Please be patient as the page loads
|
LIRB1_HUMAN
|
||||||
| SwissProt Accessions | Q8NHL6, O75024, O75025, Q8NHJ9, Q8NHK0 | Gene names | LILRB1, ILT2, LIR1, MIR7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukocyte immunoglobulin-like receptor subfamily B member 1 precursor (Leukocyte immunoglobulin-like receptor 1) (LIR-1) (Immunoglobulin- like transcript 2) (ILT-2) (Monocyte/macrophage immunoglobulin-like receptor 7) (MIR-7) (CD85j antigen). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
LIRA1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.990115 (rank : 3) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O75019, O75018 | Gene names | LILRA1, LIR6 | |||
|
Domain Architecture |

|
|||||
| Description | Leukocyte immunoglobulin-like receptor subfamily A member 1 precursor (Leukocyte immunoglobulin-like receptor 6) (LIR-6) (CD85i antigen). | |||||
|
LIRA2_HUMAN
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.985150 (rank : 8) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8N149, O75020 | Gene names | LILRA2, ILT1, LIR7 | |||
|
Domain Architecture |

|
|||||
| Description | Leukocyte immunoglobulin-like receptor subfamily A member 2 precursor (Leukocyte immunoglobulin-like receptor 7) (LIR-7) (Immunoglobulin- like transcript 1) (ILT-1) (CD85h antigen). | |||||
|
LIRA3_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.987720 (rank : 5) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q8N6C8, O15469, O15470, O75016, Q8N151, Q8N154, Q8NHJ1, Q8NHJ2, Q8NHJ3, Q8NHJ4 | Gene names | LILRA3, ILT6, LIR4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukocyte immunoglobulin-like receptor subfamily A member 3 precursor (Leukocyte immunoglobulin-like receptor 4) (LIR-4) (Immunoglobulin- like transcript 6) (ILT-6) (Monocyte inhibitory receptor HM43/HM31) (CD85e antigen). | |||||
|
LIRB1_HUMAN
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 116 | |
| SwissProt Accessions | Q8NHL6, O75024, O75025, Q8NHJ9, Q8NHK0 | Gene names | LILRB1, ILT2, LIR1, MIR7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukocyte immunoglobulin-like receptor subfamily B member 1 precursor (Leukocyte immunoglobulin-like receptor 1) (LIR-1) (Immunoglobulin- like transcript 2) (ILT-2) (Monocyte/macrophage immunoglobulin-like receptor 7) (MIR-7) (CD85j antigen). | |||||
|
LIRB2_HUMAN
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.992058 (rank : 2) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q8N423, O75017, Q8NHJ7, Q8NHJ8 | Gene names | LILRB2, ILT4, LIR2, MIR10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukocyte immunoglobulin-like receptor subfamily B member 2 precursor (Leukocyte immunoglobulin-like receptor 2) (LIR-2) (Immunoglobulin- like transcript 4) (ILT-4) (Monocyte/macrophage immunoglobulin-like receptor 10) (MIR-10) (CD85d antigen). | |||||
|
LIRB3_HUMAN
|
||||||
| θ value | 0 (rank : 6) | NC score | 0.989732 (rank : 4) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | O75022, O15471, Q86U49 | Gene names | LILRB3, ILT5, LIR3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukocyte immunoglobulin-like receptor subfamily B member 3 precursor (Leukocyte immunoglobulin-like receptor 3) (LIR-3) (Immunoglobulin- like transcript 5) (ILT-5) (Monocyte inhibitory receptor HL9) (CD85a antigen). | |||||
|
LIRB5_HUMAN
|
||||||
| θ value | 1.04281e-181 (rank : 7) | NC score | 0.986058 (rank : 6) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O75023, Q8N760 | Gene names | LILRB5, LIR8 | |||
|
Domain Architecture |

|
|||||
| Description | Leukocyte immunoglobulin-like receptor subfamily B member 5 precursor (Leukocyte immunoglobulin-like receptor 8) (LIR-8) (CD85c antigen). | |||||
|
LIRA4_HUMAN
|
||||||
| θ value | 2.04937e-153 (rank : 8) | NC score | 0.985232 (rank : 7) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P59901 | Gene names | LILRA4, ILT7 | |||
|
Domain Architecture |

|
|||||
| Description | Leukocyte immunoglobulin-like receptor subfamily A member 4 precursor (Immunoglobulin-like transcript 7) (ILT-7) (CD85g antigen). | |||||
|
LIRB4_HUMAN
|
||||||
| θ value | 7.09276e-130 (rank : 9) | NC score | 0.968267 (rank : 9) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q8NHJ6, O15468, O75021, Q8N1C7, Q8NHL5 | Gene names | LILRB4, ILT3, LIR5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukocyte immunoglobulin-like receptor subfamily B member 4 precursor (Leukocyte immunoglobulin-like receptor 5) (LIR-5) (Immunoglobulin- like transcript 3) (ILT-3) (Monocyte inhibitory receptor HM18) (CD85k antigen). | |||||
|
KI3L3_HUMAN
|
||||||
| θ value | 4.53632e-60 (rank : 10) | NC score | 0.909056 (rank : 18) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8N743, O95056, Q7Z7N4, Q8NHI2, Q8NHI3, Q9UEI4 | Gene names | KIR3DL3, CD158Z, KIR3DL7, KIRC1 | |||
|
Domain Architecture |
|
|||||
| Description | Killer cell immunoglobulin-like receptor 3DL3 precursor (Killer cell inhibitory receptor 1) (CD158z antigen). | |||||
|
KI3L2_HUMAN
|
||||||
| θ value | 9.45883e-58 (rank : 11) | NC score | 0.902574 (rank : 20) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P43630, Q13238, Q14947, Q14948, Q92684, Q95367 | Gene names | KIR3DL2, CD158K, NKAT4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Killer cell immunoglobulin-like receptor 3DL2 precursor (MHC class I NK cell receptor) (Natural killer-associated transcript 4) (NKAT-4) (p70 natural killer cell receptor clone CL-5) (CD158k antigen). | |||||
|
KI3L1_HUMAN
|
||||||
| θ value | 4.85792e-54 (rank : 12) | NC score | 0.899253 (rank : 21) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P43629, O43473, Q16541 | Gene names | KIR3DL1, NKAT3, NKB1 | |||
|
Domain Architecture |
|
|||||
| Description | Killer cell immunoglobulin-like receptor 3DL1 precursor (MHC class I NK cell receptor) (Natural killer-associated transcript 3) (NKAT-3) (p70 natural killer cell receptor clones CL-2/CL-11) (HLA-BW4-specific inhibitory NK cell receptor). | |||||
|
KI3S1_HUMAN
|
||||||
| θ value | 1.08223e-53 (rank : 13) | NC score | 0.905753 (rank : 19) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q14943 | Gene names | KIR3DS1, NKAT10 | |||
|
Domain Architecture |
|
|||||
| Description | Killer cell immunoglobulin-like receptor 3DS1 precursor (MHC class I NK cell receptor) (Natural killer-associated transcript 10) (NKAT-10). | |||||
|
KI2S2_HUMAN
|
||||||
| θ value | 2.3352e-48 (rank : 14) | NC score | 0.885026 (rank : 22) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P43631, Q14955 | Gene names | KIR2DS2, CD158J, NKAT5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Killer cell immunoglobulin-like receptor 2DS2 precursor (MHC class I NK cell receptor) (Natural killer-associated transcript 5) (NKAT-5) (p58 natural killer cell receptor clone CL-49) (p58 NK receptor) (NK receptor 183 ActI) (CD158j antigen). | |||||
|
LIRB4_MOUSE
|
||||||
| θ value | 6.7944e-48 (rank : 15) | NC score | 0.946490 (rank : 10) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q64281, Q64312 | Gene names | Lilrb4, Gp49b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukocyte immunoglobulin-like receptor subfamily B member 4 precursor (Mast cell surface glycoprotein Gp49B). | |||||
|
KI3L1_MOUSE
|
||||||
| θ value | 1.08474e-45 (rank : 16) | NC score | 0.913424 (rank : 17) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P83555 | Gene names | Kir3dl1 | |||
|
Domain Architecture |
|
|||||
| Description | Killer cell immunoglobulin-like receptor 3DL1 precursor. | |||||
|
KI2L2_HUMAN
|
||||||
| θ value | 5.38352e-45 (rank : 17) | NC score | 0.879443 (rank : 24) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P43627, Q14951 | Gene names | KIR2DL2, NKAT6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Killer cell immunoglobulin-like receptor 2DL2 precursor (MHC class I NK cell receptor) (Natural killer-associated transcript 6) (NKAT-6) (p58 natural killer cell receptor clone CL-43) (p58 NK receptor). | |||||
|
GP49A_MOUSE
|
||||||
| θ value | 1.19932e-44 (rank : 18) | NC score | 0.940275 (rank : 11) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q61450 | Gene names | Gp49a, Gp49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mast cell surface glycoprotein Gp49A precursor. | |||||
|
KI2S4_HUMAN
|
||||||
| θ value | 2.04573e-44 (rank : 19) | NC score | 0.880080 (rank : 23) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P43632 | Gene names | KIR2DS4, CD158I, KKA3, NKAT8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Killer cell immunoglobulin-like receptor 2DS4 precursor (MHC class I NK cell receptor) (Natural killer-associated transcript 8) (NKAT-8) (P58 natural killer cell receptor clone CL-39) (p58 NK receptor) (CL- 17) (CD158i antigen). | |||||
|
KI2L1_HUMAN
|
||||||
| θ value | 5.95217e-44 (rank : 20) | NC score | 0.879055 (rank : 27) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P43626, O43470 | Gene names | KIR2DL1, CD158A, NKAT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Killer cell immunoglobulin-like receptor 2DL1 precursor (MHC class I NK cell receptor) (Natural killer-associated transcript 1) (NKAT-1) (p58 natural killer cell receptor clones CL-42/47.11) (p58 NK receptor) (p58.1 MHC class-I-specific NK receptor) (CD158a antigen). | |||||
|
KI2S3_HUMAN
|
||||||
| θ value | 2.95404e-43 (rank : 21) | NC score | 0.879438 (rank : 25) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q14952, O00644 | Gene names | KIR2DS3, NKAT7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Killer cell immunoglobulin-like receptor 2DS3 precursor (MHC class I NK cell receptor) (Natural killer-associated transcript 7) (NKAT-7). | |||||
|
KI2L3_HUMAN
|
||||||
| θ value | 5.03882e-43 (rank : 22) | NC score | 0.878265 (rank : 28) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P43628, O43472, P78402, Q14944, Q14945, Q9UM51, Q9UQ70 | Gene names | KIR2DL3, CD158B, KIRCL23, NKAT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Killer cell immunoglobulin-like receptor 2DL3 precursor (MHC class I NK cell receptor) (Natural killer-associated transcript 2) (NKAT-2) (NKAT2a) (NKAT2b) (p58 natural killer cell receptor clone CL-6) (p58 NK receptor) (p58.2 MHC class-I-specific NK receptor) (Killer inhibitory receptor cl 2-3) (KIR-023GB) (CD158b antigen). | |||||
|
KI2S1_HUMAN
|
||||||
| θ value | 2.76489e-41 (rank : 23) | NC score | 0.876426 (rank : 29) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q14954, O43471 | Gene names | KIR2DS1, CD158H | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Killer cell immunoglobulin-like receptor 2DS1 precursor (MHC class I NK cell receptor Eb6 ActI) (CD158h antigen). | |||||
|
GPVI_HUMAN
|
||||||
| θ value | 7.52953e-39 (rank : 24) | NC score | 0.929360 (rank : 12) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9HCN6, Q9HCN7, Q9UIF2 | Gene names | GP6, GPVI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Platelet glycoprotein VI precursor. | |||||
|
KI2S5_HUMAN
|
||||||
| θ value | 9.83387e-39 (rank : 25) | NC score | 0.875434 (rank : 30) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q14953 | Gene names | KIR2DS5, CD158G, NKAT9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Killer cell immunoglobulin-like receptor 2DS5 precursor (MHC class I NK cell receptor) (Natural killer-associated transcript 9) (NKAT-9) (CD158g antigen). | |||||
|
GPVI_MOUSE
|
||||||
| θ value | 1.98606e-31 (rank : 26) | NC score | 0.924378 (rank : 13) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P0C191 | Gene names | Gp6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Platelet glycoprotein VI precursor (GPVI). | |||||
|
NCTR1_MOUSE
|
||||||
| θ value | 1.98606e-31 (rank : 27) | NC score | 0.915031 (rank : 15) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8C567, Q1RLN4, Q80UY6, Q9Z0Q4 | Gene names | Ncr1, Ly94 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Natural cytotoxicity triggering receptor 1 precursor (Natural killer cell p46-related protein) (NKp46) (mNKp46) (NK-p46) (Mouse-activating receptor 1) (mAR-1) (Lymphocyte antigen 94). | |||||
|
NCTR1_HUMAN
|
||||||
| θ value | 4.42448e-31 (rank : 28) | NC score | 0.921390 (rank : 14) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O76036, O76016, O76017, O76018 | Gene names | NCR1, LY94 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Natural cytotoxicity triggering receptor 1 precursor (Natural killer cell p46-related protein) (NKp46) (hNKp46) (NK-p46) (NK cell- activating receptor) (Lymphocyte antigen 94 homolog) (CD335 antigen). | |||||
|
KI2L4_HUMAN
|
||||||
| θ value | 2.19584e-30 (rank : 29) | NC score | 0.879366 (rank : 26) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q99706, O14621, O14622, O14623, O14624, O43534, P78400, P78401, Q99559, Q99560, Q99561, Q99562, Q9UQJ7 | Gene names | KIR2DL4, CD158D, KIR103AS | |||
|
Domain Architecture |
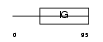
|
|||||
| Description | Killer cell immunoglobulin-like receptor 2DL4 precursor (MHC class I NK cell receptor KIR103AS) (Killer cell inhibitory receptor 103AS) (KIR-103AS) (G9P) (CD158d antigen). | |||||
|
FCAR_HUMAN
|
||||||
| θ value | 5.40856e-29 (rank : 30) | NC score | 0.914666 (rank : 16) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P24071, Q13603, Q13604, Q15727, Q15728, Q92590 | Gene names | FCAR, CD89 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Immunoglobulin alpha Fc receptor precursor (IgA Fc receptor) (CD89 antigen). | |||||
|
A1BG_HUMAN
|
||||||
| θ value | 9.24701e-21 (rank : 31) | NC score | 0.816357 (rank : 32) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P04217, Q8IYJ6, Q96P39 | Gene names | A1BG | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-1B-glycoprotein precursor (Alpha-1-B glycoprotein). | |||||
|
LAIR1_HUMAN
|
||||||
| θ value | 1.16975e-15 (rank : 32) | NC score | 0.781244 (rank : 33) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6GTX8 | Gene names | LAIR1, CD305 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukocyte-associated immunoglobulin-like receptor 1 precursor (LAIR-1) (hLAIR1) (CD305 antigen). | |||||
|
LAIR2_HUMAN
|
||||||
| θ value | 3.76295e-14 (rank : 33) | NC score | 0.827496 (rank : 31) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q6ISS4, Q6PEZ4 | Gene names | LAIR2, CD306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukocyte-associated immunoglobulin-like receptor 2 precursor (LAIR-2) (CD306 antigen). | |||||
|
CD22_MOUSE
|
||||||
| θ value | 4.92598e-06 (rank : 34) | NC score | 0.180061 (rank : 40) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P35329, Q9JHL2, Q9JJX9, Q9JJY0, Q9JJY1, Q9R056, Q9R094, Q9WU51 | Gene names | Cd22, Lyb-8 | |||
|
Domain Architecture |

