Please be patient as the page loads
|
P53_MOUSE
|
||||||
| SwissProt Accessions | P02340, Q9QUP3 | Gene names | Tp53, P53, Trp53 | |||
|
Domain Architecture |
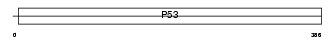
|
|||||
| Description | Cellular tumor antigen p53 (Tumor suppressor p53). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
P53_MOUSE
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P02340, Q9QUP3 | Gene names | Tp53, P53, Trp53 | |||
|
Domain Architecture |

|
|||||
| Description | Cellular tumor antigen p53 (Tumor suppressor p53). | |||||
|
P53_HUMAN
|
||||||
| θ value | 9.80574e-164 (rank : 2) | NC score | 0.989168 (rank : 2) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P04637, Q15086, Q15087, Q15088, Q16535, Q16807, Q16808, Q16809, Q16810, Q16811, Q16848, Q86UG1, Q8J016, Q99659, Q9BTM4, Q9HAQ8, Q9NP68, Q9NPJ2, Q9NZD0, Q9UBI2, Q9UQ61 | Gene names | TP53, P53 | |||
|
Domain Architecture |
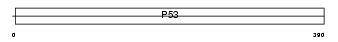
|
|||||
| Description | Cellular tumor antigen p53 (Tumor suppressor p53) (Phosphoprotein p53) (Antigen NY-CO-13). | |||||
|
P73L_HUMAN
|
||||||
| θ value | 6.7473e-72 (rank : 3) | NC score | 0.910001 (rank : 5) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H3D4, O75080, O75195, O75922, O76078, Q6VEG2, Q6VEG3, Q6VEG4, Q6VFJ1, Q6VFJ2, Q6VFJ3, Q6VH20, Q7LDI3, Q7LDI4, Q7LDI5, Q96KR0, Q9H3D2, Q9H3D3, Q9H3P8, Q9NPH7, Q9P1B4, Q9P1B5, Q9P1B6, Q9P1B7, Q9UBV9, Q9UE10, Q9UP26, Q9UP27, Q9UP28, Q9UP74 | Gene names | TP73L, KET, P63, P73H, P73L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor protein p73-like (p73L) (p63) (Tumor protein 63) (TP63) (p51) (p40) (Keratinocyte transcription factor KET) (Chronic ulcerative stomatitis protein) (CUSP). | |||||
|
P73L_MOUSE
|
||||||
| θ value | 6.7473e-72 (rank : 4) | NC score | 0.911548 (rank : 4) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O88898, O88897, O88899, O89097, Q8C826, Q9QWY9, Q9QWZ0 | Gene names | Tp73l, P63, P73l, Trp63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor protein p73-like (p73L) (p63) (Transformation-related protein 63). | |||||
|
P73_HUMAN
|
||||||
| θ value | 1.96316e-71 (rank : 5) | NC score | 0.923116 (rank : 3) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15350, O15351, Q5TBV5, Q5TBV6, Q8NHW9, Q8TDY5, Q8TDY6, Q9NTK8 | Gene names | TP73, P73 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor protein p73 (p53-like transcription factor) (p53-related protein). | |||||
|
LNK_HUMAN
|
||||||
| θ value | 0.365318 (rank : 6) | NC score | 0.032222 (rank : 8) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UQQ2, O95184 | Gene names | LNK | |||
|
Domain Architecture |
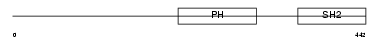
|
|||||
| Description | Lymphocyte-specific adapter protein Lnk (Signal transduction protein Lnk) (Lymphocyte adapter protein). | |||||
|
CNTRB_MOUSE
|
||||||
| θ value | 0.813845 (rank : 7) | NC score | 0.014275 (rank : 10) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 1029 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CB62, Q5NCF3 | Gene names | Cntrob, Lip8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrobin (LYST-interacting protein 8). | |||||
|
AKAP5_HUMAN
|
||||||
| θ value | 1.06291 (rank : 8) | NC score | 0.075916 (rank : 6) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P24588 | Gene names | AKAP5, AKAP79 | |||
|
Domain Architecture |
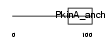
|
|||||
| Description | A-kinase anchor protein 5 (A-kinase anchor protein 79 kDa) (AKAP 79) (cAMP-dependent protein kinase regulatory subunit II high affinity- binding protein) (H21). | |||||
|
GPTC8_HUMAN
|
||||||
