Please be patient as the page loads
|
NPTXR_HUMAN
|
||||||
| SwissProt Accessions | O95502 | Gene names | NPTXR | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal pentraxin receptor. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
NPTXR_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O95502 | Gene names | NPTXR | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal pentraxin receptor. | |||||
|
NPTXR_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.988040 (rank : 2) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q99J85 | Gene names | Nptxr, Npr | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal pentraxin receptor. | |||||
|
NPTX2_MOUSE
|
||||||
| θ value | 1.48581e-111 (rank : 3) | NC score | 0.968619 (rank : 3) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O70340 | Gene names | Nptx2 | |||
|
Domain Architecture |
|
|||||
| Description | Neuronal pentraxin-2 precursor (NP2) (Neuronal pentraxin II) (NP-II). | |||||
|
NPTX2_HUMAN
|
||||||
| θ value | 7.37397e-111 (rank : 4) | NC score | 0.967804 (rank : 4) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P47972, Q86XV7, Q96G70 | Gene names | NPTX2 | |||
|
Domain Architecture |
|
|||||
| Description | Neuronal pentraxin-2 precursor (NP2) (Neuronal pentraxin II) (NP-II). | |||||
|
NPTX1_MOUSE
|
||||||
| θ value | 9.98939e-100 (rank : 5) | NC score | 0.965300 (rank : 6) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q62443 | Gene names | Nptx1 | |||
|
Domain Architecture |
|
|||||
| Description | Neuronal pentraxin-1 precursor (NP1) (Neuronal pentraxin I) (NP-I). | |||||
|
NPTX1_HUMAN
|
||||||
| θ value | 3.79596e-99 (rank : 6) | NC score | 0.967023 (rank : 5) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q15818, Q5FWE6 | Gene names | NPTX1 | |||
|
Domain Architecture |
|
|||||
| Description | Neuronal pentraxin-1 precursor (NP1) (Neuronal pentraxin I) (NP-I). | |||||
|
GP144_HUMAN
|
||||||
| θ value | 3.62785e-25 (rank : 7) | NC score | 0.423875 (rank : 13) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q7Z7M1, Q86SL4, Q8NH12 | Gene names | GPR144, PGR24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 144 (G-protein coupled receptor PGR24). | |||||
|
PTX3_MOUSE
|
||||||
| θ value | 5.60996e-18 (rank : 8) | NC score | 0.755530 (rank : 7) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P48759, Q8VDF1 | Gene names | Ptx3, Tsg14 | |||
|
Domain Architecture |
|
|||||
| Description | Pentraxin-related protein PTX3 precursor (Pentaxin-related protein PTX3) (Tumor necrosis factor-inducible protein TSG-14). | |||||
|
PTX3_HUMAN
|
||||||
| θ value | 2.7842e-17 (rank : 9) | NC score | 0.752369 (rank : 8) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P26022 | Gene names | PTX3, TSG14 | |||
|
Domain Architecture |
|
|||||
| Description | Pentraxin-related protein PTX3 precursor (Pentaxin-related protein PTX3) (Tumor necrosis factor-inducible protein TSG-14). | |||||
|
CRP_HUMAN
|
||||||
| θ value | 2.20605e-14 (rank : 10) | NC score | 0.702863 (rank : 10) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P02741, Q8WW75 | Gene names | CRP, PTX1 | |||
|
Domain Architecture |
|
|||||
| Description | C-reactive protein precursor [Contains: C-reactive protein(1-205)]. | |||||
|
CRP_MOUSE
|
||||||
| θ value | 2.20605e-14 (rank : 11) | NC score | 0.705991 (rank : 9) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P14847 | Gene names | Crp, Ptx1 | |||
|
Domain Architecture |
|
|||||
| Description | C-reactive protein precursor. | |||||
|
SAMP_HUMAN
|
||||||
| θ value | 4.16044e-13 (rank : 12) | NC score | 0.691676 (rank : 11) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P02743 | Gene names | APCS, PTX2 | |||
|
Domain Architecture |
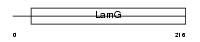
|
|||||
| Description | Serum amyloid P-component precursor (SAP) (9.5S alpha-1-glycoprotein) [Contains: Serum amyloid P-component(1-203)]. | |||||
|
SAMP_MOUSE
|
||||||
| θ value | 9.26847e-13 (rank : 13) | NC score | 0.689809 (rank : 12) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P12246 | Gene names | Apcs, Ptx2, Sap | |||
|
Domain Architecture |
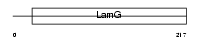
|
|||||
| Description | Serum amyloid P-component precursor (SAP). | |||||
|
GP112_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 14) | NC score | 0.119261 (rank : 14) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8IZF6, Q86SM6 | Gene names | GPR112 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 112. | |||||
|
GP126_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 15) | NC score | 0.084411 (rank : 15) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q86SQ4, Q6MZU7, Q8NC14, Q96JW0 | Gene names | GPR126 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 126 precursor. | |||||