|
|||||
| Description | B-cell receptor CD22 precursor (Leu-14) (B-lymphocyte cell adhesion molecule) (BL-CAM) (Siglec-2). | |||||
|
PGBM_MOUSE
|
||||||
| θ value | 9.29e-05 (rank : 35) | NC score | 0.076344 (rank : 60) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1073 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q05793 | Gene names | Hspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG) (Perlecan) (PLC). | |||||
|
FCRLA_MOUSE
|
||||||
| θ value | 0.000158464 (rank : 36) | NC score | 0.420250 (rank : 35) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q920A9, Q3T9Q2, Q8K3U9, Q8VHP5 | Gene names | Fcrla, Fcrl, Fcrl1, Fcrlm1, Fcrx, Freb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fc receptor-like and mucin-like 1 precursor (Fc receptor-like A) (Fc receptor homolog expressed in B-cells) (Fc receptor-related protein X) (FcRX) (Fc receptor-like protein). | |||||
|
CD22_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 37) | NC score | 0.148957 (rank : 43) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P20273, O95699, O95701, O95702, O95703, Q01665, Q92872, Q92873, Q9UQA6, Q9UQA7, Q9UQA8, Q9UQA9, Q9UQB0, Q9Y2A6 | Gene names | CD22 | |||
|
Domain Architecture |

|
|||||
| Description | B-cell receptor CD22 precursor (Leu-14) (B-lymphocyte cell adhesion molecule) (BL-CAM) (Siglec-2). | |||||
|
VCAM1_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 38) | NC score | 0.093946 (rank : 54) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P19320, Q6NUP8 | Gene names | VCAM1, L1CAM | |||
|
Domain Architecture |

|
|||||
| Description | Vascular cell adhesion protein 1 precursor (V-CAM 1) (CD106 antigen) (INCAM-100). | |||||
|
FCGR2_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 39) | NC score | 0.224818 (rank : 38) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P08101, P08102, P12316, P97917, Q60938, Q60939, Q60940, Q61170, Q61558 | Gene names | Fcgr2, Fcgr2b, Ly-17 | |||
|
Domain Architecture |
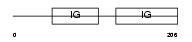
|
|||||
| Description | Low affinity immunoglobulin gamma Fc region receptor II precursor (Fc- gamma RII) (FcRII) (IgG Fc receptor II beta) (Fc gamma receptor IIB) (Fc-gamma-RIIB) (Lymphocyte antigen Ly-17) (CD32 antigen). | |||||
|
FCRLA_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 40) | NC score | 0.346845 (rank : 36) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q7L513, Q5VXA1, Q5VXA2, Q5VXA3, Q5VXB0, Q8NEW4, Q8WXH3, Q96PC6, Q96PJ0, Q96PJ1, Q96PJ2, Q96PJ4, Q9BR57 | Gene names | FCRLA, FCRL, FCRL1, FCRLM1, FCRX, FREB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fc receptor-like and mucin-like 1 precursor (Fc receptor-like A) (Fc receptor homolog expressed in B-cells) (Fc receptor-related protein X) (FcRX) (Fc receptor-like protein). | |||||
|
FCRL5_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 41) | NC score | 0.175562 (rank : 42) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q96RD9, Q495Q2, Q495Q4, Q5VYK9, Q6UY46 | Gene names | FCRL5, FCRH5, IRTA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fc receptor-like protein 5 precursor (Immunoglobulin receptor translocation-associated gene 2 protein) (BXMAS1) (CD307 antigen). | |||||
|
MAMC1_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 42) | NC score | 0.056482 (rank : 67) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P60755 | Gene names | Mamdc1, Mdga2 | |||
|
Domain Architecture |

|
|||||
| Description | MAM domain-containing protein 1 precursor (MAM domain-containing glycosylphosphatidylinositol anchor protein 2). | |||||
|
MUC18_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 43) | NC score | 0.180126 (rank : 39) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8R2Y2, Q9EPF1, Q9ESS7, Q9JHQ2, Q9JHQ3 | Gene names | Mcam, Muc18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell surface glycoprotein MUC18 precursor (Melanoma-associated antigen MUC18) (Melanoma cell adhesion molecule) (Gicerin). | |||||
|
FCGR3_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 44) | NC score | 0.228876 (rank : 37) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P08508 | Gene names | Fcgr3 | |||
|
Domain Architecture |

|
|||||
| Description | Low affinity immunoglobulin gamma Fc region receptor III precursor (IgG Fc receptor III) (Fc-gamma RIII) (FcRIII). | |||||
|
BT2A1_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 45) | NC score | 0.013968 (rank : 89) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q7KYR7, O00475, P78408, Q7KYQ7, Q7Z386, Q9NU62 | Gene names | BTN2A1, BT2.1, BTF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Butyrophilin subfamily 2 member A1 precursor. | |||||
|
FCGR1_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 46) | NC score | 0.137046 (rank : 46) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P26151 | Gene names | Fcgr1, Fcg1 | |||
|
Domain Architecture |

