Please be patient as the page loads
|
PARVG_MOUSE
|
||||||
| SwissProt Accessions | Q9ERD8, Q91X89 | Gene names | Parvg | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-parvin. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PARVG_MOUSE
|
||||||
| θ value | 6.32649e-179 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9ERD8, Q91X89 | Gene names | Parvg | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-parvin. | |||||
|
PARVG_HUMAN
|
||||||
| θ value | 1.68428e-139 (rank : 2) | NC score | 0.984222 (rank : 2) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9HBI0, Q9BQX5, Q9NSG1 | Gene names | PARVG | |||
|
Domain Architecture |
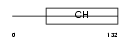
|
|||||
| Description | Gamma-parvin. | |||||
|
PARVA_HUMAN
|
||||||
| θ value | 1.45591e-66 (rank : 3) | NC score | 0.943989 (rank : 3) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9NVD7, Q96C85, Q9HA48 | Gene names | PARVA | |||
|
Domain Architecture |
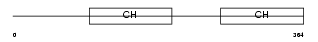
|
|||||
| Description | Alpha-parvin (Calponin-like integrin-linked kinase-binding protein) (CH-ILKBP). | |||||
|
PARVA_MOUSE
|
||||||
| θ value | 1.45591e-66 (rank : 4) | NC score | 0.943773 (rank : 4) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9EPC1, Q9JJ65 | Gene names | Parva, Actp | |||
|
Domain Architecture |
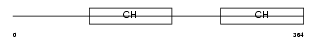
|
|||||
| Description | Alpha-parvin (Actopaxin). | |||||
|
PARVB_HUMAN
|
||||||
| θ value | 1.07972e-61 (rank : 5) | NC score | 0.943732 (rank : 5) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9HBI1, Q5TGJ5, Q86X93, Q9NSP7, Q9UGT3, Q9Y368, Q9Y3L6, Q9Y3L7 | Gene names | PARVB | |||
|
Domain Architecture |
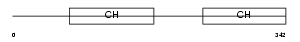
|
|||||
| Description | Beta-parvin (Affixin). | |||||
|
PARVB_MOUSE
|
||||||
| θ value | 5.0155e-59 (rank : 6) | NC score | 0.939976 (rank : 6) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9ES46 | Gene names | Parvb | |||
|
Domain Architecture |
|
|||||
| Description | Beta-parvin. | |||||
|
ACTN4_HUMAN
|
||||||
| θ value | 4.92598e-06 (rank : 7) | NC score | 0.301023 (rank : 7) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O43707, O76048 | Gene names | ACTN4 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-4 (Non-muscle alpha-actinin 4) (F-actin cross-linking protein). | |||||
|
ACTN4_MOUSE
|
||||||
| θ value | 4.92598e-06 (rank : 8) | NC score | 0.300706 (rank : 8) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P57780 | Gene names | Actn4 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-4 (Non-muscle alpha-actinin 4) (F-actin cross-linking protein). | |||||
|
ACTN1_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 9) | NC score | 0.298479 (rank : 10) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P12814, Q9BTN1 | Gene names | ACTN1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-1 (Alpha-actinin cytoskeletal isoform) (Non-muscle alpha-actinin-1) (F-actin cross-linking protein). | |||||
|
ACTN1_MOUSE
|
||||||
| θ value | 9.29e-05 (rank : 10) | NC score | 0.299062 (rank : 9) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q7TPR4 | Gene names | Actn1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-1 (Alpha-actinin cytoskeletal isoform) (Non-muscle alpha-actinin-1) (F-actin cross-linking protein). | |||||
|
ACTN2_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 11) | NC score | 0.283628 (rank : 14) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P35609, Q86TF4, Q86TI8 | Gene names | ACTN2 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-2 (Alpha actinin skeletal muscle isoform 2) (F-actin cross-linking protein). | |||||
|
ACTN2_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 12) | NC score | 0.284426 (rank : 13) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9JI91 | Gene names | Actn2 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-2 (Alpha actinin skeletal muscle isoform 2) (F-actin cross-linking protein). | |||||
|
ACTN3_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 13) | NC score | 0.287664 (rank : 11) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q08043 | Gene names | ACTN3 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-3 (Alpha actinin skeletal muscle isoform 3) (F-actin cross-linking protein). | |||||
|
ACTN3_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 14) | NC score | 0.286970 (rank : 12) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 265 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O88990 | Gene names | Actn3 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-3 (Alpha actinin skeletal muscle isoform 3) (F-actin cross-linking protein). | |||||
|
FLNA_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 15) | NC score | 0.226209 (rank : 19) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P21333 | Gene names | FLNA, FLN, FLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Filamin-A (Alpha-filamin) (Filamin-1) (Endothelial actin-binding protein) (Actin-binding protein 280) (ABP-280) (Nonmuscle filamin). | |||||
|
FLNA_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 16) | NC score | 0.225795 (rank : 20) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8BTM8, O54934, Q7TQI1, Q8BLK1, Q8BTN7, Q8VHX5, Q8VHX8, Q99KQ2 | Gene names | Flna, Fln, Fln1 | |||
|
Domain Architecture |

