Please be patient as the page loads
|
NPTXR_MOUSE
|
||||||
| SwissProt Accessions | Q99J85 | Gene names | Nptxr, Npr | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal pentraxin receptor. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
NPTXR_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.988040 (rank : 2) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O95502 | Gene names | NPTXR | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal pentraxin receptor. | |||||
|
NPTXR_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q99J85 | Gene names | Nptxr, Npr | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal pentraxin receptor. | |||||
|
NPTX2_HUMAN
|
||||||
| θ value | 2.9938e-112 (rank : 3) | NC score | 0.961820 (rank : 3) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P47972, Q86XV7, Q96G70 | Gene names | NPTX2 | |||
|
Domain Architecture |
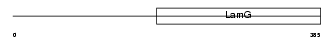
|
|||||
| Description | Neuronal pentraxin-2 precursor (NP2) (Neuronal pentraxin II) (NP-II). | |||||
|
NPTX2_MOUSE
|
||||||
| θ value | 2.9938e-112 (rank : 4) | NC score | 0.961624 (rank : 4) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O70340 | Gene names | Nptx2 | |||
|
Domain Architecture |
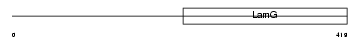
|
|||||
| Description | Neuronal pentraxin-2 precursor (NP2) (Neuronal pentraxin II) (NP-II). | |||||
|
NPTX1_HUMAN
|
||||||
| θ value | 5.85632e-100 (rank : 5) | NC score | 0.959256 (rank : 5) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q15818, Q5FWE6 | Gene names | NPTX1 | |||
|
Domain Architecture |
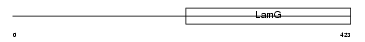
|
|||||
| Description | Neuronal pentraxin-1 precursor (NP1) (Neuronal pentraxin I) (NP-I). | |||||
|
NPTX1_MOUSE
|
||||||
| θ value | 9.98939e-100 (rank : 6) | NC score | 0.957539 (rank : 6) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q62443 | Gene names | Nptx1 | |||
|
Domain Architecture |
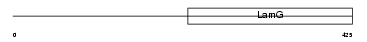
|
|||||
| Description | Neuronal pentraxin-1 precursor (NP1) (Neuronal pentraxin I) (NP-I). | |||||
|
GP144_HUMAN
|
||||||
| θ value | 3.28125e-26 (rank : 7) | NC score | 0.419557 (rank : 13) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q7Z7M1, Q86SL4, Q8NH12 | Gene names | GPR144, PGR24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 144 (G-protein coupled receptor PGR24). | |||||
|
PTX3_HUMAN
|
||||||
| θ value | 3.28887e-18 (rank : 8) | NC score | 0.745083 (rank : 8) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P26022 | Gene names | PTX3, TSG14 | |||
|
Domain Architecture |
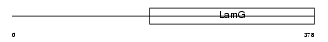
|
|||||
| Description | Pentraxin-related protein PTX3 precursor (Pentaxin-related protein PTX3) (Tumor necrosis factor-inducible protein TSG-14). | |||||
|
PTX3_MOUSE
|
||||||
| θ value | 1.058e-16 (rank : 9) | NC score | 0.747317 (rank : 7) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P48759, Q8VDF1 | Gene names | Ptx3, Tsg14 | |||
|
Domain Architecture |
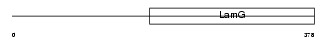
|
|||||
| Description | Pentraxin-related protein PTX3 precursor (Pentaxin-related protein PTX3) (Tumor necrosis factor-inducible protein TSG-14). | |||||
|
SAMP_HUMAN
|
||||||
| θ value | 1.68911e-14 (rank : 10) | NC score | 0.690092 (rank : 11) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P02743 | Gene names | APCS, PTX2 | |||
|
Domain Architecture |
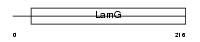
|
|||||
| Description | Serum amyloid P-component precursor (SAP) (9.5S alpha-1-glycoprotein) [Contains: Serum amyloid P-component(1-203)]. | |||||
|
CRP_HUMAN
|
||||||
| θ value | 2.88119e-14 (rank : 11) | NC score | 0.699622 (rank : 10) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P02741, Q8WW75 | Gene names | CRP, PTX1 | |||
|
Domain Architecture |
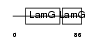
|
|||||
| Description | C-reactive protein precursor [Contains: C-reactive protein(1-205)]. | |||||
|
CRP_MOUSE
|
||||||
| θ value | 2.88119e-14 (rank : 12) | NC score | 0.702387 (rank : 9) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P14847 | Gene names | Crp, Ptx1 | |||
|
Domain Architecture |
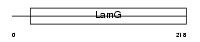
|
|||||
| Description | C-reactive protein precursor. | |||||
|
SAMP_MOUSE
|
||||||
| θ value | 8.38298e-14 (rank : 13) | NC score | 0.687839 (rank : 12) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P12246 | Gene names | Apcs, Ptx2, Sap | |||
|
Domain Architecture |
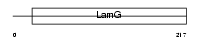
|
|||||
| Description | Serum amyloid P-component precursor (SAP). | |||||
|
GP112_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 14) | NC score | 0.114879 (rank : 14) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8IZF6, Q86SM6 | Gene names | GPR112 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 112. | |||||
|
GP126_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 15) | NC score | 0.082187 (rank : 15) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q86SQ4, Q6MZU7, Q8NC14, Q96JW0 | Gene names | GPR126 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 126 precursor. | |||||
|
MYH14_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 16) | NC score | 0.067081 (rank : 20) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 995 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q7Z406, Q5CZ75, Q6XYE4, Q76B62, Q8WV23, Q96I22, Q9BT27, Q9BW35, Q9H882 | Gene names | MYH14, KIAA2034 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
MYH10_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 17) | NC score | 0.063993 (rank : 22) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P35580, Q16087 | Gene names | MYH10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
MYH10_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 18) | NC score | 0.062860 (rank : 24) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1612 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q61879, Q5SV63 | Gene names | Myh10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
RUFY1_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 19) | NC score | 0.053826 (rank : 59) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8BIJ7, Q8BKQ4, Q8BL21, Q9EPM6 | Gene names | Rufy1, Rabip4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 1 (Rab4-interacting protein). | |||||
|
HOOK2_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 20) | NC score | 0.068591 (rank : 19) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 714 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q96ED9, O60562 | Gene names | HOOK2 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 2 (h-hook2) (hHK2). | |||||
|
CCHCR_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 21) | NC score | 0.075013 (rank : 17) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 717 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8TD31, Q9NRK8, Q9NWY9, Q9NXJ4, Q9NXK3, Q9Y6W1, Q9Y6W2 | Gene names | CCHCR1, C6orf18, HCR | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil alpha-helical rod protein 1 (Alpha helical coiled-coil rod protein) (Putative gene 8 protein) (Pg8). | |||||
|
REST_HUMAN
|
||||||
| θ value | 0.125558 (rank : 22) | NC score | 0.054145 (rank : 56) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1605 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P30622 | Gene names | RSN, CYLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Restin (Cytoplasmic linker protein 170 alpha-2) (CLIP-170) (Reed- Sternberg intermediate filament-associated protein) (Cytoplasmic linker protein 1). | |||||
|
HOOK2_MOUSE
|
||||||
| θ value | 0.163984 (rank : 23) | NC score | 0.073640 (rank : 18) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 736 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q7TMK6, Q8BY47, Q8R347, Q8VCN4, Q99LU2 | Gene names | Hook2 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 2. | |||||
|
NUMA1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 24) | NC score | 0.055395 (rank : 42) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
AKAP9_HUMAN
|
||||||
| θ value | 0.21417 (rank : 25) | NC score | 0.054669 (rank : 52) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
AMOL2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 26) | NC score | 0.056385 (rank : 37) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 603 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9Y2J4, Q53EP1, Q96F99, Q9UKB4 | Gene names | AMOTL2, KIAA0989 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiomotin-like protein 2 (Leman coiled-coil protein) (LCCP). | |||||
|
GP133_HUMAN
|
||||||
| θ value | 0.279714 (rank : 27) | NC score | 0.079078 (rank : 16) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6QNK2, Q6ZMQ1, Q7Z7M2, Q86SM4 | Gene names | GPR133, PGR25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 133 precursor (G-protein coupled receptor PGR25). | |||||
|
SPTN5_HUMAN
|
||||||
| θ value | 0.279714 (rank : 28) | NC score | 0.041534 (rank : 87) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9NRC6 | Gene names | SPTBN5, SPTBN4 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 4 (Spectrin, non-erythroid beta chain 4) (Beta-V spectrin) (BSPECV). | |||||
|
CYLN2_MOUSE
|
||||||
| θ value | 0.365318 (rank : 29) | NC score | 0.061264 (rank : 27) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1002 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9Z0H8, Q7TSI9, Q8CHU1, Q9EP81 | Gene names | Cyln2, Kiaa0291 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic linker protein 2 (Cytoplasmic linker protein 115) (CLIP- 115). | |||||
|
MRCKG_MOUSE
|
||||||
| θ value | 0.365318 (rank : 30) | NC score | 0.020376 (rank : 101) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1466 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q80UW5 | Gene names | Cdc42bpg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK gamma (EC 2.7.11.1) (CDC42- binding protein kinase gamma) (Myotonic dystrophy kinase-related CDC42-binding kinase gamma) (Myotonic dystrophy protein kinase-like alpha) (MRCK gamma) (DMPK-like gamma). | |||||
|
MYH11_HUMAN
|
||||||
| θ value | 0.365318 (rank : 31) | NC score | 0.062502 (rank : 26) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
CNTRB_HUMAN
|
||||||
| θ value | 0.47712 (rank : 32) | NC score | 0.059897 (rank : 29) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8N137, Q331K3, Q69YV7, Q8NCB8, Q8WXV3, Q96CQ7, Q9C060 | Gene names | CNTROB, LIP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrobin (LYST-interacting protein 8). | |||||
|
MYH11_MOUSE
|
||||||
| θ value | 0.62314 (rank : 33) | NC score | 0.060645 (rank : 28) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
CKAP4_HUMAN
|
||||||
| θ value | 0.813845 (rank : 34) | NC score | 0.054558 (rank : 53) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q07065, Q504S5, Q53ES6 | Gene names | CKAP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-associated protein 4 (63 kDa membrane protein) (p63). | |||||
|
CROCC_MOUSE
|
||||||
| θ value | 1.06291 (rank : 35) | NC score | 0.054903 (rank : 51) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1460 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q8CJ40, Q7TQL2, Q80U01, Q8R0B9 | Gene names | Crocc, Kiaa0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
ZFY19_HUMAN
|
||||||
| θ value | 1.06291 (rank : 36) | NC score | 0.032714 (rank : 91) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96K21, Q86WC2, Q8WU96 | Gene names | ZFYVE19, MPFYVE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger FYVE domain-containing protein 19 (MLL partner containing FYVE domain). | |||||
|
GOGA1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 37) | NC score | 0.052170 (rank : 65) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1222 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q92805, Q8IYZ9 | Gene names | GOLGA1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
HIP1R_HUMAN
|
||||||
| θ value | 1.38821 (rank : 38) | NC score | 0.046814 (rank : 82) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 809 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O75146, Q9UED9 | Gene names | HIP1R, HIP12, KIAA0655 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1-related protein (Hip1-related) (Hip 12). | |||||
|
K1543_HUMAN
|
||||||
| θ value | 1.81305 (rank : 39) | NC score | 0.018118 (rank : 102) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9P1Y5, Q8NDF1 | Gene names | KIAA1543 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1543. | |||||
|
SURF6_MOUSE
|
||||||
| θ value | 1.81305 (rank : 40) | NC score | 0.037851 (rank : 89) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P70279, Q8BPI4, Q8BRL7, Q8R0Q7 | Gene names | Surf6, Surf-6 | |||
|
Domain Architecture |
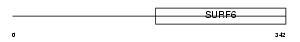
|
|||||
| Description | Surfeit locus protein 6. | |||||
|
DMD_HUMAN
|
||||||
| θ value | 2.36792 (rank : 41) | NC score | 0.046340 (rank : 83) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P11532, Q02295, Q14169, Q14170, Q5JYU0, Q7KZ48 | Gene names | DMD | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
DMD_MOUSE
|
||||||
| θ value | 2.36792 (rank : 42) | NC score | 0.048991 (rank : 80) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1111 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P11531, O35653, Q60703 | Gene names | Dmd | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
KGP1A_HUMAN
|
||||||
| θ value | 2.36792 (rank : 43) | NC score | 0.004904 (rank : 106) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 901 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q13976 | Gene names | PRKG1, PRKGR1A | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 1, alpha isozyme (EC 2.7.11.12) (CGK 1 alpha) (cGKI-alpha). | |||||
|
MRCKB_HUMAN
|
||||||
| θ value | 2.36792 (rank : 44) | NC score | 0.027140 (rank : 97) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 2239 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9Y5S2, Q2L7A5, Q86TJ1, Q9ULU5 | Gene names | CDC42BPB, KIAA1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
MRCKB_MOUSE
|
||||||
| θ value | 2.36792 (rank : 45) | NC score | 0.026084 (rank : 98) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 2180 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q7TT50, Q69ZR5, Q80W33 | Gene names | Cdc42bpb, Kiaa1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
MYO5C_HUMAN
|
||||||
| θ value | 2.36792 (rank : 46) | NC score | 0.054351 (rank : 54) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1206 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9NQX4 | Gene names | MYO5C | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5C (Myosin Vc). | |||||
|
AMOL2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 47) | NC score | 0.050921 (rank : 70) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8K371, Q3TPM1, Q7TPE4, Q8BP84, Q8BS08, Q9QUS0 | Gene names | Amotl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiomotin-like protein 2. | |||||
|
CING_MOUSE
|
||||||
| θ value | 3.0926 (rank : 48) | NC score | 0.057201 (rank : 33) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1432 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P59242 | Gene names | Cgn | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
FLIP1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 49) | NC score | 0.050577 (rank : 74) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1176 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9CS72 | Gene names | Filip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
MY18A_MOUSE
|
||||||
| θ value | 3.0926 (rank : 50) | NC score | 0.053630 (rank : 61) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
MYH1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 51) | NC score | 0.055551 (rank : 41) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1461 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q5SX40 | Gene names | Myh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1). | |||||
|
PCNT_HUMAN
|
||||||
| θ value | 3.0926 (rank : 52) | NC score | 0.062842 (rank : 25) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
RUFY1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 53) | NC score | 0.049991 (rank : 78) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 848 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q96T51, Q59FF3, Q71S93, Q9H6I3 | Gene names | RUFY1, RABIP4, ZFYVE12 | |||
|
Domain Architecture |