|
|||||
| Description | High affinity immunoglobulin gamma Fc receptor I precursor (Fc-gamma RI) (FcRI) (IgG Fc receptor I). | |||||
|
CD276_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 47) | NC score | 0.176197 (rank : 41) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q5ZPR3, Q6P5Y4, Q6UXI2, Q8NBI8, Q8NC34, Q8NCB6, Q9BXR1 | Gene names | CD276, B7H3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CD276 antigen precursor (Costimulatory molecule) (B7 homolog 3) (B7- H3) (4Ig-B7-H3). | |||||
|
CEA20_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 48) | NC score | 0.086928 (rank : 56) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q6UY09 | Gene names | CEACAM20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carcinoembryonic antigen-related cell adhesion molecule 20 precursor. | |||||
|
CEAM8_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 49) | NC score | 0.042353 (rank : 70) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P31997, O60399, Q16574 | Gene names | CEACAM8, CGM6 | |||
|
Domain Architecture |
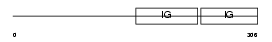
|
|||||
| Description | Carcinoembryonic antigen-related cell adhesion molecule 8 precursor (Carcinoembryonic antigen CGM6) (Nonspecific cross-reacting antigen NCA-95) (CD67 antigen) (CD66b antigen). | |||||
|
SN_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 50) | NC score | 0.108637 (rank : 50) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9BZZ2, Q96DL4, Q9GZS5, Q9H1H6, Q9H1H7, Q9H7L7 | Gene names | SN | |||
|
Domain Architecture |

|
|||||
| Description | Sialoadhesin precursor (Sialic acid-binding Ig-like lectin-1) (Siglec- 1) (CD169 antigen). | |||||
|
ICAM5_MOUSE
|
||||||
| θ value | 0.125558 (rank : 51) | NC score | 0.071241 (rank : 61) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q60625 | Gene names | Icam5, Tlcn | |||
|
Domain Architecture |

|
|||||
| Description | Intercellular adhesion molecule 5 precursor (ICAM-5) (Telencephalin). | |||||
|
VCAM1_MOUSE
|
||||||
| θ value | 0.125558 (rank : 52) | NC score | 0.115488 (rank : 48) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 320 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P29533 | Gene names | Vcam1, Vcam-1 | |||
|
Domain Architecture |

|
|||||
| Description | Vascular cell adhesion protein 1 precursor (V-CAM 1). | |||||
|
PGBM_HUMAN
|
||||||
| θ value | 0.163984 (rank : 53) | NC score | 0.078471 (rank : 59) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1113 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P98160, Q16287, Q9H3V5 | Gene names | HSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG) (Perlecan) (PLC). | |||||
|
CN155_HUMAN
|
||||||
| θ value | 0.21417 (rank : 54) | NC score | 0.031610 (rank : 74) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
PTK7_HUMAN
|
||||||
| θ value | 0.21417 (rank : 55) | NC score | 0.020308 (rank : 84) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1103 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q13308, Q13417 | Gene names | PTK7, CCK4 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase-like 7 precursor (Colon carcinoma kinase 4) (CCK-4). | |||||
|
SELPL_MOUSE
|
||||||
| θ value | 0.21417 (rank : 56) | NC score | 0.018646 (rank : 85) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q62170 | Gene names | Selplg, Selp1, Selpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand). | |||||
|
TITIN_HUMAN
|
||||||
| θ value | 0.21417 (rank : 57) | NC score | 0.028330 (rank : 78) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
PCDGF_HUMAN
|
||||||
| θ value | 0.365318 (rank : 58) | NC score | 0.003909 (rank : 116) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y5G1, Q9Y5C7 | Gene names | PCDHGB3 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma B3 precursor (PCDH-gamma-B3). | |||||
|
SN_MOUSE
|
||||||
| θ value | 0.47712 (rank : 59) | NC score | 0.102178 (rank : 53) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q62230, O55216, Q62228, Q62229 | Gene names | Sn, Sa | |||
|
Domain Architecture |

|
|||||
| Description | Sialoadhesin precursor (Sialic acid-binding Ig-like lectin-1) (Siglec- 1) (Sheep erythrocyte receptor) (SER) (CD169 antigen). | |||||
|
ATBF1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 60) | NC score | 0.004882 (rank : 110) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1547 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15911, O15101, Q13719 | Gene names | ATBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
ATBF1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 61) | NC score | 0.004029 (rank : 115) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
CCD1A_HUMAN
|
||||||
| θ value | 0.62314 (rank : 62) | NC score | 0.028160 (rank : 79) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6P1N0, Q7Z435, Q86XV0, Q8NF89, Q9H603, Q9NXI1 | Gene names | CC2D1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil and C2 domain-containing protein 1A (Five repressor element under dual repression-binding protein 1) (FRE under dual repression-binding protein 1) (Freud-1) (Putative NF-kappa-B- activating protein 023N). | |||||
|
CNOT3_HUMAN
|
||||||
| θ value | 0.62314 (rank : 63) | NC score | 0.027475 (rank : 80) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O75175, Q9NZN7, Q9UF76 | Gene names | CNOT3, KIAA0691, NOT3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
NUCL_MOUSE
|
||||||
| θ value | 0.62314 (rank : 64) | NC score | 0.014136 (rank : 88) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P09405, Q548M9, Q61991, Q99K50 | Gene names | Ncl, Nuc | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolin (Protein C23). | |||||
|
EPN3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 65) | NC score | 0.011045 (rank : 92) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H201, Q9BVN6, Q9NWK2 | Gene names | EPN3 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-3 (EPS-15-interacting protein 3). | |||||
|
JPH2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 66) | NC score | 0.013233 (rank : 90) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
CNTN1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 67) | NC score | 0.030472 (rank : 75) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q12860, Q12861, Q14030, Q8N466 | Gene names | CNTN1 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-1 precursor (Neural cell surface protein F3) (Glycoprotein gp135). | |||||
|
CNOT3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 68) | NC score | 0.022181 (rank : 83) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K0V4 | Gene names | Cnot3, Not3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
JPH2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 69) | NC score | 0.029576 (rank : 77) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9ET78, Q9ET79 | Gene names | Jph2, Jp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
LAIR1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 70) | NC score | 0.477813 (rank : 34) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BG84, Q80UN5, Q8BHB6, Q8BRR6, Q8C6N8, Q8CEP5 | Gene names | Lair1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukocyte-associated immunoglobulin-like receptor 1 precursor (LAIR-1) (mLAIR1) (CD305 antigen). | |||||
|
PJA1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 71) | NC score | 0.009619 (rank : 96) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O55176, Q8CFU2, Q99MJ1, Q99MJ2, Q99MJ3, Q9DB04 | Gene names | Pja1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin protein ligase Praja1 (EC 6.3.2.-). | |||||
|
PTN22_MOUSE
|
||||||
| θ value | 1.81305 (rank : 72) | NC score | 0.008733 (rank : 100) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P29352, Q7TMP9 | Gene names | Ptpn22, Ptpn8 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 22 (EC 3.1.3.48) (Hematopoietic cell protein-tyrosine phosphatase 70Z-PEP). | |||||
|
CD276_MOUSE
|
||||||
| θ value | 2.36792 (rank : 73) | NC score | 0.146183 (rank : 44) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8VE98 | Gene names | Cd276, B7h3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CD276 antigen precursor (Costimulatory molecule) (B7 homolog 3) (B7- H3). | |||||
|
PTPRU_HUMAN
|
||||||
| θ value | 2.36792 (rank : 74) | NC score | 0.008612 (rank : 101) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q92729 | Gene names | PTPRU, PCP2 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase U precursor (EC 3.1.3.48) (R-PTP-U) (Protein-tyrosine phosphatase J) (PTP-J) (Pancreatic carcinoma phosphatase 2) (PCP-2). | |||||
|
RIN3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 75) | NC score | 0.011041 (rank : 93) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8TB24, Q8NF30, Q8TEE8, Q8WYP4, Q9H6A5, Q9HAG1 | Gene names | RIN3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras and Rab interactor 3 (Ras interaction/interference protein 3). | |||||
|
SEM6C_HUMAN
|
||||||
| θ value | 2.36792 (rank : 76) | NC score | 0.003203 (rank : 119) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H3T2, Q5JR71, Q8WXT8, Q8WXT9, Q8WXU0, Q96JF8 | Gene names | SEMA6C, KIAA1869, SEMAY | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6C precursor (Semaphorin Y) (Sema Y). | |||||
|
UBP43_MOUSE
|
||||||
| θ value | 2.36792 (rank : 77) | NC score | 0.009805 (rank : 95) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BUM9, Q8VDP5 | Gene names | Usp43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 43 (EC 3.1.2.15) (Ubiquitin thioesterase 43) (Ubiquitin-specific-processing protease 43) (Deubiquitinating enzyme 43). | |||||
|
HMCN1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 78) | NC score | 0.034287 (rank : 73) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 983 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q96RW7, Q5TYR7, Q96DN3, Q96DN8, Q96SC3 | Gene names | HMCN1, FIBL6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hemicentin-1 precursor (Fibulin-6) (FIBL-6). | |||||
|
LR37B_HUMAN
|
||||||
| θ value | 3.0926 (rank : 79) | NC score | 0.005652 (rank : 106) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96QE4, Q5YKG6 | Gene names | LRRC37B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 37B precursor (C66 SLIT-like testicular protein). | |||||
|
CD166_MOUSE
|
||||||
| θ value | 4.03905 (rank : 80) | NC score | 0.047588 (rank : 69) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q61490, O70136, Q8CDA5, Q8R2T0 | Gene names | Alcam | |||
|
Domain Architecture |
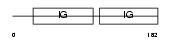
|
|||||
| Description | CD166 antigen precursor (Activated leukocyte-cell adhesion molecule) (ALCAM) (DM-GRASP protein). | |||||
|
CNTN1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 81) | NC score | 0.025569 (rank : 81) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P12960, Q6NXV7, Q8BR42, Q8C6A0 | Gene names | Cntn1 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-1 precursor (Neural cell surface protein F3). | |||||
|
NEGR1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 82) | NC score | 0.039006 (rank : 72) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q7Z3B1, Q5VT21, Q6UY06, Q8NAQ3 | Gene names | NEGR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal growth regulator 1 precursor. | |||||
|
NEGR1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 83) | NC score | 0.040554 (rank : 71) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q80Z24, Q3UHQ8, Q80T70 | Gene names | Negr1, Kiaa3001, Ntra | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal growth regulator 1 precursor (Kilon protein) (Kindred of IgLON) (Neurotractin). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 4.03905 (rank : 84) | NC score | -0.000339 (rank : 124) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
OXR1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 85) | NC score | 0.005637 (rank : 107) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N573, Q3LIB5, Q6ZVK9, Q8N8V0, Q9H266, Q9NWC7 | Gene names | OXR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oxidation resistance protein 1. | |||||
|
RTN4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 86) | NC score | 0.008777 (rank : 98) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99P72, Q5DTK9, Q7TNB7, Q80W95, Q8BGK7, Q8BGM9, Q8K290, Q8K3G8, Q9CTE3 | Gene names | Rtn4, Kiaa0886, Nogo | |||
|
Domain Architecture |