|
|||||
| Description | Filamin-A (Alpha-filamin) (Filamin-1) (Endothelial actin-binding protein) (Actin-binding protein 280) (ABP-280) (Nonmuscle filamin). | |||||
|
SPTN4_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 17) | NC score | 0.230074 (rank : 17) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9H254, Q9H1K7, Q9H1K8, Q9H1K9, Q9H3G8, Q9HCD0 | Gene names | SPTBN4, KIAA1642, SPTBN3 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 3 (Spectrin, non-erythroid beta chain 3) (Beta-IV spectrin). | |||||
|
MACF1_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 18) | NC score | 0.168729 (rank : 30) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1341 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9UPN3, O75053, Q5VW20, Q8WXY2, Q9H540, Q9UKP0, Q9ULG9 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5 (Actin cross-linking family protein 7) (Macrophin-1) (Trabeculin-alpha) (620 kDa actin-binding protein) (ABP620). | |||||
|
MACF1_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 19) | NC score | 0.173696 (rank : 29) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9QXZ0, P97394, P97395, P97396 | Gene names | Macf1, Acf7, Aclp7, Macf | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1 (Actin cross-linking family 7). | |||||
|
SPTB2_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 20) | NC score | 0.214242 (rank : 26) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q01082, O60837, Q16057 | Gene names | SPTBN1, SPTB2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
SPTB2_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 21) | NC score | 0.213332 (rank : 27) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q62261, Q5SQL8 | Gene names | Sptbn1, Spnb-2, Spnb2, Sptb2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
SPTN2_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 22) | NC score | 0.219978 (rank : 23) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 535 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O15020, O14872, O14873 | Gene names | SPTBN2, KIAA0302 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 2 (Spectrin, non-erythroid beta chain 2) (Beta-III spectrin). | |||||
|
BPA1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 23) | NC score | 0.162657 (rank : 31) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q91ZU6, Q91ZU7 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 1/2/3/4 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
SPTB1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 24) | NC score | 0.215803 (rank : 24) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P15508 | Gene names | Sptb, Spnb-1, Spnb1, Sptb1 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, erythrocyte (Beta-I spectrin). | |||||
|
SPTB1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 25) | NC score | 0.221769 (rank : 22) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 541 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P11277, Q15510, Q15519 | Gene names | SPTB, SPTB1 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, erythrocyte (Beta-I spectrin). | |||||
|
CLMN_HUMAN
|
||||||
| θ value | 0.279714 (rank : 26) | NC score | 0.232384 (rank : 16) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q96JQ2, Q9H713, Q9HA23, Q9HA57, Q9UFP4, Q9ULN2 | Gene names | CLMN, KIAA1188 | |||
|
Domain Architecture |

|
|||||
| Description | Calmin. | |||||
|
CLMN_MOUSE
|
||||||
| θ value | 0.279714 (rank : 27) | NC score | 0.240649 (rank : 15) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8C5W0, Q91V71, Q91XT7, Q91XT8, Q91XU9 | Gene names | Clmn | |||
|
Domain Architecture |

|
|||||
| Description | Calmin. | |||||
|
ASPM_MOUSE
|
||||||
| θ value | 0.62314 (rank : 28) | NC score | 0.140910 (rank : 40) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8CJ27, O88482, Q8BJI8, Q8BKT4 | Gene names | Aspm, Calmbp1, Sha1 | |||
|
Domain Architecture |

|
|||||
| Description | Abnormal spindle-like microcephaly-associated protein homolog (Calmodulin-binding protein 1) (Spindle and hydroxyurea checkpoint abnormal protein) (Calmodulin-binding protein Sha1). | |||||
|
UTRO_HUMAN
|
||||||
| θ value | 0.813845 (rank : 29) | NC score | 0.143666 (rank : 37) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P46939 | Gene names | UTRN, DMDL | |||
|
Domain Architecture |

|
|||||
| Description | Utrophin (Dystrophin-related protein 1) (DRP1) (DRP). | |||||
|
ASPM_HUMAN
|
||||||
| θ value | 1.06291 (rank : 30) | NC score | 0.142644 (rank : 39) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8IZT6, Q86UX4, Q8IUL2, Q8IZJ7, Q8IZJ8, Q8IZJ9, Q8N4D1, Q9NVS1, Q9NVT6 | Gene names | ASPM, MCPH5 | |||
|
Domain Architecture |