|
|||||
| Description | RUN and FYVE domain-containing protein 1 (FYVE-finger protein EIP1) (Zinc finger FYVE domain-containing protein 12) (La-binding protein 1) (Rab4-interacting protein). | |||||
|
ZN197_HUMAN
|
||||||
| θ value | 3.0926 (rank : 54) | NC score | -0.002306 (rank : 110) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O14709 | Gene names | ZNF197, ZNF166 | |||
|
Domain Architecture |
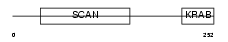
|
|||||
| Description | Zinc finger protein 197 (ZnF20). | |||||
|
A16L1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 55) | NC score | 0.024379 (rank : 100) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 776 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8C0J2, Q6KAT7, Q80U97, Q80U98, Q80U99, Q80Y53, Q9DB63 | Gene names | Atg16l1, Apg16l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Autophagy-related protein 16-1 (APG16-like 1). | |||||
|
EVPL_HUMAN
|
||||||
| θ value | 4.03905 (rank : 56) | NC score | 0.044147 (rank : 85) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 895 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q92817 | Gene names | EVPL | |||
|
Domain Architecture |

|
|||||
| Description | Envoplakin (210 kDa paraneoplastic pemphigus antigen) (p210) (210 kDa cornified envelope precursor protein). | |||||
|
KIF5A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 57) | NC score | 0.028798 (rank : 95) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 843 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q12840, Q4LE26 | Gene names | KIF5A, NKHC1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain isoform 5A (Neuronal kinesin heavy chain) (NKHC) (Kinesin heavy chain neuron-specific 1). | |||||
|
MYH4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 58) | NC score | 0.055366 (rank : 43) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
MYH9_HUMAN
|
||||||
| θ value | 4.03905 (rank : 59) | NC score | 0.063287 (rank : 23) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
MYH9_MOUSE
|
||||||
| θ value | 4.03905 (rank : 60) | NC score | 0.064403 (rank : 21) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
PALM_HUMAN
|
||||||
| θ value | 4.03905 (rank : 61) | NC score | 0.035054 (rank : 90) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O75781, O43359, O95673, Q92559, Q9UPJ4, Q9UQS2, Q9UQS3 | Gene names | PALM, KIAA0270 | |||
|
Domain Architecture |

|
|||||
| Description | Paralemmin. | |||||
|
RB6I2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 62) | NC score | 0.050243 (rank : 77) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q99MI1, Q80TK7, Q8BPL1, Q8C7Y1, Q99MI2 | Gene names | Erc1, Cast2, Kiaa1081, Rab6ip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1) (ERC1) (CAZ-associated structural protein 2) (CAST2). | |||||
|
RFWD2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 63) | NC score | 0.017349 (rank : 105) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8NHY2, Q6H103, Q9H6L7 | Gene names | RFWD2, COP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger and WD repeat domain protein 2 (EC 6.3.2.-) (Ubiquitin- protein ligase COP1) (Constitutive photomorphogenesis protein 1 homolog) (hCOP1). | |||||
|
RFWD2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 64) | NC score | 0.017379 (rank : 104) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 535 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9R1A8 | Gene names | Rfwd2, Cop1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger and WD repeat domain protein 2 (EC 6.3.2.-) (Ubiquitin- protein ligase COP1) (Constitutive photomorphogenesis protein 1 homolog) (mCOP1). | |||||
|
SCG2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 65) | NC score | 0.017640 (rank : 103) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q03517, Q80Y79, Q9CW80 | Gene names | Scg2, Chgc, Scg-2 | |||
|
Domain Architecture |

|
|||||
| Description | Secretogranin-2 precursor (Secretogranin II) (SgII) (Chromogranin C) [Contains: Secretoneurin (SN)]. | |||||
|
CP135_MOUSE
|
||||||
| θ value | 5.27518 (rank : 66) | NC score | 0.052150 (rank : 66) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1278 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q6P5D4, Q6A030 | Gene names | Cep135, Cep4, Kiaa0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
EEA1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 67) | NC score | 0.050736 (rank : 72) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
FYCO1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 68) | NC score | 0.047232 (rank : 81) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1149 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9BQS8, Q3MJE6, Q86T41, Q86TB1, Q8TEF9, Q96IV5, Q9H8P9 | Gene names | FYCO1, ZFYVE7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1 (Zinc finger FYVE domain-containing protein 7). | |||||
|
LIPA2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 69) | NC score | 0.043085 (rank : 86) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1282 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8BSS9, Q8BN73 | Gene names | Ppfia2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-2) (PTPRF-interacting protein alpha-2). | |||||
|
MYH1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 70) | NC score | 0.055696 (rank : 40) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1478 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P12882, Q9Y622 | Gene names | MYH1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1) (Myosin heavy chain IIx/d) (MyHC-IIx/d). | |||||
|
MYH4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 71) | NC score | 0.056054 (rank : 38) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1377 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9Y623 | Gene names | MYH4 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal) (Myosin heavy chain IIb) (MyHC-IIb). | |||||
|
NEB1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 72) | NC score | 0.030803 (rank : 93) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 829 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9ULJ8, O76059, Q9NXT2 | Gene names | PPP1R9A, KIAA1222 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-1 (Neurabin-I) (Neural tissue-specific F-actin-binding protein I) (Protein phosphatase 1 regulatory subunit 9A). | |||||
|
RB6I2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 73) | NC score | 0.052462 (rank : 64) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1584 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q8IUD2, Q6NVK2, Q8IUD3, Q8IUD4, Q8IUD5, Q8NAS1, Q9NXN5, Q9UIK7, Q9UPS1 | Gene names | ERC1, ELKS, KIAA1081, RAB6IP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1). | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 74) | NC score | 0.001972 (rank : 108) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
CROCC_HUMAN
|
||||||
| θ value | 6.88961 (rank : 75) | NC score | 0.056411 (rank : 36) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1409 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q5TZA2, Q2VHY3, Q66GT7, Q7Z2L4, Q7Z5D7 | Gene names | CROCC, KIAA0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
DISC1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 76) | NC score | 0.029330 (rank : 94) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9NRI5, O75045, Q5VT44, Q5VT45, Q8IXJ0, Q8IXJ1, Q9BX19, Q9NRI3, Q9NRI4 | Gene names | DISC1, KIAA0457 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disrupted in schizophrenia 1 protein. | |||||
|
EEA1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 77) | NC score | 0.049076 (rank : 79) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1637 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8BL66, Q6DIC2 | Gene names | Eea1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early endosome antigen 1. | |||||
|
KIF5A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 78) | NC score | 0.028393 (rank : 96) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 859 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P33175, Q5DTP1, Q6PDY7, Q9Z2F9 | Gene names | Kif5a, Kiaa4086, Kif5, Nkhc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin heavy chain isoform 5A (Neuronal kinesin heavy chain) (NKHC) (Kinesin heavy chain neuron-specific 1). | |||||
|
LZTS2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 79) | NC score | 0.031085 (rank : 92) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q91YU6, Q569X6 | Gene names | Lzts2, Kiaa1813 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine zipper putative tumor suppressor 2. | |||||
|
MYH8_MOUSE
|
||||||
| θ value | 6.88961 (rank : 80) | NC score | 0.054926 (rank : 50) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1600 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P13542, Q5SX36 | Gene names | Myh8, Myhsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
CENPF_HUMAN
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | 0.058715 (rank : 31) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
CORO6_MOUSE
|
||||||
| θ value | 8.99809 (rank : 82) | NC score | 0.002557 (rank : 107) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q920M5, Q920M3, Q920M4 | Gene names | Coro6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coronin-6 (Clipin E). | |||||
|
LRP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 83) | NC score | 0.000145 (rank : 109) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 617 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q07954, Q2PP12, Q8IVG8 | Gene names | LRP1, A2MR | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 1 precursor (LRP) (Alpha-2-macroglobulin receptor) (A2MR) (Apolipoprotein E receptor) (APOER) (CD91 antigen). | |||||
|
MYH2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 84) | NC score | 0.055045 (rank : 48) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1491 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9UKX2, Q14322, Q16229 | Gene names | MYH2, MYHSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, skeletal muscle, adult 2 (Myosin heavy chain IIa) (MyHC-IIa). | |||||
|
PALM_MOUSE
|
||||||
| θ value | 8.99809 (rank : 85) | NC score | 0.025867 (rank : 99) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Z0P4, Q9Z0P3 | Gene names | Palm | |||
|
Domain Architecture |

