value | 2.36792 (rank : 12) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 476 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60296, Q8WVH7, Q96NS2, Q9C0K5, Q9C0K6 | Gene names | TRAK2, ALS2CR3, KIAA0549 | |||
|
Domain Architecture |

|
|||||
| Description | Trafficking kinesin-binding protein 2 (Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 3 protein). | |||||
|
LAP4_MOUSE
|
||||||
| NC score | 0.008478 (rank : 15) | θ value | 3.0926 (rank : 14) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 590 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80U72, Q6P5H7, Q7TPH8, Q80VB1, Q8CI48, Q8VII1, Q922S3 | Gene names | Scrib, Kiaa0147, Lap4, Scrib1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP4 (Protein scribble homolog). | |||||
|
LAP4_HUMAN
|
||||||
| NC score | 0.006643 (rank : 16) | θ value | 8.99809 (rank : 18) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 608 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14160, Q6P496, Q7Z5D1, Q8WWV8, Q96C69, Q96GG1 | Gene names | SCRIB, CRIB1, KIAA0147, LAP4, SCRB1, VARTUL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP4 (Protein scribble homolog) (hScrib). | |||||
|
AATK_HUMAN
|
||||||
| NC score | 0.006110 (rank : 17) | θ value | 1.38821 (rank : 10) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 1243 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6ZMQ8, O75136, Q6ZN31, Q86X28 | Gene names | AATK, KIAA0641 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis-associated tyrosine-protein kinase (EC 2.7.10.2) (AATYK) (Brain apoptosis-associated tyrosine kinase) (CDK5-binding protein) (p35-binding protein) (p35BP). | |||||
|
PLXA1_HUMAN
|
||||||
| NC score | 0.005884 (rank : 18) | θ value | 6.88961 (rank : 17) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UIW2 | Gene names | PLXNA1, NOV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-A1 precursor (Semaphorin receptor NOV). | |||||
|
RIPK3_HUMAN
|
||||||
| NC score | 0.001224 (rank : 19) | θ value | 8.99809 (rank : 19) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y572 | Gene names | RIPK3, RIP3 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-interacting serine/threonine-protein kinase 3 (EC 2.7.11.1) (RIP-like protein kinase 3) (Receptor-interacting protein 3) (RIP-3). | |||||