|
|||||
| Description | Abnormal spindle-like microcephaly-associated protein (Abnormal spindle protein homolog) (Asp homolog). | |||||
|
KTN1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 31) | NC score | 0.040483 (rank : 62) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1324 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q61595, Q8BG49, Q8BHF4, Q8BHM8, Q8C9Y5, Q8CG51, Q8CG52, Q8CG53, Q8CG54, Q8CG55, Q8CG56, Q8CG57, Q8CG58, Q8CG59, Q8CG60, Q8CG61, Q8CG62, Q8CG63 | Gene names | Ktn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin. | |||||
|
RAD50_HUMAN
|
||||||
| θ value | 1.38821 (rank : 32) | NC score | 0.029394 (rank : 66) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q92878, O43254, Q6GMT7, Q6P5X3, Q9UP86 | Gene names | RAD50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (hRAD50). | |||||
|
DMD_HUMAN
|
||||||
| θ value | 3.0926 (rank : 33) | NC score | 0.136063 (rank : 42) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P11532, Q02295, Q14169, Q14170, Q5JYU0, Q7KZ48 | Gene names | DMD | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
MD1L1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 34) | NC score | 0.038202 (rank : 64) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1001 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9WTX8, Q9WTX9 | Gene names | Mad1l1, Mad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1). | |||||
|
AKAP9_HUMAN
|
||||||
| θ value | 4.03905 (rank : 35) | NC score | 0.036164 (rank : 65) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
KI13B_HUMAN
|
||||||
| θ value | 4.03905 (rank : 36) | NC score | 0.006198 (rank : 69) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NQT8, O75134, Q9BYJ6 | Gene names | KIF13B, GAKIN, KIAA0639 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF13B (Kinesin-like protein GAKIN). | |||||
|
CROCC_HUMAN
|
||||||
| θ value | 5.27518 (rank : 37) | NC score | 0.038376 (rank : 63) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1409 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q5TZA2, Q2VHY3, Q66GT7, Q7Z2L4, Q7Z5D7 | Gene names | CROCC, KIAA0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
PLEC1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 38) | NC score | 0.145666 (rank : 34) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
DMD_MOUSE
|
||||||
| θ value | 8.99809 (rank : 39) | NC score | 0.131768 (rank : 43) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1111 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P11531, O35653, Q60703 | Gene names | Dmd | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
SMC3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 40) | NC score | 0.028927 (rank : 67) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1186 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UQE7, O60464 | Gene names | CSPG6, BAM, BMH, SMC3, SMC3L1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome-associated polypeptide) (hCAP) (Bamacan) (Basement membrane-associated chondroitin proteoglycan). | |||||
|
SMC3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 41) | NC score | 0.028704 (rank : 68) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1187 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9CW03, O35667, Q9QUS3 | Gene names | Cspg6, Bam, Bmh, Mmip1, Smc3, Smc3l1, Smcd | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome segregation protein SmcD) (Bamacan) (Basement membrane-associated chondroitin proteoglycan) (Mad member- interacting protein 1). | |||||
|
BPAEA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 42) | NC score | 0.084371 (rank : 57) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1369 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O94833, Q5TBT0, Q8N1T8, Q8N8J3, Q8WXK9, Q96AK9, Q96DQ5, Q9H555 | Gene names | DST, BPAG1, DMH, DT, KIAA0728 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 6/9/10 (Trabeculin-beta) (Bullous pemphigoid antigen) (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
CYTSA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 43) | NC score | 0.080385 (rank : 60) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1138 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q69YQ0, O15081 | Gene names | CYTSA, KIAA0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A (NY-REN-22 antigen). | |||||
|
CYTSA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 44) | NC score | 0.078567 (rank : 61) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q2KN98, Q8CHF9 | Gene names | Cytsa, Kiaa0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A. | |||||
|
FLNB_HUMAN
|
||||||
| θ value | θ > 10 (rank : 45) | NC score | 0.215493 (rank : 25) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O75369, Q13706, Q8WXS9, Q8WXT0, Q8WXT1, Q8WXT2, Q9NRB5, Q9NT26, Q9UEV9 | Gene names | FLNB, FLN1L, FLN3, TABP, TAP | |||
|
Domain Architecture |

|
|||||
| Description | Filamin-B (FLN-B) (Beta-filamin) (Actin-binding-like protein) (Thyroid autoantigen) (Truncated actin-binding protein) (Truncated ABP) (ABP- 280 homolog) (ABP-278) (Filamin 3) (Filamin homolog 1) (Fh1). | |||||
|
FLNB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 46) | NC score | 0.228180 (rank : 18) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q80X90, Q8VHX4, Q8VHX7, Q99KY3 | Gene names | Flnb | |||
|
Domain Architecture |

|
|||||
| Description | Filamin-B (FLN-B) (Beta-filamin) (Actin-binding-like protein) (ABP- 280-like protein). | |||||
|
FLNC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 47) | NC score | 0.223587 (rank : 21) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q14315, O95303, Q07985, Q9NS12, Q9NYE5, Q9UMR8, Q9Y503 | Gene names | FLNC, ABPL, FLN2 | |||
|
Domain Architecture |

|
|||||
| Description | Filamin-C (Gamma-filamin) (Filamin-2) (Protein FLNc) (Actin-binding- like protein) (ABP-L) (ABP-280-like protein). | |||||
|
MACF4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 48) | NC score | 0.082730 (rank : 58) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1321 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96PK2, Q8WXY1 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoform 4. | |||||
|
MICA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 49) | NC score | 0.095114 (rank : 52) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8TDZ2, Q7Z633, Q8IVS9, Q96G47, Q9H6X6, Q9H7I0, Q9HAA1, Q9UFF7 | Gene names | MICAL1, MICAL, NICAL | |||
|
Domain Architecture |

|
|||||
| Description | NEDD9-interacting protein with calponin homology and LIM domains (Molecule interacting with CasL protein 1). | |||||
|
MICA1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 50) | NC score | 0.095655 (rank : 51) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8VDP3 | Gene names | Mical1, Mical, Nical | |||
|
Domain Architecture |

|
|||||
| Description | NEDD9-interacting protein with calponin homology and LIM domains (Molecule interacting with CasL protein 1). | |||||
|
MICA2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 51) | NC score | 0.122617 (rank : 47) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O94851, Q7Z3A8 | Gene names | MICAL2, KIAA0750 | |||
|
Domain Architecture |

|
|||||
| Description | Protein MICAL-2. | |||||
|
MICA2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 52) | NC score | 0.113513 (rank : 50) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BML1, Q3UPU6 | Gene names | Mical2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein MICAL-2. | |||||
|
MICA3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 53) | NC score | 0.119457 (rank : 49) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q7RTP6, Q6ICK4, Q9P2I3 | Gene names | MICAL3, KIAA1364 | |||
|
Domain Architecture |