| θ value | 4.03905 (rank : 9) | NC score | 0.019068 (rank : 9) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UKJ3, O60300 | Gene names | GPATC8, KIAA0553 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 8. | |||||
|
ZN620_HUMAN
|
||||||
| θ value | 4.03905 (rank : 10) | NC score | 0.000010 (rank : 15) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6ZNG0, Q8N223 | Gene names | ZNF620 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 620. | |||||
|
CASL_HUMAN
|
||||||
| θ value | 6.88961 (rank : 11) | NC score | 0.009331 (rank : 13) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14511 | Gene names | NEDD9, CASL | |||
|
Domain Architecture |
|
|||||
| Description | Enhancer of filamentation 1 (HEF1) (CRK-associated substrate-related protein) (CAS-L) (CasL) (p105) (Protein NEDD9) (NY-REN-12 antigen). | |||||
|
ZADH2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 12) | NC score | 0.011418 (rank : 12) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N4Q0 | Gene names | ZADH2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc-binding alcohol dehydrogenase domain-containing protein 2 (EC 1.-.-.-). | |||||
|
EPN3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 13) | NC score | 0.007159 (rank : 14) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91W69, Q9CV55 | Gene names | Epn3 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-3 (EPS-15-interacting protein 3). | |||||
|
PDLI7_HUMAN
|
||||||
| θ value | 8.99809 (rank : 14) | NC score | 0.012536 (rank : 11) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 547 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NR12, Q14250, Q5XG82, Q6NVZ5, Q96C91, Q9BXB8, Q9BXB9 | Gene names | PDLIM7, ENIGMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ and LIM domain protein 7 (LIM mineralization protein) (LMP) (Protein enigma). | |||||
|
EVI2B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 15) | NC score | 0.068325 (rank : 7) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VD58, Q3U1A3 | Gene names | Evi2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EVI2B protein precursor (Ecotropic viral integration site 2B protein). | |||||
|
P53_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P02340, Q9QUP3 | Gene names | Tp53, P53, Trp53 | |||
|
Domain Architecture |

|
|||||
| Description | Cellular tumor antigen p53 (Tumor suppressor p53). | |||||
|
P53_HUMAN
|
||||||
| NC score | 0.989168 (rank : 2) | θ value | 9.80574e-164 (rank : 2) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P04637, Q15086, Q15087, Q15088, Q16535, Q16807, Q16808, Q16809, Q16810, Q16811, Q16848, Q86UG1, Q8J016, Q99659, Q9BTM4, Q9HAQ8, Q9NP68, Q9NPJ2, Q9NZD0, Q9UBI2, Q9UQ61 | Gene names | TP53, P53 | |||
|
Domain Architecture |

|
|||||
| Description | Cellular tumor antigen p53 (Tumor suppressor p53) (Phosphoprotein p53) (Antigen NY-CO-13). | |||||
|
P73_HUMAN
|
||||||
| NC score | 0.923116 (rank : 3) | θ value | 1.96316e-71 (rank : 5) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15350, O15351, Q5TBV5, Q5TBV6, Q8NHW9, Q8TDY5, Q8TDY6, Q9NTK8 | Gene names | TP73, P73 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor protein p73 (p53-like transcription factor) (p53-related protein). | |||||
|
P73L_MOUSE
|
||||||
| NC score | 0.911548 (rank : 4) | θ value | 6.7473e-72 (rank : 4) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O88898, O88897, O88899, O89097, Q8C826, Q9QWY9, Q9QWZ0 | Gene names | Tp73l, P63, P73l, Trp63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor protein p73-like (p73L) (p63) (Transformation-related protein 63). | |||||
|
P73L_HUMAN
|
||||||