|
GP133_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 16) | NC score | 0.081831 (rank : 16) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6QNK2, Q6ZMQ1, Q7Z7M2, Q86SM4 | Gene names | GPR133, PGR25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 133 precursor (G-protein coupled receptor PGR25). | |||||
|
NIN_HUMAN
|
||||||
| θ value | 0.125558 (rank : 17) | NC score | 0.026299 (rank : 23) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1416 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8N4C6, Q6P0P6, Q9BWU6, Q9C012, Q9C013, Q9C014, Q9H5I6, Q9HAT7, Q9HBY5, Q9HCK7, Q9UH61 | Gene names | NIN, KIAA1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein (hNinein) (Glycogen synthase kinase 3 beta-interacting protein) (GSK3B-interacting protein). | |||||
|
CCHCR_HUMAN
|
||||||
| θ value | 0.21417 (rank : 18) | NC score | 0.042136 (rank : 18) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 717 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8TD31, Q9NRK8, Q9NWY9, Q9NXJ4, Q9NXK3, Q9Y6W1, Q9Y6W2 | Gene names | CCHCR1, C6orf18, HCR | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil alpha-helical rod protein 1 (Alpha helical coiled-coil rod protein) (Putative gene 8 protein) (Pg8). | |||||
|
CO9A1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 19) | NC score | 0.018649 (rank : 42) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q05722, Q61269, Q61270, Q61433, Q61940 | Gene names | Col9a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IX) chain precursor. | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 20) | NC score | 0.010433 (rank : 51) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
MYH10_HUMAN
|
||||||
| θ value | 0.813845 (rank : 21) | NC score | 0.026648 (rank : 22) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P35580, Q16087 | Gene names | MYH10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
RFWD2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 22) | NC score | 0.023509 (rank : 26) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NHY2, Q6H103, Q9H6L7 | Gene names | RFWD2, COP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger and WD repeat domain protein 2 (EC 6.3.2.-) (Ubiquitin- protein ligase COP1) (Constitutive photomorphogenesis protein 1 homolog) (hCOP1). | |||||
|
RFWD2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 23) | NC score | 0.023687 (rank : 25) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 535 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9R1A8 | Gene names | Rfwd2, Cop1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger and WD repeat domain protein 2 (EC 6.3.2.-) (Ubiquitin- protein ligase COP1) (Constitutive photomorphogenesis protein 1 homolog) (mCOP1). | |||||
|
CNTRB_HUMAN
|
||||||
| θ value | 1.06291 (rank : 24) | NC score | 0.031970 (rank : 20) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8N137, Q331K3, Q69YV7, Q8NCB8, Q8WXV3, Q96CQ7, Q9C060 | Gene names | CNTROB, LIP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrobin (LYST-interacting protein 8). | |||||
|
IQGA1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 25) | NC score | 0.021465 (rank : 38) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9JKF1 | Gene names | Iqgap1 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating-like protein IQGAP1. | |||||
|
ANR26_MOUSE
|
||||||
| θ value | 1.81305 (rank : 26) | NC score | 0.018883 (rank : 41) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1554 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q811D2, Q69ZS2, Q9CS61 | Gene names | Ankrd26, Kiaa1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
GOGA2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 27) | NC score | 0.021579 (rank : 37) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q921M4 | Gene names | Golga2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgin subfamily A member 2 (Cis-Golgi matrix protein GM130). | |||||
|
MYH10_MOUSE
|
||||||
| θ value | 1.81305 (rank : 28) | NC score | 0.025926 (rank : 24) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1612 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q61879, Q5SV63 | Gene names | Myh10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
MYH3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 29) | NC score | 0.022066 (rank : 35) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1660 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P11055, Q15492 | Gene names | MYH3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, fast skeletal muscle, embryonic (Muscle embryonic myosin heavy chain) (SMHCE). | |||||
|