|
|||||
| Description | Protein MICAL-3. | |||||
|
MICA3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 54) | NC score | 0.123225 (rank : 46) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8CJ19, Q80TE8, Q8BXB1 | Gene names | Mical3, Kiaa1364 | |||
|
Domain Architecture |

|
|||||
| Description | Protein MICAL-3. | |||||
|
MILK1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 55) | NC score | 0.144317 (rank : 36) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8N3F8, Q5TI16, Q7RTP5, Q8N3N8, Q9BVL9, Q9BY92, Q9UH43, Q9UH44, Q9UH45 | Gene names | MIRAB13, KIAA1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
MILK1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 56) | NC score | 0.130155 (rank : 44) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BGT6, Q8BJ60 | Gene names | Mirab13, D15Mit260, Kiaa1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
MILK2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 57) | NC score | 0.146388 (rank : 33) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8IY33, Q7RTP4, Q7Z655, Q8TEQ4, Q9H5F9 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | MICAL-like protein 2. | |||||
|
PLSI_HUMAN
|
||||||
| θ value | θ > 10 (rank : 58) | NC score | 0.080438 (rank : 59) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q14651 | Gene names | PLS1 | |||
|
Domain Architecture |

|
|||||
| Description | Plastin-1 (I-plastin) (Intestine-specific plastin). | |||||
|
PLSL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 59) | NC score | 0.089507 (rank : 56) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P13796 | Gene names | LCP1, PLS2 | |||
|
Domain Architecture |

|
|||||
| Description | Plastin-2 (L-plastin) (Lymphocyte cytosolic protein 1) (LCP-1) (LC64P). | |||||
|
PLSL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 60) | NC score | 0.090914 (rank : 55) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q61233, Q3UE24, Q8R1X3, Q9CV77 | Gene names | Lcp1, Pls2 | |||
|
Domain Architecture |

|
|||||
| Description | Plastin-2 (L-plastin) (Lymphocyte cytosolic protein 1) (LCP-1) (65 kDa macrophage protein) (pp65). | |||||
|
PLST_HUMAN
|
||||||
| θ value | θ > 10 (rank : 61) | NC score | 0.092943 (rank : 54) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P13797, Q86YI6 | Gene names | PLS3 | |||
|
Domain Architecture |

|
|||||
| Description | Plastin-3 (T-plastin). | |||||
|
PLST_MOUSE
|
||||||
| θ value | θ > 10 (rank : 62) | NC score | 0.093728 (rank : 53) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q99K51 | Gene names | Pls3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plastin-3 (T-plastin). | |||||
|
SMOO_HUMAN
|
||||||
| θ value | θ > 10 (rank : 63) | NC score | 0.153946 (rank : 32) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P53814, O00569, O95769, O95937, Q9P1S8, Q9UIT1, Q9UIT2 | Gene names | SMTN, SMSMO | |||
|
Domain Architecture |

|
|||||
| Description | Smoothelin. | |||||
|
SPTA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 64) | NC score | 0.138216 (rank : 41) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P02549, Q15514, Q6LDY5 | Gene names | SPTA1, SPTA | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, erythrocyte (Erythroid alpha-spectrin). | |||||
|
SPTA1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.145445 (rank : 35) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 675 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P08032, P97502 | Gene names | Spta1, Spna1, Spta | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, erythrocyte (Erythroid alpha-spectrin). | |||||
|
SPTA2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.143403 (rank : 38) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 888 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q13813, Q13186, Q15324, Q16606, Q59EF1, Q5VXV5, Q5VXV6, Q7Z6M5, Q9P0V0 | Gene names | SPTAN1, SPTA2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, brain (Spectrin, non-erythroid alpha chain) (Alpha-II spectrin) (Fodrin alpha chain). | |||||
|
SPTN5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.181277 (rank : 28) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9NRC6 | Gene names | SPTBN5, SPTBN4 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 4 (Spectrin, non-erythroid beta chain 4) (Beta-V spectrin) (BSPECV). | |||||
|
SYNE1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.121806 (rank : 48) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
SYNE2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.123403 (rank : 45) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
PARVG_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 6.32649e-179 (rank : 1) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9ERD8, Q91X89 | Gene names | Parvg | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-parvin. | |||||
|
PARVG_HUMAN
|
||||||
| NC score | 0.984222 (rank : 2) | θ value | 1.68428e-139 (rank : 2) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9HBI0, Q9BQX5, Q9NSG1 | Gene names | PARVG | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-parvin. | |||||
|
PARVA_HUMAN
|
||||||
| NC score | 0.943989 (rank : 3) | θ value | 1.45591e-66 (rank : 3) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9NVD7, Q96C85, Q9HA48 | Gene names | PARVA | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-parvin (Calponin-like integrin-linked kinase-binding protein) (CH-ILKBP). | |||||
|
PARVA_MOUSE
|
||||||
| NC score | 0.943773 (rank : 4) | θ value | 1.45591e-66 (rank : 4) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9EPC1, Q9JJ65 | Gene names | Parva, Actp | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-parvin (Actopaxin). | |||||
|
PARVB_HUMAN
|
||||||
| NC score | 0.943732 (rank : 5) | θ value | 1.07972e-61 (rank : 5) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9HBI1, Q5TGJ5, Q86X93, Q9NSP7, Q9UGT3, Q9Y368, Q9Y3L6, Q9Y3L7 | Gene names | PARVB | |||
|
Domain Architecture |