| NC score | 0.910001 (rank : 5) | θ value | 6.7473e-72 (rank : 3) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H3D4, O75080, O75195, O75922, O76078, Q6VEG2, Q6VEG3, Q6VEG4, Q6VFJ1, Q6VFJ2, Q6VFJ3, Q6VH20, Q7LDI3, Q7LDI4, Q7LDI5, Q96KR0, Q9H3D2, Q9H3D3, Q9H3P8, Q9NPH7, Q9P1B4, Q9P1B5, Q9P1B6, Q9P1B7, Q9UBV9, Q9UE10, Q9UP26, Q9UP27, Q9UP28, Q9UP74 | Gene names | TP73L, KET, P63, P73H, P73L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor protein p73-like (p73L) (p63) (Tumor protein 63) (TP63) (p51) (p40) (Keratinocyte transcription factor KET) (Chronic ulcerative stomatitis protein) (CUSP). | |||||
|
AKAP5_HUMAN
|
||||||
| NC score | 0.075916 (rank : 6) | θ value | 1.06291 (rank : 8) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P24588 | Gene names | AKAP5, AKAP79 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 5 (A-kinase anchor protein 79 kDa) (AKAP 79) (cAMP-dependent protein kinase regulatory subunit II high affinity- binding protein) (H21). | |||||
|
EVI2B_MOUSE
|
||||||
| NC score | 0.068325 (rank : 7) | θ value | θ > 10 (rank : 15) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VD58, Q3U1A3 | Gene names | Evi2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EVI2B protein precursor (Ecotropic viral integration site 2B protein). | |||||
|
LNK_HUMAN
|
||||||
| NC score | 0.032222 (rank : 8) | θ value | 0.365318 (rank : 6) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UQQ2, O95184 | Gene names | LNK | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte-specific adapter protein Lnk (Signal transduction protein Lnk) (Lymphocyte adapter protein). | |||||
|
GPTC8_HUMAN
|
||||||
| NC score | 0.019068 (rank : 9) | θ value | 4.03905 (rank : 9) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UKJ3, O60300 | Gene names | GPATC8, KIAA0553 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 8. | |||||
|
CNTRB_MOUSE
|
||||||
| NC score | 0.014275 (rank : 10) | θ value | 0.813845 (rank : 7) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 1029 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CB62, Q5NCF3 | Gene names | Cntrob, Lip8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrobin (LYST-interacting protein 8). | |||||
|
PDLI7_HUMAN
|
||||||
| NC score | 0.012536 (rank : 11) | θ value | 8.99809 (rank : 14) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 547 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NR12, Q14250, Q5XG82, Q6NVZ5, Q96C91, Q9BXB8, Q9BXB9 | Gene names | PDLIM7, ENIGMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ and LIM domain protein 7 (LIM mineralization protein) (LMP) (Protein enigma). | |||||
|
ZADH2_HUMAN
|
||||||
| NC score | 0.011418 (rank : 12) | θ value | 6.88961 (rank : 12) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N4Q0 | Gene names | ZADH2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc-binding alcohol dehydrogenase domain-containing protein 2 (EC 1.-.-.-). | |||||
|
CASL_HUMAN
|
||||||
| NC score | 0.009331 (rank : 13) | θ value | 6.88961 (rank : 11) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14511 | Gene names | NEDD9, CASL | |||
|
Domain Architecture |

|
|||||
| Description | Enhancer of filamentation 1 (HEF1) (CRK-associated substrate-related protein) (CAS-L) (CasL) (p105) (Protein NEDD9) (NY-REN-12 antigen). | |||||
|
EPN3_MOUSE
|
||||||
| NC score | 0.007159 (rank : 14) | θ value | 8.99809 (rank : 13) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91W69, Q9CV55 | Gene names | Epn3 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-3 (EPS-15-interacting protein 3). | |||||
|
ZN620_HUMAN
|
||||||
| NC score | 0.000010 (rank : 15) | θ value | 4.03905 (rank : 10) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6ZNG0, Q8N223 | Gene names | ZNF620 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 620. | |||||