CO4A2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 30) | NC score | 0.014536 (rank : 47) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P08122, Q61375 | Gene names | Col4a2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(IV) chain precursor. | |||||
|
MYH14_HUMAN
|
||||||
| θ value | 2.36792 (rank : 31) | NC score | 0.029179 (rank : 21) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 995 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q7Z406, Q5CZ75, Q6XYE4, Q76B62, Q8WV23, Q96I22, Q9BT27, Q9BW35, Q9H882 | Gene names | MYH14, KIAA2034 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
CO4A2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 32) | NC score | 0.014553 (rank : 46) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 454 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P08572 | Gene names | COL4A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(IV) chain precursor. | |||||
|
CROCC_HUMAN
|
||||||
| θ value | 3.0926 (rank : 33) | NC score | 0.023431 (rank : 27) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1409 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q5TZA2, Q2VHY3, Q66GT7, Q7Z2L4, Q7Z5D7 | Gene names | CROCC, KIAA0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
MYO5C_HUMAN
|
||||||
| θ value | 3.0926 (rank : 34) | NC score | 0.022936 (rank : 29) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1206 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9NQX4 | Gene names | MYO5C | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5C (Myosin Vc). | |||||
|
RIPK2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 35) | NC score | 0.001008 (rank : 61) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 776 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P58801 | Gene names | Ripk2 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-interacting serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
CO9A1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 36) | NC score | 0.016311 (rank : 44) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P20849, Q13699, Q13700, Q5TF52, Q6P467, Q96BM8, Q99225, Q9H151, Q9H152, Q9Y6P2, Q9Y6P3 | Gene names | COL9A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IX) chain precursor. | |||||
|
HOOK2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 37) | NC score | 0.035300 (rank : 19) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 736 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q7TMK6, Q8BY47, Q8R347, Q8VCN4, Q99LU2 | Gene names | Hook2 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 2. | |||||
|
KGP1A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 38) | NC score | 0.002677 (rank : 60) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 901 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13976 | Gene names | PRKG1, PRKGR1A | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 1, alpha isozyme (EC 2.7.11.12) (CGK 1 alpha) (cGKI-alpha). | |||||
|
LETM1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 39) | NC score | 0.019337 (rank : 40) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O95202, Q9UF65 | Gene names | LETM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper-EF-hand-containing transmembrane protein 1, mitochondrial precursor. | |||||
|
MYH4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 40) | NC score | 0.023206 (rank : 28) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1377 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Y623 | Gene names | MYH4 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal) (Myosin heavy chain IIb) (MyHC-IIb). | |||||
|
AKAP9_HUMAN
|
||||||
| θ value | 5.27518 (rank : 41) | NC score | 0.021588 (rank : 36) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
CO5A2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 42) | NC score | 0.012236 (rank : 50) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P05997 | Gene names | COL5A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(V) chain precursor. | |||||
|
EEA1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 43) | NC score | 0.017245 (rank : 43) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
FLIP1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 44) | NC score | 0.020151 (rank : 39) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q7Z7B0, Q5VUL6, Q86TC3, Q8N8B9, Q96SK6, Q9NVI8, Q9ULE5 | Gene names | FILIP1, KIAA1275 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
FLIP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 45) | NC score | 0.022739 (rank : 31) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1176 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9CS72 | Gene names | Filip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
MRCKG_MOUSE
|
||||||