|
|||||
| Description | Beta-parvin (Affixin). | |||||
|
PARVB_MOUSE
|
||||||
| NC score | 0.939976 (rank : 6) | θ value | 5.0155e-59 (rank : 6) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9ES46 | Gene names | Parvb | |||
|
Domain Architecture |

|
|||||
| Description | Beta-parvin. | |||||
|
ACTN4_HUMAN
|
||||||
| NC score | 0.301023 (rank : 7) | θ value | 4.92598e-06 (rank : 7) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O43707, O76048 | Gene names | ACTN4 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-4 (Non-muscle alpha-actinin 4) (F-actin cross-linking protein). | |||||
|
ACTN4_MOUSE
|
||||||
| NC score | 0.300706 (rank : 8) | θ value | 4.92598e-06 (rank : 8) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P57780 | Gene names | Actn4 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-4 (Non-muscle alpha-actinin 4) (F-actin cross-linking protein). | |||||
|
ACTN1_MOUSE
|
||||||
| NC score | 0.299062 (rank : 9) | θ value | 9.29e-05 (rank : 10) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q7TPR4 | Gene names | Actn1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-1 (Alpha-actinin cytoskeletal isoform) (Non-muscle alpha-actinin-1) (F-actin cross-linking protein). | |||||
|
ACTN1_HUMAN
|
||||||
| NC score | 0.298479 (rank : 10) | θ value | 9.29e-05 (rank : 9) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P12814, Q9BTN1 | Gene names | ACTN1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-1 (Alpha-actinin cytoskeletal isoform) (Non-muscle alpha-actinin-1) (F-actin cross-linking protein). | |||||
|
ACTN3_HUMAN
|
||||||
| NC score | 0.287664 (rank : 11) | θ value | 0.00228821 (rank : 13) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q08043 | Gene names | ACTN3 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-3 (Alpha actinin skeletal muscle isoform 3) (F-actin cross-linking protein). | |||||
|
ACTN3_MOUSE
|
||||||
| NC score | 0.286970 (rank : 12) | θ value | 0.00228821 (rank : 14) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 265 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O88990 | Gene names | Actn3 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-3 (Alpha actinin skeletal muscle isoform 3) (F-actin cross-linking protein). | |||||
|
ACTN2_MOUSE
|
||||||
| NC score | 0.284426 (rank : 13) | θ value | 0.00228821 (rank : 12) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9JI91 | Gene names | Actn2 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-2 (Alpha actinin skeletal muscle isoform 2) (F-actin cross-linking protein). | |||||
|
ACTN2_HUMAN
|
||||||
| NC score | 0.283628 (rank : 14) | θ value | 0.00228821 (rank : 11) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P35609, Q86TF4, Q86TI8 | Gene names | ACTN2 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-2 (Alpha actinin skeletal muscle isoform 2) (F-actin cross-linking protein). | |||||
|
CLMN_MOUSE
|
||||||
| NC score | 0.240649 (rank : 15) | θ value | 0.279714 (rank : 27) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8C5W0, Q91V71, Q91XT7, Q91XT8, Q91XU9 | Gene names | Clmn | |||
|
Domain Architecture |

|
|||||
| Description | Calmin. | |||||
|
CLMN_HUMAN
|
||||||
| NC score | 0.232384 (rank : 16) | θ value | 0.279714 (rank : 26) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q96JQ2, Q9H713, Q9HA23, Q9HA57, Q9UFP4, Q9ULN2 | Gene names | CLMN, KIAA1188 | |||
|
Domain Architecture |

|
|||||
| Description | Calmin. | |||||
|
SPTN4_HUMAN
|
||||||
| NC score | 0.230074 (rank : 17) | θ value | 0.0113563 (rank : 17) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9H254, Q9H1K7, Q9H1K8, Q9H1K9, Q9H3G8, Q9HCD0 | Gene names | SPTBN4, KIAA1642, SPTBN3 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 3 (Spectrin, non-erythroid beta chain 3) (Beta-IV spectrin). | |||||
|
FLNB_MOUSE
|
||||||
| NC score | 0.228180 (rank : 18) | θ value | θ > 10 (rank : 46) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q80X90, Q8VHX4, Q8VHX7, Q99KY3 | Gene names | Flnb | |||
|
Domain Architecture |

|
|||||
| Description | Filamin-B (FLN-B) (Beta-filamin) (Actin-binding-like protein) (ABP- 280-like protein). | |||||
|
FLNA_HUMAN
|
||||||
| NC score | 0.226209 (rank : 19) | θ value | 0.0113563 (rank : 15) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P21333 | Gene names | FLNA, FLN, FLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Filamin-A (Alpha-filamin) (Filamin-1) (Endothelial actin-binding protein) (Actin-binding protein 280) (ABP-280) (Nonmuscle filamin). | |||||
|
FLNA_MOUSE
|
||||||
| NC score | 0.225795 (rank : 20) | θ value | 0.0113563 (rank : 16) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8BTM8, O54934, Q7TQI1, Q8BLK1, Q8BTN7, Q8VHX5, Q8VHX8, Q99KQ2 | Gene names | Flna, Fln, Fln1 | |||
|
Domain Architecture |