|
|||||
| Description | Paralemmin. | |||||
|
PEPL_HUMAN
|
||||||
| θ value | 8.99809 (rank : 86) | NC score | 0.044207 (rank : 84) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1141 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O60437, O60314, O60454 | Gene names | PPL, KIAA0568 | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin (195 kDa cornified envelope precursor protein) (190 kDa paraneoplastic pemphigus antigen). | |||||
|
TACC1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 87) | NC score | 0.039179 (rank : 88) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q6Y685, Q6Y686 | Gene names | Tacc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transforming acidic coiled-coil-containing protein 1. | |||||
|
CCD13_HUMAN
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.051626 (rank : 67) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 721 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8IYE1 | Gene names | CCDC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 13. | |||||
|
CCD39_HUMAN
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.050507 (rank : 75) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 868 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9UFE4 | Gene names | CCDC39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 39. | |||||
|
CCHCR_MOUSE
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.054288 (rank : 55) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 932 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8K2I2 | Gene names | Cchcr1, Hcr | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil alpha-helical rod protein 1 (Alpha helical coiled-coil rod protein). | |||||
|
CF060_HUMAN
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.055098 (rank : 47) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1270 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8NB25, Q96GY8, Q9H851 | Gene names | C6orf60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf60. | |||||
|
CING_HUMAN
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.053730 (rank : 60) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9P2M7, Q5T386, Q9NR25 | Gene names | CGN, KIAA1319 | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
CP250_MOUSE
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.055042 (rank : 49) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
CYLN2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.056870 (rank : 34) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1004 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9UDT6, O14527, O43611 | Gene names | CYLN2, KIAA0291, WBSCR4, WSCR4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic linker protein 2 (Cytoplasmic linker protein 115) (CLIP- 115) (Williams-Beuren syndrome chromosome region 4 protein). | |||||
|
GCC2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.053985 (rank : 57) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
GOGA1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.051112 (rank : 69) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 994 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9CW79, Q80YB0 | Gene names | Golga1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
GOGA6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.057558 (rank : 32) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9NYA3, Q9NYA7 | Gene names | GOLGA6, GLP | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 6 (Golgin linked to PML) (Golgin-like protein). | |||||
|
GPR98_MOUSE
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.050700 (rank : 73) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VHN7, Q6ZQ69, Q810D2, Q810D3, Q91ZS0, Q91ZS1 | Gene names | Gpr98, Kiaa0686, Mass1, Vlgr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor 98 precursor (Monogenic audiogenic seizure susceptibility protein 1) (Very large G-protein coupled receptor 1) (Neurepin). | |||||
|
MY18A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.052567 (rank : 63) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
MY18B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.050753 (rank : 71) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1299 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8IUG5, Q8NDI8, Q8TE65, Q8WWS0, Q96KH2, Q96KR8, Q96KR9 | Gene names | MYO18B | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18B (Myosin XVIIIb). | |||||
|
MYH13_HUMAN
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.053379 (rank : 62) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1504 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9UKX3, O95252 | Gene names | MYH13 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-13 (Myosin heavy chain, skeletal muscle, extraocular) (MyHC- eo). | |||||
|
MYH14_MOUSE
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.058719 (rank : 30) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1225 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q6URW6, Q80V64, Q80ZE6 | Gene names | Myh14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
MYH3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.055815 (rank : 39) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1660 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P11055, Q15492 | Gene names | MYH3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, fast skeletal muscle, embryonic (Muscle embryonic myosin heavy chain) (SMHCE). | |||||
|
MYH6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.055110 (rank : 46) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1426 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P13533, Q13943, Q14906, Q14907 | Gene names | MYH6, MYHCA | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
MYH6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.055343 (rank : 44) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1501 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q02566, Q64258, Q64738 | Gene names | Myh6, Myhca | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
MYH7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.056426 (rank : 35) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P12883, Q14836, Q14837, Q14904, Q16579, Q92679, Q9H1D5, Q9UMM8 | Gene names | MYH7, MYHCB | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle beta isoform (MyHC-beta). | |||||
|
MYH8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.055327 (rank : 45) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1610 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P13535, Q14910 | Gene names | MYH8 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
NIN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.050350 (rank : 76) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1416 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8N4C6, Q6P0P6, Q9BWU6, Q9C012, Q9C013, Q9C014, Q9H5I6, Q9HAT7, Q9HBY5, Q9HCK7, Q9UH61 | Gene names | NIN, KIAA1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein (hNinein) (Glycogen synthase kinase 3 beta-interacting protein) (GSK3B-interacting protein). | |||||
|
REST_MOUSE
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.053861 (rank : 58) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1506 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q922J3, Q571L7 | Gene names | Rsn, Kiaa4046 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin. | |||||
|
RFIP4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.051301 (rank : 68) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q86YS3, Q8N829, Q8NDT7, Q969D8 | Gene names | RAB11FIP4, KIAA1821 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 4 (Rab11-FIP4). | |||||
|
NPTXR_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q99J85 | Gene names | Nptxr, Npr | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal pentraxin receptor. | |||||
|
NPTXR_HUMAN
|
||||||
| NC score | 0.988040 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O95502 | Gene names | NPTXR | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal pentraxin receptor. | |||||
|
NPTX2_HUMAN
|
||||||
| NC score | 0.961820 (rank : 3) | θ value | 2.9938e-112 (rank : 3) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P47972, Q86XV7, Q96G70 | Gene names | NPTX2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal pentraxin-2 precursor (NP2) (Neuronal pentraxin II) (NP-II). | |||||
|
NPTX2_MOUSE
|
||||||
| NC score | 0.961624 (rank : 4) | θ value | 2.9938e-112 (rank : 4) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O70340 | Gene names | Nptx2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal pentraxin-2 precursor (NP2) (Neuronal pentraxin II) (NP-II). | |||||
|
NPTX1_HUMAN
|
||||||
| NC score | 0.959256 (rank : 5) | θ value | 5.85632e-100 (rank : 5) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q15818, Q5FWE6 | Gene names | NPTX1 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal pentraxin-1 precursor (NP1) (Neuronal pentraxin I) (NP-I). | |||||
|
NPTX1_MOUSE
|
||||||
| NC score | 0.957539 (rank : 6) | θ value | 9.98939e-100 (rank : 6) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q62443 | Gene names | Nptx1 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal pentraxin-1 precursor (NP1) (Neuronal pentraxin I) (NP-I). | |||||
|
PTX3_MOUSE
|
||||||
| NC score | 0.747317 (rank : 7) | θ value | 1.058e-16 (rank : 9) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P48759, Q8VDF1 | Gene names | Ptx3, Tsg14 | |||
|
Domain Architecture |