|
|||||
| Description | Reticulon-4 (Neurite outgrowth inhibitor) (Nogo protein). | |||||
|
CCNK_MOUSE
|
||||||
| θ value | 5.27518 (rank : 87) | NC score | 0.010939 (rank : 94) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O88874, Q8R068 | Gene names | Ccnk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-K. | |||||
|
CRUM3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 88) | NC score | 0.022279 (rank : 82) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8QZT4, Q3TJV6 | Gene names | Crb3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Crumbs protein homolog 3 precursor. | |||||
|
EXOC8_HUMAN
|
||||||
| θ value | 5.27518 (rank : 89) | NC score | 0.011360 (rank : 91) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IYI6, Q5TE82 | Gene names | EXOC8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Exocyst complex component 8 (Exocyst complex 84 kDa subunit). | |||||
|
FCRL2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 90) | NC score | 0.121083 (rank : 47) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q96LA5, Q6NTA1, Q9BZI4, Q9BZI5, Q9BZI6 | Gene names | FCRL2, FCRH2, IFGP4, IRTA4, SPAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fc receptor-like protein 2 precursor (SH2 domain-containing phosphatase anchor protein 1) (Fc receptor homolog 2) (FcRH2) (Immunoglobulin receptor translocation-associated 4 protein). | |||||
|
MUC7_HUMAN
|
||||||
| θ value | 5.27518 (rank : 91) | NC score | 0.017103 (rank : 87) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
PFTA_MOUSE
|
||||||
| θ value | 5.27518 (rank : 92) | NC score | 0.008757 (rank : 99) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61239, Q921F7 | Gene names | Fnta | |||
|
Domain Architecture |

|
|||||
| Description | Protein farnesyltransferase/geranylgeranyltransferase type I alpha subunit (EC 2.5.1.58) (EC 2.5.1.59) (CAAX farnesyltransferase alpha subunit) (Ras proteins prenyltransferase alpha) (FTase-alpha) (Type I protein geranyl-geranyltransferase alpha subunit) (GGTase-I-alpha). | |||||
|
PI5PA_MOUSE
|
||||||
| θ value | 5.27518 (rank : 93) | NC score | 0.007332 (rank : 103) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P59644 | Gene names | Pib5pa | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
CNTP4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 94) | NC score | 0.004691 (rank : 111) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9C0A0 | Gene names | CNTNAP4, CASPR4, KIAA1763 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein-like 4 precursor (Cell recognition molecule Caspr4). | |||||
|
FCGR1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 95) | NC score | 0.144037 (rank : 45) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P12314, P12315, Q92495, Q92663 | Gene names | FCGR1A, FCG1, FCGR1, IGFR1 | |||
|
Domain Architecture |
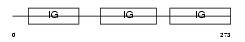
|
|||||
| Description | High affinity immunoglobulin gamma Fc receptor I precursor (Fc-gamma RI) (FcRI) (IgG Fc receptor I) (CD64 antigen). | |||||
|
KIF12_MOUSE
|
||||||
| θ value | 6.88961 (rank : 96) | NC score | 0.000962 (rank : 123) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D2Z8, O35061 | Gene names | Kif12 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF12. | |||||
|
LRC24_HUMAN
|
||||||
| θ value | 6.88961 (rank : 97) | NC score | 0.008938 (rank : 97) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 404 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q50LG9 | Gene names | LRRC24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 24 precursor. | |||||
|
LRRC4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 98) | NC score | 0.004548 (rank : 113) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q99PH1, Q8VI35 | Gene names | Lrrc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing 4 protein precursor (Brain tumor- associated protein MBAG1). | |||||
|
MADCA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 99) | NC score | 0.018200 (rank : 86) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13477, O60222, O75867 | Gene names | MADCAM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucosal addressin cell adhesion molecule 1 precursor (MAdCAM-1) (hMAdCAM-1). | |||||
|
MCAT_HUMAN
|
||||||
| θ value | 6.88961 (rank : 100) | NC score | 0.002126 (rank : 120) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O43772, Q9UIQ2 | Gene names | SLC25A20, CAC, CACT | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial carnitine/acylcarnitine carrier protein (Carnitine/acylcarnitine translocase) (CAC) (Solute carrier family 25 member 20). | |||||
|
MDGA1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 101) | NC score | 0.052047 (rank : 68) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8NFP4, Q8NBE3 | Gene names | MDGA1, MAMDC3 | |||
|
Domain Architecture |

|
|||||
| Description | MAM domain-containing glycosylphosphatidylinositol anchor protein 1 precursor (MAM domain-containing protein 3) (GPI and MAM protein) (Glycosylphosphatidylinositol-MAM) (GPIM). | |||||
|
MINT_MOUSE
|
||||||
| θ value | 6.88961 (rank : 102) | NC score | 0.008564 (rank : 102) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
PKD1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 103) | NC score | 0.003451 (rank : 118) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P98161, Q15140, Q15141 | Gene names | PKD1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-1 precursor (Autosomal dominant polycystic kidney disease protein 1). | |||||
|
RIMB1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 104) | NC score | -0.002919 (rank : 126) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7TNF8, Q5NCP7, Q80TV9, Q8BIH5 | Gene names | Bzrap1, Kiaa0612, Rbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
RIMS2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 105) | NC score | 0.006076 (rank : 104) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UQ26, O43413, Q86XL9, Q8IWV9, Q8IWW1 | Gene names | RIMS2, KIAA0751, RIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 2 (Rab3-interacting molecule 2) (RIM 2). | |||||
|
RIMS2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 106) | NC score | 0.005908 (rank : 105) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9EQZ7, Q8C433, Q8CCK2 | Gene names | Rims2, Rab3ip2, Rim2 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 2 (Rab3-interacting molecule 2) (RIM 2) (Rab3-interacting protein 2). | |||||
|
SLAP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 107) | NC score | 0.001901 (rank : 121) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q60898, Q8C9Q8, Q8CAT0, Q8CBE9, Q8QZX8 | Gene names | Sla, Slap, Slap1 | |||
|
Domain Architecture |
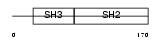
|
|||||
| Description | SRC-like-adapter (Src-like-adapter protein 1) (mSLAP). | |||||
|
ATCAY_MOUSE
|
||||||
| θ value | 8.99809 (rank : 108) | NC score | 0.003613 (rank : 117) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BHE3, Q3TR94 | Gene names | Atcay | |||
|
Domain Architecture |
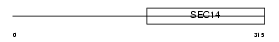
|
|||||
| Description | Caytaxin. | |||||
|
DNL1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 109) | NC score | 0.004044 (rank : 114) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P18858 | Gene names | LIG1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA ligase 1 (EC 6.5.1.1) (DNA ligase I) (Polydeoxyribonucleotide synthase [ATP] 1). | |||||
|
DOK7_HUMAN
|
||||||
| θ value | 8.99809 (rank : 110) | NC score | 0.005554 (rank : 108) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q18PE1, Q6P6A6, Q86XG5, Q8N2J3, Q8NBC1 | Gene names | DOK7, C4orf25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Dok-7 (Downstream of tyrosine kinase 7). | |||||
|
EP400_MOUSE
|
||||||
| θ value | 8.99809 (rank : 111) | NC score | 0.001408 (rank : 122) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
LRRC4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 112) | NC score | 0.004628 (rank : 112) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9HBW1, Q6ZMI8, Q96A85 | Gene names | LRRC4, BAG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 4 precursor (Brain tumor- associated protein LRRC4) (NAG14). | |||||
|
M4K2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 113) | NC score | -0.003790 (rank : 127) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 850 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q12851 | Gene names | MAP4K2, GCK, RAB8IP | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 2 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 2) (MEK kinase kinase 2) (MEKKK 2) (Germinal center kinase) (GC kinase) (Rab8-interacting protein) (B lymphocyte serine/threonine-protein kinase). | |||||
|
NR1H2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 114) | NC score | -0.000369 (rank : 125) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q60644 | Gene names | Nr1h2, Lxrb, Rip15, Unr, Unr2 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterols receptor LXR-beta (Liver X receptor beta) (Nuclear orphan receptor LXR-beta) (Ubiquitously-expressed nuclear receptor) (Retinoid X receptor-interacting protein No.15). | |||||
|
PSG8_HUMAN
|
||||||
| θ value | 8.99809 (rank : 115) | NC score | 0.030400 (rank : 76) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9UQ74, O60410 | Gene names | PSG8 | |||
|
Domain Architecture |
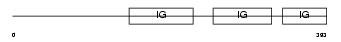
|
|||||
| Description | Pregnancy-specific beta-1-glycoprotein 8 precursor (PSBG-8). | |||||
|
RBM6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 116) | NC score | 0.005402 (rank : 109) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P78332, O60549, O75524 | Gene names | RBM6, DEF3 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 6 (RNA-binding motif protein 6) (RNA-binding protein DEF-3) (Lung cancer antigen NY-LU-12) (Protein G16). | |||||
|
CEAM1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.061664 (rank : 65) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P13688, O60430, Q13858, Q15600, Q9UQV9 | Gene names | CEACAM1, BGP, BGP1 | |||
|
Domain Architecture |
|
|||||
| Description | Carcinoembryonic antigen-related cell adhesion molecule 1 precursor (Biliary glycoprotein 1) (BGP-1) (CD66 antigen) (CD66a antigen). | |||||
|
CEAM5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.064422 (rank : 63) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P06731 | Gene names | CEACAM5, CEA | |||
|
Domain Architecture |