|
|||||
| Description | Filamin-A (Alpha-filamin) (Filamin-1) (Endothelial actin-binding protein) (Actin-binding protein 280) (ABP-280) (Nonmuscle filamin). | |||||
|
FLNC_HUMAN
|
||||||
| NC score | 0.223587 (rank : 21) | θ value | θ > 10 (rank : 47) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q14315, O95303, Q07985, Q9NS12, Q9NYE5, Q9UMR8, Q9Y503 | Gene names | FLNC, ABPL, FLN2 | |||
|
Domain Architecture |

|
|||||
| Description | Filamin-C (Gamma-filamin) (Filamin-2) (Protein FLNc) (Actin-binding- like protein) (ABP-L) (ABP-280-like protein). | |||||
|
SPTB1_HUMAN
|
||||||
| NC score | 0.221769 (rank : 22) | θ value | 0.21417 (rank : 25) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 541 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P11277, Q15510, Q15519 | Gene names | SPTB, SPTB1 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, erythrocyte (Beta-I spectrin). | |||||
|
SPTN2_HUMAN
|
||||||
| NC score | 0.219978 (rank : 23) | θ value | 0.0961366 (rank : 22) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 535 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O15020, O14872, O14873 | Gene names | SPTBN2, KIAA0302 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 2 (Spectrin, non-erythroid beta chain 2) (Beta-III spectrin). | |||||
|
SPTB1_MOUSE
|
||||||
| NC score | 0.215803 (rank : 24) | θ value | 0.163984 (rank : 24) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P15508 | Gene names | Sptb, Spnb-1, Spnb1, Sptb1 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, erythrocyte (Beta-I spectrin). | |||||
|
FLNB_HUMAN
|
||||||
| NC score | 0.215493 (rank : 25) | θ value | θ > 10 (rank : 45) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O75369, Q13706, Q8WXS9, Q8WXT0, Q8WXT1, Q8WXT2, Q9NRB5, Q9NT26, Q9UEV9 | Gene names | FLNB, FLN1L, FLN3, TABP, TAP | |||
|
Domain Architecture |

|
|||||
| Description | Filamin-B (FLN-B) (Beta-filamin) (Actin-binding-like protein) (Thyroid autoantigen) (Truncated actin-binding protein) (Truncated ABP) (ABP- 280 homolog) (ABP-278) (Filamin 3) (Filamin homolog 1) (Fh1). | |||||
|
SPTB2_HUMAN
|
||||||
| NC score | 0.214242 (rank : 26) | θ value | 0.0193708 (rank : 20) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q01082, O60837, Q16057 | Gene names | SPTBN1, SPTB2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
SPTB2_MOUSE
|
||||||
| NC score | 0.213332 (rank : 27) | θ value | 0.0193708 (rank : 21) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q62261, Q5SQL8 | Gene names | Sptbn1, Spnb-2, Spnb2, Sptb2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
SPTN5_HUMAN
|
||||||
| NC score | 0.181277 (rank : 28) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9NRC6 | Gene names | SPTBN5, SPTBN4 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 4 (Spectrin, non-erythroid beta chain 4) (Beta-V spectrin) (BSPECV). | |||||
|
MACF1_MOUSE
|
||||||
| NC score | 0.173696 (rank : 29) | θ value | 0.0148317 (rank : 19) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9QXZ0, P97394, P97395, P97396 | Gene names | Macf1, Acf7, Aclp7, Macf | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1 (Actin cross-linking family 7). | |||||
|
MACF1_HUMAN
|
||||||
| NC score | 0.168729 (rank : 30) | θ value | 0.0148317 (rank : 18) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1341 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9UPN3, O75053, Q5VW20, Q8WXY2, Q9H540, Q9UKP0, Q9ULG9 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5 (Actin cross-linking family protein 7) (Macrophin-1) (Trabeculin-alpha) (620 kDa actin-binding protein) (ABP620). | |||||
|
BPA1_MOUSE
|
||||||
| NC score | 0.162657 (rank : 31) | θ value | 0.163984 (rank : 23) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q91ZU6, Q91ZU7 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 1/2/3/4 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
SMOO_HUMAN
|
||||||
| NC score | 0.153946 (rank : 32) | θ value | θ > 10 (rank : 63) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P53814, O00569, O95769, O95937, Q9P1S8, Q9UIT1, Q9UIT2 | Gene names | SMTN, SMSMO | |||
|
Domain Architecture |

|
|||||
| Description | Smoothelin. | |||||
|
MILK2_HUMAN
|
||||||
| NC score | 0.146388 (rank : 33) | θ value | θ > 10 (rank : 57) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8IY33, Q7RTP4, Q7Z655, Q8TEQ4, Q9H5F9 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | MICAL-like protein 2. | |||||
|
PLEC1_HUMAN
|
||||||
| NC score | 0.145666 (rank : 34) | θ value | 6.88961 (rank : 38) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
SPTA1_MOUSE
|
||||||
| NC score | 0.145445 (rank : 35) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 675 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P08032, P97502 | Gene names | Spta1, Spna1, Spta | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, erythrocyte (Erythroid alpha-spectrin). | |||||
|
MILK1_HUMAN
|
||||||
| NC score | 0.144317 (rank : 36) | θ value | θ > 10 (rank : 55) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8N3F8, Q5TI16, Q7RTP5, Q8N3N8, Q9BVL9, Q9BY92, Q9UH43, Q9UH44, Q9UH45 | Gene names | MIRAB13, KIAA1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
UTRO_HUMAN
|
||||||
| NC score | 0.143666 (rank : 37) | θ value | 0.813845 (rank : 29) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P46939 | Gene names | UTRN, DMDL | |||
|
Domain Architecture |