| θ value | 6.88961 (rank : 46) | NC score | 0.006778 (rank : 58) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1466 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q80UW5 | Gene names | Cdc42bpg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK gamma (EC 2.7.11.1) (CDC42- binding protein kinase gamma) (Myotonic dystrophy kinase-related CDC42-binding kinase gamma) (Myotonic dystrophy protein kinase-like alpha) (MRCK gamma) (DMPK-like gamma). | |||||
|
MYH1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 47) | NC score | 0.022690 (rank : 32) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1478 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P12882, Q9Y622 | Gene names | MYH1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1) (Myosin heavy chain IIx/d) (MyHC-IIx/d). | |||||
|
MYH1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 48) | NC score | 0.022875 (rank : 30) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1461 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q5SX40 | Gene names | Myh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1). | |||||
|
PHLPP_HUMAN
|
||||||
| θ value | 6.88961 (rank : 49) | NC score | 0.003868 (rank : 59) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 604 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O60346, Q641Q7, Q6P4C4, Q6PJI6, Q86TN6, Q96FK2, Q9NUY1 | Gene names | PHLPP, KIAA0606, PLEKHE1, SCOP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein) (hSCOP). | |||||
|
RUNX3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 50) | NC score | 0.007868 (rank : 56) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q64131, Q99P92, Q9R199 | Gene names | Runx3, Aml2, Cbfa3, Pebp2a3 | |||
|
Domain Architecture |
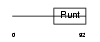
|
|||||
| Description | Runt-related transcription factor 3 (Core-binding factor, alpha 3 subunit) (CBF-alpha 3) (Acute myeloid leukemia 2 protein) (Oncogene AML-2) (Polyomavirus enhancer-binding protein 2 alpha C subunit) (PEBP2-alpha C) (PEA2-alpha C) (SL3-3 enhancer factor 1 alpha C subunit) (SL3/AKV core-binding factor alpha C subunit). | |||||
|
ARMX2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 51) | NC score | 0.007433 (rank : 57) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6A058, Q9CXI9 | Gene names | Armcx2, Kiaa0512 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 2. | |||||
|
CO4A1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 52) | NC score | 0.013307 (rank : 49) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P02462 | Gene names | COL4A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IV) chain precursor. | |||||
|
EMIL1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 53) | NC score | 0.013854 (rank : 48) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y6C2, Q96G58, Q9UG76 | Gene names | EMILIN1, EMI | |||
|
Domain Architecture |

|
|||||
| Description | EMILIN-1 precursor (Elastin microfibril interface-located protein 1) (Elastin microfibril interfacer 1). | |||||
|
GSCR1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 54) | NC score | 0.008496 (rank : 55) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
IRBP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 55) | NC score | 0.009535 (rank : 52) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P49194, Q6B4R6, Q8VD34, Q9R0H8 | Gene names | Rbp3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interphotoreceptor retinoid-binding protein precursor (IRBP) (Interstitial retinol-binding protein). | |||||
|
KCD18_HUMAN
|
||||||
| θ value | 8.99809 (rank : 56) | NC score | 0.008854 (rank : 54) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6PI47, Q53T21, Q6NW26, Q6PCD8, Q8N9B7, Q96N73 | Gene names | KCTD18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD18. | |||||
|
MYH2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 57) | NC score | 0.022176 (rank : 34) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1491 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UKX2, Q14322, Q16229 | Gene names | MYH2, MYHSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, skeletal muscle, adult 2 (Myosin heavy chain IIa) (MyHC-IIa). | |||||
|
MYH4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 58) | NC score | 0.022652 (rank : 33) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
SON_HUMAN
|
||||||
| θ value | 8.99809 (rank : 59) | NC score | 0.008950 (rank : 53) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
TPM4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 60) | NC score | 0.014578 (rank : 45) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 703 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6IRU2 | Gene names | Tpm4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tropomyosin alpha-4 chain (Tropomyosin-4). | |||||
|
GPR98_MOUSE
|
||||||