|
|||||
| Description | Pentraxin-related protein PTX3 precursor (Pentaxin-related protein PTX3) (Tumor necrosis factor-inducible protein TSG-14). | |||||
|
PTX3_HUMAN
|
||||||
| NC score | 0.745083 (rank : 8) | θ value | 3.28887e-18 (rank : 8) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P26022 | Gene names | PTX3, TSG14 | |||
|
Domain Architecture |

|
|||||
| Description | Pentraxin-related protein PTX3 precursor (Pentaxin-related protein PTX3) (Tumor necrosis factor-inducible protein TSG-14). | |||||
|
CRP_MOUSE
|
||||||
| NC score | 0.702387 (rank : 9) | θ value | 2.88119e-14 (rank : 12) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P14847 | Gene names | Crp, Ptx1 | |||
|
Domain Architecture |

|
|||||
| Description | C-reactive protein precursor. | |||||
|
CRP_HUMAN
|
||||||
| NC score | 0.699622 (rank : 10) | θ value | 2.88119e-14 (rank : 11) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P02741, Q8WW75 | Gene names | CRP, PTX1 | |||
|
Domain Architecture |

|
|||||
| Description | C-reactive protein precursor [Contains: C-reactive protein(1-205)]. | |||||
|
SAMP_HUMAN
|
||||||
| NC score | 0.690092 (rank : 11) | θ value | 1.68911e-14 (rank : 10) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P02743 | Gene names | APCS, PTX2 | |||
|
Domain Architecture |

|
|||||
| Description | Serum amyloid P-component precursor (SAP) (9.5S alpha-1-glycoprotein) [Contains: Serum amyloid P-component(1-203)]. | |||||
|
SAMP_MOUSE
|
||||||
| NC score | 0.687839 (rank : 12) | θ value | 8.38298e-14 (rank : 13) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P12246 | Gene names | Apcs, Ptx2, Sap | |||
|
Domain Architecture |