|
|||||
| Description | Carcinoembryonic antigen-related cell adhesion molecule 5 precursor (Carcinoembryonic antigen) (CEA) (Meconium antigen 100) (CD66e antigen). | |||||
|
FCERA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.085358 (rank : 57) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P12319 | Gene names | FCER1A, FCE1A | |||
|
Domain Architecture |
|
|||||
| Description | High affinity immunoglobulin epsilon receptor alpha-subunit precursor (FcERI) (IgE Fc receptor, alpha-subunit) (Fc-epsilon RI-alpha). | |||||
|
FCG2A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.078602 (rank : 58) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P12318 | Gene names | FCGR2A, CD32, FCG2, IGFR2 | |||
|
Domain Architecture |
|
|||||
| Description | Low affinity immunoglobulin gamma Fc region receptor II-a precursor (Fc-gamma RII-a) (FcRII-a) (IgG Fc receptor II-a) (Fc-gamma-RIIa) (CD32 antigen) (CDw32). | |||||
|
FCG2B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.109241 (rank : 49) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P31994, O95649, Q53X85, Q5VXA9, Q8NIA1 | Gene names | FCGR2B, CD32, FCG2, IGFR2 | |||
|
Domain Architecture |
|
|||||
| Description | Low affinity immunoglobulin gamma Fc region receptor II-b precursor (Fc-gamma RII-b) (FcRII-b) (IgG Fc receptor II-b) (Fc-gamma-RIIb) (CD32 antigen) (CDw32). | |||||
|
FCG2C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.107555 (rank : 51) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P31995, O00523, O00524, O00525 | Gene names | FCGR2C, CD32, FCG2, IGFR2 | |||
|
Domain Architecture |
|
|||||
| Description | Low affinity immunoglobulin gamma Fc region receptor II-c precursor (Fc-gamma RII-c) (FcRII-c) (IgG Fc receptor II-c) (Fc-gamma-RIIc) (CD32 antigen) (CDw32). | |||||
|
FCG3A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.066159 (rank : 62) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P08637 | Gene names | FCGR3A, CD16A, FCG3, FCGR3, IGFR3 | |||
|
Domain Architecture |
|
|||||
| Description | Low affinity immunoglobulin gamma Fc region receptor III-A precursor (IgG Fc receptor III-2) (Fc-gamma RIII-alpha) (Fc-gamma RIIIa) (FcRIIIa) (Fc-gamma RIII) (FcRIII) (FcR-10) (CD16a antigen). | |||||
|
FCG3B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.061590 (rank : 66) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O75015 | Gene names | FCGR3B, CD16B, FCG3, FCGR3, IGFR3 | |||
|
Domain Architecture |
|
|||||
| Description | Low affinity immunoglobulin gamma Fc region receptor III-B precursor (IgG Fc receptor III-1) (Fc-gamma RIII-beta) (Fc-gamma RIIIb) (FcRIIIb) (Fc-gamma RIII) (FcRIII) (FcR-10) (CD16b antigen). | |||||
|
MUC18_HUMAN
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.088992 (rank : 55) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P43121, O95812, Q59E86, Q6PHR3, Q6ZTR2 | Gene names | MCAM, MUC18 | |||
|
Domain Architecture |

|
|||||
| Description | Cell surface glycoprotein MUC18 precursor (Melanoma-associated antigen MUC18) (Melanoma cell adhesion molecule) (Melanoma-associated antigen A32) (S-endo 1 endothelial-associated antigen) (Cell surface glycoprotein P1H12) (CD146 antigen). | |||||
|
PECA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.104142 (rank : 52) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P16284, Q6LDA9, Q8TBH1, Q96RF5, Q96RF6, Q9NP65, Q9NPB7, Q9NPG9, Q9NQS9, Q9NQT0, Q9NQT1, Q9NQT2 | Gene names | PECAM1 | |||
|
Domain Architecture |

|
|||||
| Description | Platelet endothelial cell adhesion molecule precursor (PECAM-1) (EndoCAM) (GPIIA') (CD31 antigen). | |||||
|
PECA1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.064316 (rank : 64) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q08481 | Gene names | Pecam1, Pecam, Pecam-1 | |||
|
Domain Architecture |

|
|||||
| Description | Platelet endothelial cell adhesion molecule precursor (PECAM-1) (CD31 antigen). | |||||
|
LIRB1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 116 | |
| SwissProt Accessions | Q8NHL6, O75024, O75025, Q8NHJ9, Q8NHK0 | Gene names | LILRB1, ILT2, LIR1, MIR7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukocyte immunoglobulin-like receptor subfamily B member 1 precursor (Leukocyte immunoglobulin-like receptor 1) (LIR-1) (Immunoglobulin- like transcript 2) (ILT-2) (Monocyte/macrophage immunoglobulin-like receptor 7) (MIR-7) (CD85j antigen). | |||||
|
LIRB2_HUMAN
|
||||||
| NC score | 0.992058 (rank : 2) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q8N423, O75017, Q8NHJ7, Q8NHJ8 | Gene names | LILRB2, ILT4, LIR2, MIR10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukocyte immunoglobulin-like receptor subfamily B member 2 precursor (Leukocyte immunoglobulin-like receptor 2) (LIR-2) (Immunoglobulin- like transcript 4) (ILT-4) (Monocyte/macrophage immunoglobulin-like receptor 10) (MIR-10) (CD85d antigen). | |||||
|
LIRA1_HUMAN
|
||||||
| NC score | 0.990115 (rank : 3) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O75019, O75018 | Gene names | LILRA1, LIR6 | |||
|
Domain Architecture |

|
|||||
| Description | Leukocyte immunoglobulin-like receptor subfamily A member 1 precursor (Leukocyte immunoglobulin-like receptor 6) (LIR-6) (CD85i antigen). | |||||
|
LIRB3_HUMAN
|
||||||
| NC score | 0.989732 (rank : 4) | θ value | 0 (rank : 6) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | O75022, O15471, Q86U49 | Gene names | LILRB3, ILT5, LIR3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukocyte immunoglobulin-like receptor subfamily B member 3 precursor (Leukocyte immunoglobulin-like receptor 3) (LIR-3) (Immunoglobulin- like transcript 5) (ILT-5) (Monocyte inhibitory receptor HL9) (CD85a antigen). | |||||
|
LIRA3_HUMAN
|
||||||
| NC score | 0.987720 (rank : 5) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q8N6C8, O15469, O15470, O75016, Q8N151, Q8N154, Q8NHJ1, Q8NHJ2, Q8NHJ3, Q8NHJ4 | Gene names | LILRA3, ILT6, LIR4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukocyte immunoglobulin-like receptor subfamily A member 3 precursor (Leukocyte immunoglobulin-like receptor 4) (LIR-4) (Immunoglobulin- like transcript 6) (ILT-6) (Monocyte inhibitory receptor HM43/HM31) (CD85e antigen). | |||||
|
LIRB5_HUMAN
|
||||||
| NC score | 0.986058 (rank : 6) | θ value | 1.04281e-181 (rank : 7) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O75023, Q8N760 | Gene names | LILRB5, LIR8 | |||
|
Domain Architecture |

|
|||||
| Description | Leukocyte immunoglobulin-like receptor subfamily B member 5 precursor (Leukocyte immunoglobulin-like receptor 8) (LIR-8) (CD85c antigen). | |||||
|
LIRA4_HUMAN
|
||||||
| NC score | 0.985232 (rank : 7) | θ value | 2.04937e-153 (rank : 8) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P59901 | Gene names | LILRA4, ILT7 | |||
|
Domain Architecture |

|
|||||
| Description | Leukocyte immunoglobulin-like receptor subfamily A member 4 precursor (Immunoglobulin-like transcript 7) (ILT-7) (CD85g antigen). | |||||
|
LIRA2_HUMAN
|
||||||
| NC score | 0.985150 (rank : 8) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8N149, O75020 | Gene names | LILRA2, ILT1, LIR7 | |||
|
Domain Architecture |

|
|||||
| Description | Leukocyte immunoglobulin-like receptor subfamily A member 2 precursor (Leukocyte immunoglobulin-like receptor 7) (LIR-7) (Immunoglobulin- like transcript 1) (ILT-1) (CD85h antigen). | |||||
|
LIRB4_HUMAN
|
||||||
| NC score | 0.968267 (rank : 9) | θ value | 7.09276e-130 (rank : 9) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q8NHJ6, O15468, O75021, Q8N1C7, Q8NHL5 | Gene names | LILRB4, ILT3, LIR5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukocyte immunoglobulin-like receptor subfamily B member 4 precursor (Leukocyte immunoglobulin-like receptor 5) (LIR-5) (Immunoglobulin- like transcript 3) (ILT-3) (Monocyte inhibitory receptor HM18) (CD85k antigen). | |||||
|
LIRB4_MOUSE
|
||||||
| NC score | 0.946490 (rank : 10) | θ value | 6.7944e-48 (rank : 15) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q64281, Q64312 | Gene names | Lilrb4, Gp49b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukocyte immunoglobulin-like receptor subfamily B member 4 precursor (Mast cell surface glycoprotein Gp49B). | |||||
|
GP49A_MOUSE
|
||||||
| NC score | 0.940275 (rank : 11) | θ value | 1.19932e-44 (rank : 18) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q61450 | Gene names | Gp49a, Gp49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mast cell surface glycoprotein Gp49A precursor. | |||||
|
GPVI_HUMAN
|
||||||
| NC score | 0.929360 (rank : 12) | θ value | 7.52953e-39 (rank : 24) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9HCN6, Q9HCN7, Q9UIF2 | Gene names | GP6, GPVI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Platelet glycoprotein VI precursor. | |||||
|
GPVI_MOUSE
|
||||||
| NC score | 0.924378 (rank : 13) | θ value | 1.98606e-31 (rank : 26) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P0C191 | Gene names | Gp6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Platelet glycoprotein VI precursor (GPVI). | |||||
|
NCTR1_HUMAN
|
||||||
| NC score | 0.921390 (rank : 14) | θ value | 4.42448e-31 (rank : 28) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O76036, O76016, O76017, O76018 | Gene names | NCR1, LY94 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Natural cytotoxicity triggering receptor 1 precursor (Natural killer cell p46-related protein) (NKp46) (hNKp46) (NK-p46) (NK cell- activating receptor) (Lymphocyte antigen 94 homolog) (CD335 antigen). | |||||
|
NCTR1_MOUSE
|
||||||
| NC score | 0.915031 (rank : 15) | θ value | 1.98606e-31 (rank : 27) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8C567, Q1RLN4, Q80UY6, Q9Z0Q4 | Gene names | Ncr1, Ly94 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Natural cytotoxicity triggering receptor 1 precursor (Natural killer cell p46-related protein) (NKp46) (mNKp46) (NK-p46) (Mouse-activating receptor 1) (mAR-1) (Lymphocyte antigen 94). | |||||
|
FCAR_HUMAN
|
||||||
| NC score | 0.914666 (rank : 16) | θ value | 5.40856e-29 (rank : 30) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P24071, Q13603, Q13604, Q15727, Q15728, Q92590 | Gene names | FCAR, CD89 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Immunoglobulin alpha Fc receptor precursor (IgA Fc receptor) (CD89 antigen). | |||||
|
KI3L1_MOUSE
|
||||||
| NC score | 0.913424 (rank : 17) | θ value | 1.08474e-45 (rank : 16) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P83555 | Gene names | Kir3dl1 | |||
|
Domain Architecture |