| θ value | θ > 10 (rank : 61) | NC score | 0.052376 (rank : 17) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8VHN7, Q6ZQ69, Q810D2, Q810D3, Q91ZS0, Q91ZS1 | Gene names | Gpr98, Kiaa0686, Mass1, Vlgr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor 98 precursor (Monogenic audiogenic seizure susceptibility protein 1) (Very large G-protein coupled receptor 1) (Neurepin). | |||||
|
NPTXR_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O95502 | Gene names | NPTXR | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal pentraxin receptor. | |||||
|
NPTXR_MOUSE
|
||||||
| NC score | 0.988040 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q99J85 | Gene names | Nptxr, Npr | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal pentraxin receptor. | |||||
|
NPTX2_MOUSE
|
||||||
| NC score | 0.968619 (rank : 3) | θ value | 1.48581e-111 (rank : 3) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O70340 | Gene names | Nptx2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal pentraxin-2 precursor (NP2) (Neuronal pentraxin II) (NP-II). | |||||
|
NPTX2_HUMAN
|
||||||
| NC score | 0.967804 (rank : 4) | θ value | 7.37397e-111 (rank : 4) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P47972, Q86XV7, Q96G70 | Gene names | NPTX2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal pentraxin-2 precursor (NP2) (Neuronal pentraxin II) (NP-II). | |||||
|
NPTX1_HUMAN
|
||||||
| NC score | 0.967023 (rank : 5) | θ value | 3.79596e-99 (rank : 6) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q15818, Q5FWE6 | Gene names | NPTX1 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal pentraxin-1 precursor (NP1) (Neuronal pentraxin I) (NP-I). | |||||
|
NPTX1_MOUSE
|
||||||
| NC score | 0.965300 (rank : 6) | θ value | 9.98939e-100 (rank : 5) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q62443 | Gene names | Nptx1 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal pentraxin-1 precursor (NP1) (Neuronal pentraxin I) (NP-I). | |||||
|
PTX3_MOUSE
|
||||||
| NC score | 0.755530 (rank : 7) | θ value | 5.60996e-18 (rank : 8) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P48759, Q8VDF1 | Gene names | Ptx3, Tsg14 | |||
|
Domain Architecture |

|
|||||
| Description | Pentraxin-related protein PTX3 precursor (Pentaxin-related protein PTX3) (Tumor necrosis factor-inducible protein TSG-14). | |||||
|
PTX3_HUMAN
|
||||||
| NC score | 0.752369 (rank : 8) | θ value | 2.7842e-17 (rank : 9) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P26022 | Gene names | PTX3, TSG14 | |||
|
Domain Architecture |

|
|||||
| Description | Pentraxin-related protein PTX3 precursor (Pentaxin-related protein PTX3) (Tumor necrosis factor-inducible protein TSG-14). | |||||
|
CRP_MOUSE
|
||||||
| NC score | 0.705991 (rank : 9) | θ value | 2.20605e-14 (rank : 11) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P14847 | Gene names | Crp, Ptx1 | |||
|
Domain Architecture |

|
|||||
| Description | C-reactive protein precursor. | |||||
|
CRP_HUMAN
|
||||||
| NC score | 0.702863 (rank : 10) | θ value | 2.20605e-14 (rank : 10) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P02741, Q8WW75 | Gene names | CRP, PTX1 | |||
|
Domain Architecture |

|
|||||
| Description | C-reactive protein precursor [Contains: C-reactive protein(1-205)]. | |||||
|
SAMP_HUMAN
|
||||||
| NC score | 0.691676 (rank : 11) | θ value | 4.16044e-13 (rank : 12) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P02743 | Gene names | APCS, PTX2 | |||
|
Domain Architecture |

|
|||||
| Description | Serum amyloid P-component precursor (SAP) (9.5S alpha-1-glycoprotein) [Contains: Serum amyloid P-component(1-203)]. | |||||
|
SAMP_MOUSE
|
||||||
| NC score | 0.689809 (rank : 12) | θ value | 9.26847e-13 (rank : 13) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P12246 | Gene names | Apcs, Ptx2, Sap | |||
|
Domain Architecture |

|
|||||
| Description | Serum amyloid P-component precursor (SAP). | |||||
|
GP144_HUMAN
|
||||||
| NC score | 0.423875 (rank : 13) | θ value | 3.62785e-25 (rank : 7) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q7Z7M1, Q86SL4, Q8NH12 | Gene names | GPR144, PGR24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 144 (G-protein coupled receptor PGR24). | |||||
|
GP112_HUMAN
|
||||||
| NC score | 0.119261 (rank : 14) | θ value | 0.00102713 (rank : 14) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8IZF6, Q86SM6 | Gene names | GPR112 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 112. | |||||
|
GP126_HUMAN
|
||||||
| NC score | 0.084411 (rank : 15) | θ value | 0.00869519 (rank : 15) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q86SQ4, Q6MZU7, Q8NC14, Q96JW0 | Gene names | GPR126 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 126 precursor. | |||||
|