|
|||||
| Description | Serum amyloid P-component precursor (SAP). | |||||
|
GP144_HUMAN
|
||||||
| NC score | 0.419557 (rank : 13) | θ value | 3.28125e-26 (rank : 7) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q7Z7M1, Q86SL4, Q8NH12 | Gene names | GPR144, PGR24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 144 (G-protein coupled receptor PGR24). | |||||
|
GP112_HUMAN
|
||||||
| NC score | 0.114879 (rank : 14) | θ value | 0.00298849 (rank : 14) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8IZF6, Q86SM6 | Gene names | GPR112 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 112. | |||||
|
GP126_HUMAN
|
||||||
| NC score | 0.082187 (rank : 15) | θ value | 0.00665767 (rank : 15) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q86SQ4, Q6MZU7, Q8NC14, Q96JW0 | Gene names | GPR126 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 126 precursor. | |||||
|
GP133_HUMAN
|
||||||
| NC score | 0.079078 (rank : 16) | θ value | 0.279714 (rank : 27) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6QNK2, Q6ZMQ1, Q7Z7M2, Q86SM4 | Gene names | GPR133, PGR25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 133 precursor (G-protein coupled receptor PGR25). | |||||
|
CCHCR_HUMAN
|
||||||
| NC score | 0.075013 (rank : 17) | θ value | 0.0961366 (rank : 21) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 717 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8TD31, Q9NRK8, Q9NWY9, Q9NXJ4, Q9NXK3, Q9Y6W1, Q9Y6W2 | Gene names | CCHCR1, C6orf18, HCR | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil alpha-helical rod protein 1 (Alpha helical coiled-coil rod protein) (Putative gene 8 protein) (Pg8). | |||||
|
HOOK2_MOUSE
|
||||||
| NC score | 0.073640 (rank : 18) | θ value | 0.163984 (rank : 23) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 736 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q7TMK6, Q8BY47, Q8R347, Q8VCN4, Q99LU2 | Gene names | Hook2 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 2. | |||||
|
HOOK2_HUMAN
|
||||||
| NC score | 0.068591 (rank : 19) | θ value | 0.0563607 (rank : 20) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 714 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q96ED9, O60562 | Gene names | HOOK2 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 2 (h-hook2) (hHK2). | |||||
|
MYH14_HUMAN
|
||||||
| NC score | 0.067081 (rank : 20) | θ value | 0.00665767 (rank : 16) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 995 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q7Z406, Q5CZ75, Q6XYE4, Q76B62, Q8WV23, Q96I22, Q9BT27, Q9BW35, Q9H882 | Gene names | MYH14, KIAA2034 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
MYH9_MOUSE
|
||||||
| NC score | 0.064403 (rank : 21) | θ value | 4.03905 (rank : 60) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
MYH10_HUMAN
|
||||||
| NC score | 0.063993 (rank : 22) | θ value | 0.0193708 (rank : 17) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P35580, Q16087 | Gene names | MYH10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
MYH9_HUMAN
|
||||||
| NC score | 0.063287 (rank : 23) | θ value | 4.03905 (rank : 59) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
MYH10_MOUSE
|
||||||
| NC score | 0.062860 (rank : 24) | θ value | 0.0330416 (rank : 18) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1612 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q61879, Q5SV63 | Gene names | Myh10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
PCNT_HUMAN
|
||||||
| NC score | 0.062842 (rank : 25) | θ value | 3.0926 (rank : 52) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
MYH11_HUMAN
|
||||||
| NC score | 0.062502 (rank : 26) | θ value | 0.365318 (rank : 31) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
CYLN2_MOUSE
|
||||||
| NC score | 0.061264 (rank : 27) | θ value | 0.365318 (rank : 29) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1002 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9Z0H8, Q7TSI9, Q8CHU1, Q9EP81 | Gene names | Cyln2, Kiaa0291 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic linker protein 2 (Cytoplasmic linker protein 115) (CLIP- 115). | |||||
|
MYH11_MOUSE
|
||||||
| NC score | 0.060645 (rank : 28) | θ value | 0.62314 (rank : 33) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
CNTRB_HUMAN
|
||||||
| NC score | 0.059897 (rank : 29) | θ value | 0.47712 (rank : 32) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8N137, Q331K3, Q69YV7, Q8NCB8, Q8WXV3, Q96CQ7, Q9C060 | Gene names | CNTROB, LIP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrobin (LYST-interacting protein 8). | |||||
|
MYH14_MOUSE
|
||||||
| NC score | 0.058719 (rank : 30) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1225 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q6URW6, Q80V64, Q80ZE6 | Gene names | Myh14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
CENPF_HUMAN
|
||||||
| NC score | 0.058715 (rank : 31) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
GOGA6_HUMAN
|
||||||
| NC score | 0.057558 (rank : 32) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9NYA3, Q9NYA7 | Gene names | GOLGA6, GLP | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 6 (Golgin linked to PML) (Golgin-like protein). | |||||
|
CING_MOUSE
|
||||||
| NC score | 0.057201 (rank : 33) | θ value | 3.0926 (rank : 48) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1432 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P59242 | Gene names | Cgn | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
CYLN2_HUMAN
|
||||||
| NC score | 0.056870 (rank : 34) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1004 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9UDT6, O14527, O43611 | Gene names | CYLN2, KIAA0291, WBSCR4, WSCR4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic linker protein 2 (Cytoplasmic linker protein 115) (CLIP- 115) (Williams-Beuren syndrome chromosome region 4 protein). | |||||
|
MYH7_HUMAN
|
||||||
| NC score | 0.056426 (rank : 35) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P12883, Q14836, Q14837, Q14904, Q16579, Q92679, Q9H1D5, Q9UMM8 | Gene names | MYH7, MYHCB | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle beta isoform (MyHC-beta). | |||||
|
CROCC_HUMAN
|
||||||
| NC score | 0.056411 (rank : 36) | θ value | 6.88961 (rank : 75) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1409 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q5TZA2, Q2VHY3, Q66GT7, Q7Z2L4, Q7Z5D7 | Gene names | CROCC, KIAA0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
AMOL2_HUMAN
|
||||||
| NC score | 0.056385 (rank : 37) | θ value | 0.279714 (rank : 26) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 603 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9Y2J4, Q53EP1, Q96F99, Q9UKB4 | Gene names | AMOTL2, KIAA0989 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiomotin-like protein 2 (Leman coiled-coil protein) (LCCP). | |||||
|
MYH4_HUMAN
|
||||||
| NC score | 0.056054 (rank : 38) | θ value | 5.27518 (rank : 71) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1377 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9Y623 | Gene names | MYH4 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal) (Myosin heavy chain IIb) (MyHC-IIb). | |||||
|
MYH3_HUMAN
|
||||||
| NC score | 0.055815 (rank : 39) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1660 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P11055, Q15492 | Gene names | MYH3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, fast skeletal muscle, embryonic (Muscle embryonic myosin heavy chain) (SMHCE). | |||||
|
MYH1_HUMAN
|
||||||
| NC score | 0.055696 (rank : 40) | θ value | 5.27518 (rank : 70) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1478 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P12882, Q9Y622 | Gene names | MYH1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1) (Myosin heavy chain IIx/d) (MyHC-IIx/d). | |||||
|
MYH1_MOUSE
|
||||||
| NC score | 0.055551 (rank : 41) | θ value | 3.0926 (rank : 51) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1461 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q5SX40 | Gene names | Myh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1). | |||||
|
NUMA1_HUMAN
|
||||||
| NC score | 0.055395 (rank : 42) | θ value | 0.163984 (rank : 24) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
MYH4_MOUSE
|
||||||
| NC score | 0.055366 (rank : 43) | θ value | 4.03905 (rank : 58) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
MYH6_MOUSE
|
||||||
| NC score | 0.055343 (rank : 44) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1501 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q02566, Q64258, Q64738 | Gene names | Myh6, Myhca | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
MYH8_HUMAN
|
||||||
| NC score | 0.055327 (rank : 45) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1610 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P13535, Q14910 | Gene names | MYH8 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
MYH6_HUMAN
|
||||||