|
|||||
| Description | Killer cell immunoglobulin-like receptor 3DL1 precursor. | |||||
|
KI3L3_HUMAN
|
||||||
| NC score | 0.909056 (rank : 18) | θ value | 4.53632e-60 (rank : 10) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8N743, O95056, Q7Z7N4, Q8NHI2, Q8NHI3, Q9UEI4 | Gene names | KIR3DL3, CD158Z, KIR3DL7, KIRC1 | |||
|
Domain Architecture |

|
|||||
| Description | Killer cell immunoglobulin-like receptor 3DL3 precursor (Killer cell inhibitory receptor 1) (CD158z antigen). | |||||
|
KI3S1_HUMAN
|
||||||
| NC score | 0.905753 (rank : 19) | θ value | 1.08223e-53 (rank : 13) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q14943 | Gene names | KIR3DS1, NKAT10 | |||
|
Domain Architecture |

|
|||||
| Description | Killer cell immunoglobulin-like receptor 3DS1 precursor (MHC class I NK cell receptor) (Natural killer-associated transcript 10) (NKAT-10). | |||||
|
KI3L2_HUMAN
|
||||||
| NC score | 0.902574 (rank : 20) | θ value | 9.45883e-58 (rank : 11) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P43630, Q13238, Q14947, Q14948, Q92684, Q95367 | Gene names | KIR3DL2, CD158K, NKAT4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Killer cell immunoglobulin-like receptor 3DL2 precursor (MHC class I NK cell receptor) (Natural killer-associated transcript 4) (NKAT-4) (p70 natural killer cell receptor clone CL-5) (CD158k antigen). | |||||
|
KI3L1_HUMAN
|
||||||
| NC score | 0.899253 (rank : 21) | θ value | 4.85792e-54 (rank : 12) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P43629, O43473, Q16541 | Gene names | KIR3DL1, NKAT3, NKB1 | |||
|
Domain Architecture |

|
|||||
| Description | Killer cell immunoglobulin-like receptor 3DL1 precursor (MHC class I NK cell receptor) (Natural killer-associated transcript 3) (NKAT-3) (p70 natural killer cell receptor clones CL-2/CL-11) (HLA-BW4-specific inhibitory NK cell receptor). | |||||
|
KI2S2_HUMAN
|
||||||
| NC score | 0.885026 (rank : 22) | θ value | 2.3352e-48 (rank : 14) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P43631, Q14955 | Gene names | KIR2DS2, CD158J, NKAT5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Killer cell immunoglobulin-like receptor 2DS2 precursor (MHC class I NK cell receptor) (Natural killer-associated transcript 5) (NKAT-5) (p58 natural killer cell receptor clone CL-49) (p58 NK receptor) (NK receptor 183 ActI) (CD158j antigen). | |||||
|
KI2S4_HUMAN
|
||||||
| NC score | 0.880080 (rank : 23) | θ value | 2.04573e-44 (rank : 19) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P43632 | Gene names | KIR2DS4, CD158I, KKA3, NKAT8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Killer cell immunoglobulin-like receptor 2DS4 precursor (MHC class I NK cell receptor) (Natural killer-associated transcript 8) (NKAT-8) (P58 natural killer cell receptor clone CL-39) (p58 NK receptor) (CL- 17) (CD158i antigen). | |||||
|
KI2L2_HUMAN
|
||||||
| NC score | 0.879443 (rank : 24) | θ value | 5.38352e-45 (rank : 17) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P43627, Q14951 | Gene names | KIR2DL2, NKAT6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Killer cell immunoglobulin-like receptor 2DL2 precursor (MHC class I NK cell receptor) (Natural killer-associated transcript 6) (NKAT-6) (p58 natural killer cell receptor clone CL-43) (p58 NK receptor). | |||||
|
KI2S3_HUMAN
|
||||||
| NC score | 0.879438 (rank : 25) | θ value | 2.95404e-43 (rank : 21) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q14952, O00644 | Gene names | KIR2DS3, NKAT7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Killer cell immunoglobulin-like receptor 2DS3 precursor (MHC class I NK cell receptor) (Natural killer-associated transcript 7) (NKAT-7). | |||||
|
KI2L4_HUMAN
|
||||||
| NC score | 0.879366 (rank : 26) | θ value | 2.19584e-30 (rank : 29) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q99706, O14621, O14622, O14623, O14624, O43534, P78400, P78401, Q99559, Q99560, Q99561, Q99562, Q9UQJ7 | Gene names | KIR2DL4, CD158D, KIR103AS | |||
|
Domain Architecture |

|
|||||
| Description | Killer cell immunoglobulin-like receptor 2DL4 precursor (MHC class I NK cell receptor KIR103AS) (Killer cell inhibitory receptor 103AS) (KIR-103AS) (G9P) (CD158d antigen). | |||||
|
KI2L1_HUMAN
|
||||||
| NC score | 0.879055 (rank : 27) | θ value | 5.95217e-44 (rank : 20) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P43626, O43470 | Gene names | KIR2DL1, CD158A, NKAT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Killer cell immunoglobulin-like receptor 2DL1 precursor (MHC class I NK cell receptor) (Natural killer-associated transcript 1) (NKAT-1) (p58 natural killer cell receptor clones CL-42/47.11) (p58 NK receptor) (p58.1 MHC class-I-specific NK receptor) (CD158a antigen). | |||||
|
KI2L3_HUMAN
|
||||||
| NC score | 0.878265 (rank : 28) | θ value | 5.03882e-43 (rank : 22) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P43628, O43472, P78402, Q14944, Q14945, Q9UM51, Q9UQ70 | Gene names | KIR2DL3, CD158B, KIRCL23, NKAT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Killer cell immunoglobulin-like receptor 2DL3 precursor (MHC class I NK cell receptor) (Natural killer-associated transcript 2) (NKAT-2) (NKAT2a) (NKAT2b) (p58 natural killer cell receptor clone CL-6) (p58 NK receptor) (p58.2 MHC class-I-specific NK receptor) (Killer inhibitory receptor cl 2-3) (KIR-023GB) (CD158b antigen). | |||||
|
KI2S1_HUMAN
|
||||||
| NC score | 0.876426 (rank : 29) | θ value | 2.76489e-41 (rank : 23) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q14954, O43471 | Gene names | KIR2DS1, CD158H | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Killer cell immunoglobulin-like receptor 2DS1 precursor (MHC class I NK cell receptor Eb6 ActI) (CD158h antigen). | |||||
|
KI2S5_HUMAN
|
||||||
| NC score | 0.875434 (rank : 30) | θ value | 9.83387e-39 (rank : 25) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q14953 | Gene names | KIR2DS5, CD158G, NKAT9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Killer cell immunoglobulin-like receptor 2DS5 precursor (MHC class I NK cell receptor) (Natural killer-associated transcript 9) (NKAT-9) (CD158g antigen). | |||||
|
LAIR2_HUMAN
|
||||||
| NC score | 0.827496 (rank : 31) | θ value | 3.76295e-14 (rank : 33) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q6ISS4, Q6PEZ4 | Gene names | LAIR2, CD306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukocyte-associated immunoglobulin-like receptor 2 precursor (LAIR-2) (CD306 antigen). | |||||
|
A1BG_HUMAN
|
||||||
| NC score | 0.816357 (rank : 32) | θ value | 9.24701e-21 (rank : 31) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P04217, Q8IYJ6, Q96P39 | Gene names | A1BG | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-1B-glycoprotein precursor (Alpha-1-B glycoprotein). | |||||
|
LAIR1_HUMAN
|
||||||
| NC score | 0.781244 (rank : 33) | θ value | 1.16975e-15 (rank : 32) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6GTX8 | Gene names | LAIR1, CD305 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukocyte-associated immunoglobulin-like receptor 1 precursor (LAIR-1) (hLAIR1) (CD305 antigen). | |||||
|
LAIR1_MOUSE
|
||||||
| NC score | 0.477813 (rank : 34) | θ value | 1.81305 (rank : 70) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BG84, Q80UN5, Q8BHB6, Q8BRR6, Q8C6N8, Q8CEP5 | Gene names | Lair1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukocyte-associated immunoglobulin-like receptor 1 precursor (LAIR-1) (mLAIR1) (CD305 antigen). | |||||
|
FCRLA_MOUSE
|
||||||
| NC score | 0.420250 (rank : 35) | θ value | 0.000158464 (rank : 36) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q920A9, Q3T9Q2, Q8K3U9, Q8VHP5 | Gene names | Fcrla, Fcrl, Fcrl1, Fcrlm1, Fcrx, Freb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fc receptor-like and mucin-like 1 precursor (Fc receptor-like A) (Fc receptor homolog expressed in B-cells) (Fc receptor-related protein X) (FcRX) (Fc receptor-like protein). | |||||
|
FCRLA_HUMAN
|
||||||
| NC score | 0.346845 (rank : 36) | θ value | 0.00298849 (rank : 40) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q7L513, Q5VXA1, Q5VXA2, Q5VXA3, Q5VXB0, Q8NEW4, Q8WXH3, Q96PC6, Q96PJ0, Q96PJ1, Q96PJ2, Q96PJ4, Q9BR57 | Gene names | FCRLA, FCRL, FCRL1, FCRLM1, FCRX, FREB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fc receptor-like and mucin-like 1 precursor (Fc receptor-like A) (Fc receptor homolog expressed in B-cells) (Fc receptor-related protein X) (FcRX) (Fc receptor-like protein). | |||||
|
FCGR3_MOUSE
|
||||||
| NC score | 0.228876 (rank : 37) | θ value | 0.0148317 (rank : 44) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P08508 | Gene names | Fcgr3 | |||
|
Domain Architecture |