GP133_HUMAN
|
||||||
| NC score | 0.081831 (rank : 16) | θ value | 0.0961366 (rank : 16) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6QNK2, Q6ZMQ1, Q7Z7M2, Q86SM4 | Gene names | GPR133, PGR25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 133 precursor (G-protein coupled receptor PGR25). | |||||
|
GPR98_MOUSE
|
||||||
| NC score | 0.052376 (rank : 17) | θ value | θ > 10 (rank : 61) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8VHN7, Q6ZQ69, Q810D2, Q810D3, Q91ZS0, Q91ZS1 | Gene names | Gpr98, Kiaa0686, Mass1, Vlgr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor 98 precursor (Monogenic audiogenic seizure susceptibility protein 1) (Very large G-protein coupled receptor 1) (Neurepin). | |||||
|
CCHCR_HUMAN
|
||||||
| NC score | 0.042136 (rank : 18) | θ value | 0.21417 (rank : 18) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 717 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8TD31, Q9NRK8, Q9NWY9, Q9NXJ4, Q9NXK3, Q9Y6W1, Q9Y6W2 | Gene names | CCHCR1, C6orf18, HCR | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil alpha-helical rod protein 1 (Alpha helical coiled-coil rod protein) (Putative gene 8 protein) (Pg8). | |||||
|
HOOK2_MOUSE
|
||||||
| NC score | 0.035300 (rank : 19) | θ value | 4.03905 (rank : 37) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 736 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q7TMK6, Q8BY47, Q8R347, Q8VCN4, Q99LU2 | Gene names | Hook2 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 2. | |||||
|
CNTRB_HUMAN
|
||||||
| NC score | 0.031970 (rank : 20) | θ value | 1.06291 (rank : 24) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8N137, Q331K3, Q69YV7, Q8NCB8, Q8WXV3, Q96CQ7, Q9C060 | Gene names | CNTROB, LIP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrobin (LYST-interacting protein 8). | |||||
|
MYH14_HUMAN
|
||||||
| NC score | 0.029179 (rank : 21) | θ value | 2.36792 (rank : 31) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 995 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q7Z406, Q5CZ75, Q6XYE4, Q76B62, Q8WV23, Q96I22, Q9BT27, Q9BW35, Q9H882 | Gene names | MYH14, KIAA2034 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
MYH10_HUMAN
|
||||||
| NC score | 0.026648 (rank : 22) | θ value | 0.813845 (rank : 21) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P35580, Q16087 | Gene names | MYH10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
NIN_HUMAN
|
||||||
| NC score | 0.026299 (rank : 23) | θ value | 0.125558 (rank : 17) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1416 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8N4C6, Q6P0P6, Q9BWU6, Q9C012, Q9C013, Q9C014, Q9H5I6, Q9HAT7, Q9HBY5, Q9HCK7, Q9UH61 | Gene names | NIN, KIAA1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein (hNinein) (Glycogen synthase kinase 3 beta-interacting protein) (GSK3B-interacting protein). | |||||
|
MYH10_MOUSE
|
||||||
| NC score | 0.025926 (rank : 24) | θ value | 1.81305 (rank : 28) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1612 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q61879, Q5SV63 | Gene names | Myh10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
RFWD2_MOUSE
|
||||||
| NC score | 0.023687 (rank : 25) | θ value | 0.813845 (rank : 23) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 535 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9R1A8 | Gene names | Rfwd2, Cop1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger and WD repeat domain protein 2 (EC 6.3.2.-) (Ubiquitin- protein ligase COP1) (Constitutive photomorphogenesis protein 1 homolog) (mCOP1). | |||||
|
RFWD2_HUMAN
|
||||||
| NC score | 0.023509 (rank : 26) | θ value | 0.813845 (rank : 22) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NHY2, Q6H103, Q9H6L7 | Gene names | RFWD2, COP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger and WD repeat domain protein 2 (EC 6.3.2.-) (Ubiquitin- protein ligase COP1) (Constitutive photomorphogenesis protein 1 homolog) (hCOP1). | |||||
|
CROCC_HUMAN
|
||||||
| NC score | 0.023431 (rank : 27) | θ value | 3.0926 (rank : 33) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1409 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q5TZA2, Q2VHY3, Q66GT7, Q7Z2L4, Q7Z5D7 | Gene names | CROCC, KIAA0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
MYH4_HUMAN
|
||||||
| NC score | 0.023206 (rank : 28) | θ value | 4.03905 (rank : 40) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1377 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Y623 | Gene names | MYH4 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal) (Myosin heavy chain IIb) (MyHC-IIb). | |||||
|
MYO5C_HUMAN
|
||||||