| NC score | 0.055110 (rank : 46) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1426 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P13533, Q13943, Q14906, Q14907 | Gene names | MYH6, MYHCA | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
CF060_HUMAN
|
||||||
| NC score | 0.055098 (rank : 47) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1270 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8NB25, Q96GY8, Q9H851 | Gene names | C6orf60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf60. | |||||
|
MYH2_HUMAN
|
||||||
| NC score | 0.055045 (rank : 48) | θ value | 8.99809 (rank : 84) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1491 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9UKX2, Q14322, Q16229 | Gene names | MYH2, MYHSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, skeletal muscle, adult 2 (Myosin heavy chain IIa) (MyHC-IIa). | |||||
|
CP250_MOUSE
|
||||||
| NC score | 0.055042 (rank : 49) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
MYH8_MOUSE
|
||||||
| NC score | 0.054926 (rank : 50) | θ value | 6.88961 (rank : 80) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1600 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P13542, Q5SX36 | Gene names | Myh8, Myhsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
CROCC_MOUSE
|
||||||
| NC score | 0.054903 (rank : 51) | θ value | 1.06291 (rank : 35) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1460 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q8CJ40, Q7TQL2, Q80U01, Q8R0B9 | Gene names | Crocc, Kiaa0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
AKAP9_HUMAN
|
||||||
| NC score | 0.054669 (rank : 52) | θ value | 0.21417 (rank : 25) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
CKAP4_HUMAN
|
||||||
| NC score | 0.054558 (rank : 53) | θ value | 0.813845 (rank : 34) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q07065, Q504S5, Q53ES6 | Gene names | CKAP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-associated protein 4 (63 kDa membrane protein) (p63). | |||||
|
MYO5C_HUMAN
|
||||||
| NC score | 0.054351 (rank : 54) | θ value | 2.36792 (rank : 46) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1206 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9NQX4 | Gene names | MYO5C | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5C (Myosin Vc). | |||||
|
CCHCR_MOUSE
|
||||||
| NC score | 0.054288 (rank : 55) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 932 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8K2I2 | Gene names | Cchcr1, Hcr | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil alpha-helical rod protein 1 (Alpha helical coiled-coil rod protein). | |||||
|
REST_HUMAN
|
||||||
| NC score | 0.054145 (rank : 56) | θ value | 0.125558 (rank : 22) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1605 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P30622 | Gene names | RSN, CYLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Restin (Cytoplasmic linker protein 170 alpha-2) (CLIP-170) (Reed- Sternberg intermediate filament-associated protein) (Cytoplasmic linker protein 1). | |||||
|
GCC2_MOUSE
|
||||||
| NC score | 0.053985 (rank : 57) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
REST_MOUSE
|
||||||
| NC score | 0.053861 (rank : 58) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1506 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q922J3, Q571L7 | Gene names | Rsn, Kiaa4046 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin. | |||||
|
RUFY1_MOUSE
|
||||||
| NC score | 0.053826 (rank : 59) | θ value | 0.0431538 (rank : 19) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8BIJ7, Q8BKQ4, Q8BL21, Q9EPM6 | Gene names | Rufy1, Rabip4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 1 (Rab4-interacting protein). | |||||
|
CING_HUMAN
|
||||||
| NC score | 0.053730 (rank : 60) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9P2M7, Q5T386, Q9NR25 | Gene names | CGN, KIAA1319 | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
MY18A_MOUSE
|
||||||
| NC score | 0.053630 (rank : 61) | θ value | 3.0926 (rank : 50) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
MYH13_HUMAN
|
||||||
| NC score | 0.053379 (rank : 62) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1504 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9UKX3, O95252 | Gene names | MYH13 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-13 (Myosin heavy chain, skeletal muscle, extraocular) (MyHC- eo). | |||||
|
MY18A_HUMAN
|
||||||
| NC score | 0.052567 (rank : 63) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
RB6I2_HUMAN
|
||||||
| NC score | 0.052462 (rank : 64) | θ value | 5.27518 (rank : 73) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1584 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q8IUD2, Q6NVK2, Q8IUD3, Q8IUD4, Q8IUD5, Q8NAS1, Q9NXN5, Q9UIK7, Q9UPS1 | Gene names | ERC1, ELKS, KIAA1081, RAB6IP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1). | |||||
|
GOGA1_HUMAN
|
||||||
| NC score | 0.052170 (rank : 65) | θ value | 1.38821 (rank : 37) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1222 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q92805, Q8IYZ9 | Gene names | GOLGA1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
CP135_MOUSE
|
||||||
| NC score | 0.052150 (rank : 66) | θ value | 5.27518 (rank : 66) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1278 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q6P5D4, Q6A030 | Gene names | Cep135, Cep4, Kiaa0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
CCD13_HUMAN
|
||||||
| NC score | 0.051626 (rank : 67) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 721 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8IYE1 | Gene names | CCDC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 13. | |||||
|
RFIP4_HUMAN
|
||||||
| NC score | 0.051301 (rank : 68) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q86YS3, Q8N829, Q8NDT7, Q969D8 | Gene names | RAB11FIP4, KIAA1821 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 4 (Rab11-FIP4). | |||||
|
GOGA1_MOUSE
|
||||||
| NC score | 0.051112 (rank : 69) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 994 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9CW79, Q80YB0 | Gene names | Golga1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
AMOL2_MOUSE
|
||||||
| NC score | 0.050921 (rank : 70) | θ value | 3.0926 (rank : 47) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8K371, Q3TPM1, Q7TPE4, Q8BP84, Q8BS08, Q9QUS0 | Gene names | Amotl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiomotin-like protein 2. | |||||
|
MY18B_HUMAN
|
||||||
| NC score | 0.050753 (rank : 71) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1299 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8IUG5, Q8NDI8, Q8TE65, Q8WWS0, Q96KH2, Q96KR8, Q96KR9 | Gene names | MYO18B | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18B (Myosin XVIIIb). | |||||
|
EEA1_HUMAN
|
||||||
| NC score | 0.050736 (rank : 72) | θ value | 5.27518 (rank : 67) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
GPR98_MOUSE
|
||||||
| NC score | 0.050700 (rank : 73) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VHN7, Q6ZQ69, Q810D2, Q810D3, Q91ZS0, Q91ZS1 | Gene names | Gpr98, Kiaa0686, Mass1, Vlgr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor 98 precursor (Monogenic audiogenic seizure susceptibility protein 1) (Very large G-protein coupled receptor 1) (Neurepin). | |||||
|
FLIP1_MOUSE
|
||||||
| NC score | 0.050577 (rank : 74) | θ value | 3.0926 (rank : 49) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1176 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9CS72 | Gene names | Filip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
CCD39_HUMAN
|
||||||
| NC score | 0.050507 (rank : 75) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 868 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9UFE4 | Gene names | CCDC39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 39. | |||||
|
NIN_HUMAN
|
||||||
| NC score | 0.050350 (rank : 76) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1416 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8N4C6, Q6P0P6, Q9BWU6, Q9C012, Q9C013, Q9C014, Q9H5I6, Q9HAT7, Q9HBY5, Q9HCK7, Q9UH61 | Gene names | NIN, KIAA1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein (hNinein) (Glycogen synthase kinase 3 beta-interacting protein) (GSK3B-interacting protein). | |||||
|
RB6I2_MOUSE
|
||||||
| NC score | 0.050243 (rank : 77) | θ value | 4.03905 (rank : 62) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q99MI1, Q80TK7, Q8BPL1, Q8C7Y1, Q99MI2 | Gene names | Erc1, Cast2, Kiaa1081, Rab6ip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1) (ERC1) (CAZ-associated structural protein 2) (CAST2). | |||||
|
RUFY1_HUMAN
|
||||||
| NC score | 0.049991 (rank : 78) | θ value | 3.0926 (rank : 53) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 848 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q96T51, Q59FF3, Q71S93, Q9H6I3 | Gene names | RUFY1, RABIP4, ZFYVE12 | |||
|
Domain Architecture |