|
|||||
| Description | Low affinity immunoglobulin gamma Fc region receptor III precursor (IgG Fc receptor III) (Fc-gamma RIII) (FcRIII). | |||||
|
FCGR2_MOUSE
|
||||||
| NC score | 0.224818 (rank : 38) | θ value | 0.00298849 (rank : 39) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P08101, P08102, P12316, P97917, Q60938, Q60939, Q60940, Q61170, Q61558 | Gene names | Fcgr2, Fcgr2b, Ly-17 | |||
|
Domain Architecture |

|
|||||
| Description | Low affinity immunoglobulin gamma Fc region receptor II precursor (Fc- gamma RII) (FcRII) (IgG Fc receptor II beta) (Fc gamma receptor IIB) (Fc-gamma-RIIB) (Lymphocyte antigen Ly-17) (CD32 antigen). | |||||
|
MUC18_MOUSE
|
||||||
| NC score | 0.180126 (rank : 39) | θ value | 0.0113563 (rank : 43) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8R2Y2, Q9EPF1, Q9ESS7, Q9JHQ2, Q9JHQ3 | Gene names | Mcam, Muc18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell surface glycoprotein MUC18 precursor (Melanoma-associated antigen MUC18) (Melanoma cell adhesion molecule) (Gicerin). | |||||
|
CD22_MOUSE
|
||||||
| NC score | 0.180061 (rank : 40) | θ value | 4.92598e-06 (rank : 34) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P35329, Q9JHL2, Q9JJX9, Q9JJY0, Q9JJY1, Q9R056, Q9R094, Q9WU51 | Gene names | Cd22, Lyb-8 | |||
|
Domain Architecture |

|
|||||
| Description | B-cell receptor CD22 precursor (Leu-14) (B-lymphocyte cell adhesion molecule) (BL-CAM) (Siglec-2). | |||||
|
CD276_HUMAN
|
||||||
| NC score | 0.176197 (rank : 41) | θ value | 0.0563607 (rank : 47) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q5ZPR3, Q6P5Y4, Q6UXI2, Q8NBI8, Q8NC34, Q8NCB6, Q9BXR1 | Gene names | CD276, B7H3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CD276 antigen precursor (Costimulatory molecule) (B7 homolog 3) (B7- H3) (4Ig-B7-H3). | |||||
|
FCRL5_HUMAN
|
||||||
| NC score | 0.175562 (rank : 42) | θ value | 0.00509761 (rank : 41) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q96RD9, Q495Q2, Q495Q4, Q5VYK9, Q6UY46 | Gene names | FCRL5, FCRH5, IRTA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fc receptor-like protein 5 precursor (Immunoglobulin receptor translocation-associated gene 2 protein) (BXMAS1) (CD307 antigen). | |||||
|
CD22_HUMAN
|
||||||
| NC score | 0.148957 (rank : 43) | θ value | 0.000786445 (rank : 37) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P20273, O95699, O95701, O95702, O95703, Q01665, Q92872, Q92873, Q9UQA6, Q9UQA7, Q9UQA8, Q9UQA9, Q9UQB0, Q9Y2A6 | Gene names | CD22 | |||
|
Domain Architecture |

|
|||||
| Description | B-cell receptor CD22 precursor (Leu-14) (B-lymphocyte cell adhesion molecule) (BL-CAM) (Siglec-2). | |||||
|
CD276_MOUSE
|
||||||
| NC score | 0.146183 (rank : 44) | θ value | 2.36792 (rank : 73) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8VE98 | Gene names | Cd276, B7h3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CD276 antigen precursor (Costimulatory molecule) (B7 homolog 3) (B7- H3). | |||||
|
FCGR1_HUMAN
|
||||||
| NC score | 0.144037 (rank : 45) | θ value | 6.88961 (rank : 95) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P12314, P12315, Q92495, Q92663 | Gene names | FCGR1A, FCG1, FCGR1, IGFR1 | |||
|
Domain Architecture |

|
|||||
| Description | High affinity immunoglobulin gamma Fc receptor I precursor (Fc-gamma RI) (FcRI) (IgG Fc receptor I) (CD64 antigen). | |||||
|
FCGR1_MOUSE
|
||||||
| NC score | 0.137046 (rank : 46) | θ value | 0.0252991 (rank : 46) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P26151 | Gene names | Fcgr1, Fcg1 | |||
|
Domain Architecture |

|
|||||
| Description | High affinity immunoglobulin gamma Fc receptor I precursor (Fc-gamma RI) (FcRI) (IgG Fc receptor I). | |||||
|
FCRL2_HUMAN
|
||||||
| NC score | 0.121083 (rank : 47) | θ value | 5.27518 (rank : 90) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q96LA5, Q6NTA1, Q9BZI4, Q9BZI5, Q9BZI6 | Gene names | FCRL2, FCRH2, IFGP4, IRTA4, SPAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fc receptor-like protein 2 precursor (SH2 domain-containing phosphatase anchor protein 1) (Fc receptor homolog 2) (FcRH2) (Immunoglobulin receptor translocation-associated 4 protein). | |||||
|
VCAM1_MOUSE
|
||||||
| NC score | 0.115488 (rank : 48) | θ value | 0.125558 (rank : 52) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 320 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P29533 | Gene names | Vcam1, Vcam-1 | |||
|
Domain Architecture |

|
|||||
| Description | Vascular cell adhesion protein 1 precursor (V-CAM 1). | |||||
|
FCG2B_HUMAN
|
||||||
| NC score | 0.109241 (rank : 49) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P31994, O95649, Q53X85, Q5VXA9, Q8NIA1 | Gene names | FCGR2B, CD32, FCG2, IGFR2 | |||
|
Domain Architecture |

|
|||||
| Description | Low affinity immunoglobulin gamma Fc region receptor II-b precursor (Fc-gamma RII-b) (FcRII-b) (IgG Fc receptor II-b) (Fc-gamma-RIIb) (CD32 antigen) (CDw32). | |||||
|
SN_HUMAN
|
||||||
| NC score | 0.108637 (rank : 50) | θ value | 0.0961366 (rank : 50) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9BZZ2, Q96DL4, Q9GZS5, Q9H1H6, Q9H1H7, Q9H7L7 | Gene names | SN | |||
|
Domain Architecture |

|
|||||
| Description | Sialoadhesin precursor (Sialic acid-binding Ig-like lectin-1) (Siglec- 1) (CD169 antigen). | |||||
|
FCG2C_HUMAN
|
||||||
| NC score | 0.107555 (rank : 51) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P31995, O00523, O00524, O00525 | Gene names | FCGR2C, CD32, FCG2, IGFR2 | |||
|
Domain Architecture |

|
|||||
| Description | Low affinity immunoglobulin gamma Fc region receptor II-c precursor (Fc-gamma RII-c) (FcRII-c) (IgG Fc receptor II-c) (Fc-gamma-RIIc) (CD32 antigen) (CDw32). | |||||
|
PECA1_HUMAN
|
||||||
| NC score | 0.104142 (rank : 52) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P16284, Q6LDA9, Q8TBH1, Q96RF5, Q96RF6, Q9NP65, Q9NPB7, Q9NPG9, Q9NQS9, Q9NQT0, Q9NQT1, Q9NQT2 | Gene names | PECAM1 | |||
|
Domain Architecture |

|
|||||
| Description | Platelet endothelial cell adhesion molecule precursor (PECAM-1) (EndoCAM) (GPIIA') (CD31 antigen). | |||||
|
SN_MOUSE
|
||||||
| NC score | 0.102178 (rank : 53) | θ value | 0.47712 (rank : 59) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q62230, O55216, Q62228, Q62229 | Gene names | Sn, Sa | |||
|
Domain Architecture |

|
|||||
| Description | Sialoadhesin precursor (Sialic acid-binding Ig-like lectin-1) (Siglec- 1) (Sheep erythrocyte receptor) (SER) (CD169 antigen). | |||||
|
VCAM1_HUMAN
|
||||||
| NC score | 0.093946 (rank : 54) | θ value | 0.00228821 (rank : 38) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P19320, Q6NUP8 | Gene names | VCAM1, L1CAM | |||
|
Domain Architecture |

|
|||||
| Description | Vascular cell adhesion protein 1 precursor (V-CAM 1) (CD106 antigen) (INCAM-100). | |||||
|
MUC18_HUMAN
|
||||||
| NC score | 0.088992 (rank : 55) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P43121, O95812, Q59E86, Q6PHR3, Q6ZTR2 | Gene names | MCAM, MUC18 | |||
|
Domain Architecture |

|
|||||
| Description | Cell surface glycoprotein MUC18 precursor (Melanoma-associated antigen MUC18) (Melanoma cell adhesion molecule) (Melanoma-associated antigen A32) (S-endo 1 endothelial-associated antigen) (Cell surface glycoprotein P1H12) (CD146 antigen). | |||||
|
CEA20_HUMAN
|
||||||
| NC score | 0.086928 (rank : 56) | θ value | 0.0563607 (rank : 48) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q6UY09 | Gene names | CEACAM20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carcinoembryonic antigen-related cell adhesion molecule 20 precursor. | |||||
|
FCERA_HUMAN
|
||||||
| NC score | 0.085358 (rank : 57) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P12319 | Gene names | FCER1A, FCE1A | |||
|
Domain Architecture |

|
|||||
| Description | High affinity immunoglobulin epsilon receptor alpha-subunit precursor (FcERI) (IgE Fc receptor, alpha-subunit) (Fc-epsilon RI-alpha). | |||||
|
FCG2A_HUMAN
|
||||||
| NC score | 0.078602 (rank : 58) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P12318 | Gene names | FCGR2A, CD32, FCG2, IGFR2 | |||
|
Domain Architecture |