|
|||||
| Description | Utrophin (Dystrophin-related protein 1) (DRP1) (DRP). | |||||
|
SPTA2_HUMAN
|
||||||
| NC score | 0.143403 (rank : 38) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 888 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q13813, Q13186, Q15324, Q16606, Q59EF1, Q5VXV5, Q5VXV6, Q7Z6M5, Q9P0V0 | Gene names | SPTAN1, SPTA2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, brain (Spectrin, non-erythroid alpha chain) (Alpha-II spectrin) (Fodrin alpha chain). | |||||
|
ASPM_HUMAN
|
||||||
| NC score | 0.142644 (rank : 39) | θ value | 1.06291 (rank : 30) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8IZT6, Q86UX4, Q8IUL2, Q8IZJ7, Q8IZJ8, Q8IZJ9, Q8N4D1, Q9NVS1, Q9NVT6 | Gene names | ASPM, MCPH5 | |||
|
Domain Architecture |

|
|||||
| Description | Abnormal spindle-like microcephaly-associated protein (Abnormal spindle protein homolog) (Asp homolog). | |||||
|
ASPM_MOUSE
|
||||||
| NC score | 0.140910 (rank : 40) | θ value | 0.62314 (rank : 28) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8CJ27, O88482, Q8BJI8, Q8BKT4 | Gene names | Aspm, Calmbp1, Sha1 | |||
|
Domain Architecture |

|
|||||
| Description | Abnormal spindle-like microcephaly-associated protein homolog (Calmodulin-binding protein 1) (Spindle and hydroxyurea checkpoint abnormal protein) (Calmodulin-binding protein Sha1). | |||||
|
SPTA1_HUMAN
|
||||||
| NC score | 0.138216 (rank : 41) | θ value | θ > 10 (rank : 64) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P02549, Q15514, Q6LDY5 | Gene names | SPTA1, SPTA | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, erythrocyte (Erythroid alpha-spectrin). | |||||
|
DMD_HUMAN
|
||||||
| NC score | 0.136063 (rank : 42) | θ value | 3.0926 (rank : 33) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P11532, Q02295, Q14169, Q14170, Q5JYU0, Q7KZ48 | Gene names | DMD | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
DMD_MOUSE
|
||||||
| NC score | 0.131768 (rank : 43) | θ value | 8.99809 (rank : 39) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1111 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P11531, O35653, Q60703 | Gene names | Dmd | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
MILK1_MOUSE
|
||||||
| NC score | 0.130155 (rank : 44) | θ value | θ > 10 (rank : 56) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BGT6, Q8BJ60 | Gene names | Mirab13, D15Mit260, Kiaa1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
SYNE2_HUMAN
|
||||||
| NC score | 0.123403 (rank : 45) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
MICA3_MOUSE
|
||||||
| NC score | 0.123225 (rank : 46) | θ value | θ > 10 (rank : 54) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8CJ19, Q80TE8, Q8BXB1 | Gene names | Mical3, Kiaa1364 | |||
|
Domain Architecture |

|
|||||
| Description | Protein MICAL-3. | |||||
|
MICA2_HUMAN
|
||||||
| NC score | 0.122617 (rank : 47) | θ value | θ > 10 (rank : 51) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O94851, Q7Z3A8 | Gene names | MICAL2, KIAA0750 | |||
|
Domain Architecture |

|
|||||
| Description | Protein MICAL-2. | |||||
|
SYNE1_HUMAN
|
||||||
| NC score | 0.121806 (rank : 48) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
MICA3_HUMAN
|
||||||
| NC score | 0.119457 (rank : 49) | θ value | θ > 10 (rank : 53) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q7RTP6, Q6ICK4, Q9P2I3 | Gene names | MICAL3, KIAA1364 | |||
|
Domain Architecture |

|
|||||
| Description | Protein MICAL-3. | |||||
|
MICA2_MOUSE
|
||||||
| NC score | 0.113513 (rank : 50) | θ value | θ > 10 (rank : 52) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BML1, Q3UPU6 | Gene names | Mical2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein MICAL-2. | |||||
|
MICA1_MOUSE
|
||||||
| NC score | 0.095655 (rank : 51) | θ value | θ > 10 (rank : 50) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8VDP3 | Gene names | Mical1, Mical, Nical | |||
|
Domain Architecture |

|
|||||
| Description | NEDD9-interacting protein with calponin homology and LIM domains (Molecule interacting with CasL protein 1). | |||||
|
MICA1_HUMAN
|
||||||
| NC score | 0.095114 (rank : 52) | θ value | θ > 10 (rank : 49) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8TDZ2, Q7Z633, Q8IVS9, Q96G47, Q9H6X6, Q9H7I0, Q9HAA1, Q9UFF7 | Gene names | MICAL1, MICAL, NICAL | |||
|
Domain Architecture |