| NC score | 0.022936 (rank : 29) | θ value | 3.0926 (rank : 34) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1206 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9NQX4 | Gene names | MYO5C | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5C (Myosin Vc). | |||||
|
MYH1_MOUSE
|
||||||
| NC score | 0.022875 (rank : 30) | θ value | 6.88961 (rank : 48) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1461 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q5SX40 | Gene names | Myh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1). | |||||
|
FLIP1_MOUSE
|
||||||
| NC score | 0.022739 (rank : 31) | θ value | 6.88961 (rank : 45) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1176 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9CS72 | Gene names | Filip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
MYH1_HUMAN
|
||||||
| NC score | 0.022690 (rank : 32) | θ value | 6.88961 (rank : 47) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1478 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P12882, Q9Y622 | Gene names | MYH1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1) (Myosin heavy chain IIx/d) (MyHC-IIx/d). | |||||
|
MYH4_MOUSE
|
||||||
| NC score | 0.022652 (rank : 33) | θ value | 8.99809 (rank : 58) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
MYH2_HUMAN
|
||||||
| NC score | 0.022176 (rank : 34) | θ value | 8.99809 (rank : 57) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1491 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UKX2, Q14322, Q16229 | Gene names | MYH2, MYHSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, skeletal muscle, adult 2 (Myosin heavy chain IIa) (MyHC-IIa). | |||||
|
MYH3_HUMAN
|
||||||
| NC score | 0.022066 (rank : 35) | θ value | 1.81305 (rank : 29) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1660 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P11055, Q15492 | Gene names | MYH3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, fast skeletal muscle, embryonic (Muscle embryonic myosin heavy chain) (SMHCE). | |||||
|
AKAP9_HUMAN
|
||||||
| NC score | 0.021588 (rank : 36) | θ value | 5.27518 (rank : 41) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
GOGA2_MOUSE
|
||||||
| NC score | 0.021579 (rank : 37) | θ value | 1.81305 (rank : 27) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q921M4 | Gene names | Golga2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgin subfamily A member 2 (Cis-Golgi matrix protein GM130). | |||||
|
IQGA1_MOUSE
|
||||||
| NC score | 0.021465 (rank : 38) | θ value | 1.06291 (rank : 25) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9JKF1 | Gene names | Iqgap1 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating-like protein IQGAP1. | |||||
|
FLIP1_HUMAN
|
||||||
| NC score | 0.020151 (rank : 39) | θ value | 5.27518 (rank : 44) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q7Z7B0, Q5VUL6, Q86TC3, Q8N8B9, Q96SK6, Q9NVI8, Q9ULE5 | Gene names | FILIP1, KIAA1275 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
LETM1_HUMAN
|
||||||
| NC score | 0.019337 (rank : 40) | θ value | 4.03905 (rank : 39) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O95202, Q9UF65 | Gene names | LETM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper-EF-hand-containing transmembrane protein 1, mitochondrial precursor. | |||||
|
ANR26_MOUSE
|
||||||
| NC score | 0.018883 (rank : 41) | θ value | 1.81305 (rank : 26) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1554 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q811D2, Q69ZS2, Q9CS61 | Gene names | Ankrd26, Kiaa1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
CO9A1_MOUSE
|
||||||
| NC score | 0.018649 (rank : 42) | θ value | 0.279714 (rank : 19) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q05722, Q61269, Q61270, Q61433, Q61940 | Gene names | Col9a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IX) chain precursor. | |||||
|
EEA1_HUMAN
|
||||||
| NC score | 0.017245 (rank : 43) | θ value | 5.27518 (rank : 43) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
CO9A1_HUMAN
|
||||||
| NC score | 0.016311 (rank : 44) | θ value | 4.03905 (rank : 36) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P20849, Q13699, Q13700, Q5TF52, Q6P467, Q96BM8, Q99225, Q9H151, Q9H152, Q9Y6P2, Q9Y6P3 | Gene names | COL9A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IX) chain precursor. | |||||
|
TPM4_MOUSE
|
||||||
| NC score | 0.014578 (rank : 45) | θ value | 8.99809 (rank : 60) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 703 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6IRU2 | Gene names | Tpm4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tropomyosin alpha-4 chain (Tropomyosin-4). | |||||
|
CO4A2_HUMAN
|
||||||