|
|||||
| Description | RUN and FYVE domain-containing protein 1 (FYVE-finger protein EIP1) (Zinc finger FYVE domain-containing protein 12) (La-binding protein 1) (Rab4-interacting protein). | |||||
|
EEA1_MOUSE
|
||||||
| NC score | 0.049076 (rank : 79) | θ value | 6.88961 (rank : 77) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1637 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8BL66, Q6DIC2 | Gene names | Eea1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early endosome antigen 1. | |||||
|
DMD_MOUSE
|
||||||
| NC score | 0.048991 (rank : 80) | θ value | 2.36792 (rank : 42) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1111 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P11531, O35653, Q60703 | Gene names | Dmd | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
FYCO1_HUMAN
|
||||||
| NC score | 0.047232 (rank : 81) | θ value | 5.27518 (rank : 68) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1149 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9BQS8, Q3MJE6, Q86T41, Q86TB1, Q8TEF9, Q96IV5, Q9H8P9 | Gene names | FYCO1, ZFYVE7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1 (Zinc finger FYVE domain-containing protein 7). | |||||
|
HIP1R_HUMAN
|
||||||
| NC score | 0.046814 (rank : 82) | θ value | 1.38821 (rank : 38) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 809 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O75146, Q9UED9 | Gene names | HIP1R, HIP12, KIAA0655 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1-related protein (Hip1-related) (Hip 12). | |||||
|
DMD_HUMAN
|
||||||
| NC score | 0.046340 (rank : 83) | θ value | 2.36792 (rank : 41) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P11532, Q02295, Q14169, Q14170, Q5JYU0, Q7KZ48 | Gene names | DMD | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
PEPL_HUMAN
|
||||||
| NC score | 0.044207 (rank : 84) | θ value | 8.99809 (rank : 86) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1141 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O60437, O60314, O60454 | Gene names | PPL, KIAA0568 | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin (195 kDa cornified envelope precursor protein) (190 kDa paraneoplastic pemphigus antigen). | |||||
|
EVPL_HUMAN
|
||||||
| NC score | 0.044147 (rank : 85) | θ value | 4.03905 (rank : 56) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 895 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q92817 | Gene names | EVPL | |||
|
Domain Architecture |

|
|||||
| Description | Envoplakin (210 kDa paraneoplastic pemphigus antigen) (p210) (210 kDa cornified envelope precursor protein). | |||||
|
LIPA2_MOUSE
|
||||||
| NC score | 0.043085 (rank : 86) | θ value | 5.27518 (rank : 69) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1282 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8BSS9, Q8BN73 | Gene names | Ppfia2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-2) (PTPRF-interacting protein alpha-2). | |||||
|
SPTN5_HUMAN
|
||||||
| NC score | 0.041534 (rank : 87) | θ value | 0.279714 (rank : 28) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9NRC6 | Gene names | SPTBN5, SPTBN4 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 4 (Spectrin, non-erythroid beta chain 4) (Beta-V spectrin) (BSPECV). | |||||
|
TACC1_MOUSE
|
||||||
| NC score | 0.039179 (rank : 88) | θ value | 8.99809 (rank : 87) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q6Y685, Q6Y686 | Gene names | Tacc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transforming acidic coiled-coil-containing protein 1. | |||||
|
SURF6_MOUSE
|
||||||
| NC score | 0.037851 (rank : 89) | θ value | 1.81305 (rank : 40) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P70279, Q8BPI4, Q8BRL7, Q8R0Q7 | Gene names | Surf6, Surf-6 | |||
|
Domain Architecture |