|
|||||
| Description | NEDD9-interacting protein with calponin homology and LIM domains (Molecule interacting with CasL protein 1). | |||||
|
PLST_MOUSE
|
||||||
| NC score | 0.093728 (rank : 53) | θ value | θ > 10 (rank : 62) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q99K51 | Gene names | Pls3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plastin-3 (T-plastin). | |||||
|
PLST_HUMAN
|
||||||
| NC score | 0.092943 (rank : 54) | θ value | θ > 10 (rank : 61) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P13797, Q86YI6 | Gene names | PLS3 | |||
|
Domain Architecture |

|
|||||
| Description | Plastin-3 (T-plastin). | |||||
|
PLSL_MOUSE
|
||||||
| NC score | 0.090914 (rank : 55) | θ value | θ > 10 (rank : 60) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q61233, Q3UE24, Q8R1X3, Q9CV77 | Gene names | Lcp1, Pls2 | |||
|
Domain Architecture |

|
|||||
| Description | Plastin-2 (L-plastin) (Lymphocyte cytosolic protein 1) (LCP-1) (65 kDa macrophage protein) (pp65). | |||||
|
PLSL_HUMAN
|
||||||
| NC score | 0.089507 (rank : 56) | θ value | θ > 10 (rank : 59) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P13796 | Gene names | LCP1, PLS2 | |||
|
Domain Architecture |

|
|||||
| Description | Plastin-2 (L-plastin) (Lymphocyte cytosolic protein 1) (LCP-1) (LC64P). | |||||
|
BPAEA_HUMAN
|
||||||
| NC score | 0.084371 (rank : 57) | θ value | θ > 10 (rank : 42) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1369 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O94833, Q5TBT0, Q8N1T8, Q8N8J3, Q8WXK9, Q96AK9, Q96DQ5, Q9H555 | Gene names | DST, BPAG1, DMH, DT, KIAA0728 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 6/9/10 (Trabeculin-beta) (Bullous pemphigoid antigen) (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
MACF4_HUMAN
|
||||||
| NC score | 0.082730 (rank : 58) | θ value | θ > 10 (rank : 48) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1321 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96PK2, Q8WXY1 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoform 4. | |||||
|
PLSI_HUMAN
|
||||||
| NC score | 0.080438 (rank : 59) | θ value | θ > 10 (rank : 58) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q14651 | Gene names | PLS1 | |||
|
Domain Architecture |

|
|||||
| Description | Plastin-1 (I-plastin) (Intestine-specific plastin). | |||||
|
CYTSA_HUMAN
|
||||||
| NC score | 0.080385 (rank : 60) | θ value | θ > 10 (rank : 43) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1138 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q69YQ0, O15081 | Gene names | CYTSA, KIAA0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A (NY-REN-22 antigen). | |||||
|
CYTSA_MOUSE
|
||||||
| NC score | 0.078567 (rank : 61) | θ value | θ > 10 (rank : 44) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q2KN98, Q8CHF9 | Gene names | Cytsa, Kiaa0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A. | |||||
|
KTN1_MOUSE
|
||||||
| NC score | 0.040483 (rank : 62) | θ value | 1.38821 (rank : 31) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1324 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q61595, Q8BG49, Q8BHF4, Q8BHM8, Q8C9Y5, Q8CG51, Q8CG52, Q8CG53, Q8CG54, Q8CG55, Q8CG56, Q8CG57, Q8CG58, Q8CG59, Q8CG60, Q8CG61, Q8CG62, Q8CG63 | Gene names | Ktn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin. | |||||
|
CROCC_HUMAN
|
||||||
| NC score | 0.038376 (rank : 63) | θ value | 5.27518 (rank : 37) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1409 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q5TZA2, Q2VHY3, Q66GT7, Q7Z2L4, Q7Z5D7 | Gene names | CROCC, KIAA0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
MD1L1_MOUSE
|
||||||
| NC score | 0.038202 (rank : 64) | θ value | 3.0926 (rank : 34) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1001 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9WTX8, Q9WTX9 | Gene names | Mad1l1, Mad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1). | |||||
|
AKAP9_HUMAN
|
||||||
| NC score | 0.036164 (rank : 65) | θ value | 4.03905 (rank : 35) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
RAD50_HUMAN
|
||||||
| NC score | 0.029394 (rank : 66) | θ value | 1.38821 (rank : 32) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q92878, O43254, Q6GMT7, Q6P5X3, Q9UP86 | Gene names | RAD50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (hRAD50). | |||||
|
SMC3_HUMAN
|
||||||
| NC score | 0.028927 (rank : 67) | θ value | 8.99809 (rank : 40) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1186 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UQE7, O60464 | Gene names | CSPG6, BAM, BMH, SMC3, SMC3L1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome-associated polypeptide) (hCAP) (Bamacan) (Basement membrane-associated chondroitin proteoglycan). | |||||
|
SMC3_MOUSE
|
||||||
| NC score | 0.028704 (rank : 68) | θ value | 8.99809 (rank : 41) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1187 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9CW03, O35667, Q9QUS3 | Gene names | Cspg6, Bam, Bmh, Mmip1, Smc3, Smc3l1, Smcd | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome segregation protein SmcD) (Bamacan) (Basement membrane-associated chondroitin proteoglycan) (Mad member- interacting protein 1). | |||||
|
KI13B_HUMAN
|
||||||
| NC score | 0.006198 (rank : 69) | θ value | 4.03905 (rank : 36) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NQT8, O75134, Q9BYJ6 | Gene names | KIF13B, GAKIN, KIAA0639 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF13B (Kinesin-like protein GAKIN). | |||||