| NC score | 0.014553 (rank : 46) | θ value | 3.0926 (rank : 32) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 454 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P08572 | Gene names | COL4A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(IV) chain precursor. | |||||
|
CO4A2_MOUSE
|
||||||
| NC score | 0.014536 (rank : 47) | θ value | 2.36792 (rank : 30) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P08122, Q61375 | Gene names | Col4a2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(IV) chain precursor. | |||||
|
EMIL1_HUMAN
|
||||||
| NC score | 0.013854 (rank : 48) | θ value | 8.99809 (rank : 53) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y6C2, Q96G58, Q9UG76 | Gene names | EMILIN1, EMI | |||
|
Domain Architecture |

|
|||||
| Description | EMILIN-1 precursor (Elastin microfibril interface-located protein 1) (Elastin microfibril interfacer 1). | |||||
|
CO4A1_HUMAN
|
||||||
| NC score | 0.013307 (rank : 49) | θ value | 8.99809 (rank : 52) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P02462 | Gene names | COL4A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IV) chain precursor. | |||||
|
CO5A2_HUMAN
|
||||||
| NC score | 0.012236 (rank : 50) | θ value | 5.27518 (rank : 42) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P05997 | Gene names | COL5A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(V) chain precursor. | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.010433 (rank : 51) | θ value | 0.813845 (rank : 20) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
IRBP_MOUSE
|
||||||
| NC score | 0.009535 (rank : 52) | θ value | 8.99809 (rank : 55) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P49194, Q6B4R6, Q8VD34, Q9R0H8 | Gene names | Rbp3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interphotoreceptor retinoid-binding protein precursor (IRBP) (Interstitial retinol-binding protein). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.008950 (rank : 53) | θ value | 8.99809 (rank : 59) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
KCD18_HUMAN
|
||||||
| NC score | 0.008854 (rank : 54) | θ value | 8.99809 (rank : 56) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6PI47, Q53T21, Q6NW26, Q6PCD8, Q8N9B7, Q96N73 | Gene names | KCTD18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD18. | |||||
|
GSCR1_HUMAN
|
||||||
| NC score | 0.008496 (rank : 55) | θ value | 8.99809 (rank : 54) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
RUNX3_MOUSE
|
||||||
| NC score | 0.007868 (rank : 56) | θ value | 6.88961 (rank : 50) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q64131, Q99P92, Q9R199 | Gene names | Runx3, Aml2, Cbfa3, Pebp2a3 | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 3 (Core-binding factor, alpha 3 subunit) (CBF-alpha 3) (Acute myeloid leukemia 2 protein) (Oncogene AML-2) (Polyomavirus enhancer-binding protein 2 alpha C subunit) (PEBP2-alpha C) (PEA2-alpha C) (SL3-3 enhancer factor 1 alpha C subunit) (SL3/AKV core-binding factor alpha C subunit). | |||||
|
ARMX2_MOUSE
|
||||||
| NC score | 0.007433 (rank : 57) | θ value | 8.99809 (rank : 51) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6A058, Q9CXI9 | Gene names | Armcx2, Kiaa0512 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 2. | |||||
|
MRCKG_MOUSE
|
||||||
| NC score | 0.006778 (rank : 58) | θ value | 6.88961 (rank : 46) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1466 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q80UW5 | Gene names | Cdc42bpg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK gamma (EC 2.7.11.1) (CDC42- binding protein kinase gamma) (Myotonic dystrophy kinase-related CDC42-binding kinase gamma) (Myotonic dystrophy protein kinase-like alpha) (MRCK gamma) (DMPK-like gamma). | |||||
|
PHLPP_HUMAN
|
||||||
| NC score | 0.003868 (rank : 59) | θ value | 6.88961 (rank : 49) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 604 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O60346, Q641Q7, Q6P4C4, Q6PJI6, Q86TN6, Q96FK2, Q9NUY1 | Gene names | PHLPP, KIAA0606, PLEKHE1, SCOP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein) (hSCOP). | |||||
|
KGP1A_HUMAN
|
||||||
| NC score | 0.002677 (rank : 60) | θ value | 4.03905 (rank : 38) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 901 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13976 | Gene names | PRKG1, PRKGR1A | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 1, alpha isozyme (EC 2.7.11.12) (CGK 1 alpha) (cGKI-alpha). | |||||
|
RIPK2_MOUSE
|
||||||
| NC score | 0.001008 (rank : 61) | θ value | 3.0926 (rank : 35) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 776 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P58801 | Gene names | Ripk2 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-interacting serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||