|
|||||
| Description | Surfeit locus protein 6. | |||||
|
PALM_HUMAN
|
||||||
| NC score | 0.035054 (rank : 90) | θ value | 4.03905 (rank : 61) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O75781, O43359, O95673, Q92559, Q9UPJ4, Q9UQS2, Q9UQS3 | Gene names | PALM, KIAA0270 | |||
|
Domain Architecture |

|
|||||
| Description | Paralemmin. | |||||
|
ZFY19_HUMAN
|
||||||
| NC score | 0.032714 (rank : 91) | θ value | 1.06291 (rank : 36) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96K21, Q86WC2, Q8WU96 | Gene names | ZFYVE19, MPFYVE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger FYVE domain-containing protein 19 (MLL partner containing FYVE domain). | |||||
|
LZTS2_MOUSE
|
||||||
| NC score | 0.031085 (rank : 92) | θ value | 6.88961 (rank : 79) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q91YU6, Q569X6 | Gene names | Lzts2, Kiaa1813 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine zipper putative tumor suppressor 2. | |||||
|
NEB1_HUMAN
|
||||||
| NC score | 0.030803 (rank : 93) | θ value | 5.27518 (rank : 72) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 829 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9ULJ8, O76059, Q9NXT2 | Gene names | PPP1R9A, KIAA1222 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-1 (Neurabin-I) (Neural tissue-specific F-actin-binding protein I) (Protein phosphatase 1 regulatory subunit 9A). | |||||
|
DISC1_HUMAN
|
||||||
| NC score | 0.029330 (rank : 94) | θ value | 6.88961 (rank : 76) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9NRI5, O75045, Q5VT44, Q5VT45, Q8IXJ0, Q8IXJ1, Q9BX19, Q9NRI3, Q9NRI4 | Gene names | DISC1, KIAA0457 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disrupted in schizophrenia 1 protein. | |||||
|
KIF5A_HUMAN
|
||||||
| NC score | 0.028798 (rank : 95) | θ value | 4.03905 (rank : 57) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 843 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q12840, Q4LE26 | Gene names | KIF5A, NKHC1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain isoform 5A (Neuronal kinesin heavy chain) (NKHC) (Kinesin heavy chain neuron-specific 1). | |||||
|
KIF5A_MOUSE
|
||||||
| NC score | 0.028393 (rank : 96) | θ value | 6.88961 (rank : 78) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 859 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P33175, Q5DTP1, Q6PDY7, Q9Z2F9 | Gene names | Kif5a, Kiaa4086, Kif5, Nkhc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin heavy chain isoform 5A (Neuronal kinesin heavy chain) (NKHC) (Kinesin heavy chain neuron-specific 1). | |||||
|
MRCKB_HUMAN
|
||||||
| NC score | 0.027140 (rank : 97) | θ value | 2.36792 (rank : 44) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 2239 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9Y5S2, Q2L7A5, Q86TJ1, Q9ULU5 | Gene names | CDC42BPB, KIAA1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
MRCKB_MOUSE
|
||||||
| NC score | 0.026084 (rank : 98) | θ value | 2.36792 (rank : 45) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 2180 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q7TT50, Q69ZR5, Q80W33 | Gene names | Cdc42bpb, Kiaa1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
PALM_MOUSE
|
||||||
| NC score | 0.025867 (rank : 99) | θ value | 8.99809 (rank : 85) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Z0P4, Q9Z0P3 | Gene names | Palm | |||
|
Domain Architecture |

|
|||||
| Description | Paralemmin. | |||||
|
A16L1_MOUSE
|
||||||
| NC score | 0.024379 (rank : 100) | θ value | 4.03905 (rank : 55) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 776 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8C0J2, Q6KAT7, Q80U97, Q80U98, Q80U99, Q80Y53, Q9DB63 | Gene names | Atg16l1, Apg16l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Autophagy-related protein 16-1 (APG16-like 1). | |||||
|
MRCKG_MOUSE
|
||||||
| NC score | 0.020376 (rank : 101) | θ value | 0.365318 (rank : 30) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1466 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q80UW5 | Gene names | Cdc42bpg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK gamma (EC 2.7.11.1) (CDC42- binding protein kinase gamma) (Myotonic dystrophy kinase-related CDC42-binding kinase gamma) (Myotonic dystrophy protein kinase-like alpha) (MRCK gamma) (DMPK-like gamma). | |||||
|
K1543_HUMAN
|
||||||
| NC score | 0.018118 (rank : 102) | θ value | 1.81305 (rank : 39) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9P1Y5, Q8NDF1 | Gene names | KIAA1543 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1543. | |||||
|
SCG2_MOUSE
|
||||||
| NC score | 0.017640 (rank : 103) | θ value | 4.03905 (rank : 65) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q03517, Q80Y79, Q9CW80 | Gene names | Scg2, Chgc, Scg-2 | |||
|
Domain Architecture |

|
|||||
| Description | Secretogranin-2 precursor (Secretogranin II) (SgII) (Chromogranin C) [Contains: Secretoneurin (SN)]. | |||||
|
RFWD2_MOUSE
|
||||||
| NC score | 0.017379 (rank : 104) | θ value | 4.03905 (rank : 64) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 535 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9R1A8 | Gene names | Rfwd2, Cop1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger and WD repeat domain protein 2 (EC 6.3.2.-) (Ubiquitin- protein ligase COP1) (Constitutive photomorphogenesis protein 1 homolog) (mCOP1). | |||||
|
RFWD2_HUMAN
|
||||||
| NC score | 0.017349 (rank : 105) | θ value | 4.03905 (rank : 63) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8NHY2, Q6H103, Q9H6L7 | Gene names | RFWD2, COP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger and WD repeat domain protein 2 (EC 6.3.2.-) (Ubiquitin- protein ligase COP1) (Constitutive photomorphogenesis protein 1 homolog) (hCOP1). | |||||
|
KGP1A_HUMAN
|
||||||
| NC score | 0.004904 (rank : 106) | θ value | 2.36792 (rank : 43) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 901 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q13976 | Gene names | PRKG1, PRKGR1A | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 1, alpha isozyme (EC 2.7.11.12) (CGK 1 alpha) (cGKI-alpha). | |||||
|
CORO6_MOUSE
|
||||||
| NC score | 0.002557 (rank : 107) | θ value | 8.99809 (rank : 82) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q920M5, Q920M3, Q920M4 | Gene names | Coro6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coronin-6 (Clipin E). | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.001972 (rank : 108) | θ value | 5.27518 (rank : 74) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
LRP1_HUMAN
|
||||||
| NC score | 0.000145 (rank : 109) | θ value | 8.99809 (rank : 83) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 617 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q07954, Q2PP12, Q8IVG8 | Gene names | LRP1, A2MR | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 1 precursor (LRP) (Alpha-2-macroglobulin receptor) (A2MR) (Apolipoprotein E receptor) (APOER) (CD91 antigen). | |||||
|
ZN197_HUMAN
|
||||||
| NC score | -0.002306 (rank : 110) | θ value | 3.0926 (rank : 54) | |||
| Query Neighborhood Hits | 87 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O14709 | Gene names | ZNF197, ZNF166 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 197 (ZnF20). | |||||