|
|||||
| Description | Low affinity immunoglobulin gamma Fc region receptor II-a precursor (Fc-gamma RII-a) (FcRII-a) (IgG Fc receptor II-a) (Fc-gamma-RIIa) (CD32 antigen) (CDw32). | |||||
|
PGBM_HUMAN
|
||||||
| NC score | 0.078471 (rank : 59) | θ value | 0.163984 (rank : 53) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1113 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P98160, Q16287, Q9H3V5 | Gene names | HSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG) (Perlecan) (PLC). | |||||
|
PGBM_MOUSE
|
||||||
| NC score | 0.076344 (rank : 60) | θ value | 9.29e-05 (rank : 35) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1073 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q05793 | Gene names | Hspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG) (Perlecan) (PLC). | |||||
|
ICAM5_MOUSE
|
||||||
| NC score | 0.071241 (rank : 61) | θ value | 0.125558 (rank : 51) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q60625 | Gene names | Icam5, Tlcn | |||
|
Domain Architecture |

|
|||||
| Description | Intercellular adhesion molecule 5 precursor (ICAM-5) (Telencephalin). | |||||
|
FCG3A_HUMAN
|
||||||
| NC score | 0.066159 (rank : 62) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P08637 | Gene names | FCGR3A, CD16A, FCG3, FCGR3, IGFR3 | |||
|
Domain Architecture |

|
|||||
| Description | Low affinity immunoglobulin gamma Fc region receptor III-A precursor (IgG Fc receptor III-2) (Fc-gamma RIII-alpha) (Fc-gamma RIIIa) (FcRIIIa) (Fc-gamma RIII) (FcRIII) (FcR-10) (CD16a antigen). | |||||
|
CEAM5_HUMAN
|
||||||
| NC score | 0.064422 (rank : 63) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P06731 | Gene names | CEACAM5, CEA | |||
|
Domain Architecture |

|
|||||
| Description | Carcinoembryonic antigen-related cell adhesion molecule 5 precursor (Carcinoembryonic antigen) (CEA) (Meconium antigen 100) (CD66e antigen). | |||||
|
PECA1_MOUSE
|
||||||
| NC score | 0.064316 (rank : 64) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q08481 | Gene names | Pecam1, Pecam, Pecam-1 | |||
|
Domain Architecture |

|
|||||
| Description | Platelet endothelial cell adhesion molecule precursor (PECAM-1) (CD31 antigen). | |||||
|
CEAM1_HUMAN
|
||||||
| NC score | 0.061664 (rank : 65) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P13688, O60430, Q13858, Q15600, Q9UQV9 | Gene names | CEACAM1, BGP, BGP1 | |||
|
Domain Architecture |

|
|||||
| Description | Carcinoembryonic antigen-related cell adhesion molecule 1 precursor (Biliary glycoprotein 1) (BGP-1) (CD66 antigen) (CD66a antigen). | |||||
|
FCG3B_HUMAN
|
||||||
| NC score | 0.061590 (rank : 66) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O75015 | Gene names | FCGR3B, CD16B, FCG3, FCGR3, IGFR3 | |||
|
Domain Architecture |

|
|||||
| Description | Low affinity immunoglobulin gamma Fc region receptor III-B precursor (IgG Fc receptor III-1) (Fc-gamma RIII-beta) (Fc-gamma RIIIb) (FcRIIIb) (Fc-gamma RIII) (FcRIII) (FcR-10) (CD16b antigen). | |||||
|
MAMC1_MOUSE
|
||||||
| NC score | 0.056482 (rank : 67) | θ value | 0.00665767 (rank : 42) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P60755 | Gene names | Mamdc1, Mdga2 | |||
|
Domain Architecture |

|
|||||
| Description | MAM domain-containing protein 1 precursor (MAM domain-containing glycosylphosphatidylinositol anchor protein 2). | |||||
|
MDGA1_HUMAN
|
||||||
| NC score | 0.052047 (rank : 68) | θ value | 6.88961 (rank : 101) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8NFP4, Q8NBE3 | Gene names | MDGA1, MAMDC3 | |||
|
Domain Architecture |

|
|||||
| Description | MAM domain-containing glycosylphosphatidylinositol anchor protein 1 precursor (MAM domain-containing protein 3) (GPI and MAM protein) (Glycosylphosphatidylinositol-MAM) (GPIM). | |||||
|
CD166_MOUSE
|
||||||
| NC score | 0.047588 (rank : 69) | θ value | 4.03905 (rank : 80) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q61490, O70136, Q8CDA5, Q8R2T0 | Gene names | Alcam | |||
|
Domain Architecture |

|
|||||
| Description | CD166 antigen precursor (Activated leukocyte-cell adhesion molecule) (ALCAM) (DM-GRASP protein). | |||||
|
CEAM8_HUMAN
|
||||||
| NC score | 0.042353 (rank : 70) | θ value | 0.0563607 (rank : 49) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P31997, O60399, Q16574 | Gene names | CEACAM8, CGM6 | |||
|
Domain Architecture |

|
|||||
| Description | Carcinoembryonic antigen-related cell adhesion molecule 8 precursor (Carcinoembryonic antigen CGM6) (Nonspecific cross-reacting antigen NCA-95) (CD67 antigen) (CD66b antigen). | |||||
|
NEGR1_MOUSE
|
||||||
| NC score | 0.040554 (rank : 71) | θ value | 4.03905 (rank : 83) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q80Z24, Q3UHQ8, Q80T70 | Gene names | Negr1, Kiaa3001, Ntra | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal growth regulator 1 precursor (Kilon protein) (Kindred of IgLON) (Neurotractin). | |||||
|
NEGR1_HUMAN
|
||||||
| NC score | 0.039006 (rank : 72) | θ value | 4.03905 (rank : 82) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q7Z3B1, Q5VT21, Q6UY06, Q8NAQ3 | Gene names | NEGR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal growth regulator 1 precursor. | |||||
|
HMCN1_HUMAN
|
||||||
| NC score | 0.034287 (rank : 73) | θ value | 3.0926 (rank : 78) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 983 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q96RW7, Q5TYR7, Q96DN3, Q96DN8, Q96SC3 | Gene names | HMCN1, FIBL6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hemicentin-1 precursor (Fibulin-6) (FIBL-6). | |||||
|
CN155_HUMAN
|
||||||
| NC score | 0.031610 (rank : 74) | θ value | 0.21417 (rank : 54) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
CNTN1_HUMAN
|
||||||
| NC score | 0.030472 (rank : 75) | θ value | 1.06291 (rank : 67) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q12860, Q12861, Q14030, Q8N466 | Gene names | CNTN1 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-1 precursor (Neural cell surface protein F3) (Glycoprotein gp135). | |||||
|
PSG8_HUMAN
|
||||||
| NC score | 0.030400 (rank : 76) | θ value | 8.99809 (rank : 115) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9UQ74, O60410 | Gene names | PSG8 | |||
|
Domain Architecture |

|
|||||
| Description | Pregnancy-specific beta-1-glycoprotein 8 precursor (PSBG-8). | |||||
|
JPH2_MOUSE
|
||||||
| NC score | 0.029576 (rank : 77) | θ value | 1.81305 (rank : 69) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9ET78, Q9ET79 | Gene names | Jph2, Jp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
TITIN_HUMAN
|
||||||
| NC score | 0.028330 (rank : 78) | θ value | 0.21417 (rank : 57) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
CCD1A_HUMAN
|
||||||
| NC score | 0.028160 (rank : 79) | θ value | 0.62314 (rank : 62) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6P1N0, Q7Z435, Q86XV0, Q8NF89, Q9H603, Q9NXI1 | Gene names | CC2D1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil and C2 domain-containing protein 1A (Five repressor element under dual repression-binding protein 1) (FRE under dual repression-binding protein 1) (Freud-1) (Putative NF-kappa-B- activating protein 023N). | |||||
|
CNOT3_HUMAN
|
||||||
| NC score | 0.027475 (rank : 80) | θ value | 0.62314 (rank : 63) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O75175, Q9NZN7, Q9UF76 | Gene names | CNOT3, KIAA0691, NOT3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
CNTN1_MOUSE
|
||||||
| NC score | 0.025569 (rank : 81) | θ value | 4.03905 (rank : 81) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P12960, Q6NXV7, Q8BR42, Q8C6A0 | Gene names | Cntn1 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-1 precursor (Neural cell surface protein F3). | |||||
|
CRUM3_MOUSE
|
||||||
| NC score | 0.022279 (rank : 82) | θ value | 5.27518 (rank : 88) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8QZT4, Q3TJV6 | Gene names | Crb3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Crumbs protein homolog 3 precursor. | |||||
|
CNOT3_MOUSE
|
||||||
| NC score | 0.022181 (rank : 83) | θ value | 1.38821 (rank : 68) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K0V4 | Gene names | Cnot3, Not3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
PTK7_HUMAN
|
||||||
| NC score | 0.020308 (rank : 84) | θ value | 0.21417 (rank : 55) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1103 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q13308, Q13417 | Gene names | PTK7, CCK4 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase-like 7 precursor (Colon carcinoma kinase 4) (CCK-4). | |||||
|
SELPL_MOUSE
|
||||||
| NC score | 0.018646 (rank : 85) | θ value | 0.21417 (rank : 56) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q62170 | Gene names | Selplg, Selp1, Selpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand). | |||||
|
MADCA_HUMAN
|
||||||
| NC score | 0.018200 (rank : 86) | θ value | 6.88961 (rank : 99) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13477, O60222, O75867 | Gene names | MADCAM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucosal addressin cell adhesion molecule 1 precursor (MAdCAM-1) (hMAdCAM-1). | |||||
|
MUC7_HUMAN
|
||||||
| NC score | 0.017103 (rank : 87) | θ value | 5.27518 (rank : 91) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
NUCL_MOUSE
|
||||||
| NC score | 0.014136 (rank : 88) | θ value | 0.62314 (rank : 64) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P09405, Q548M9, Q61991, Q99K50 | Gene names | Ncl, Nuc | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolin (Protein C23). | |||||
|
BT2A1_HUMAN
|
||||||
| NC score | 0.013968 (rank : 89) | θ value | 0.0252991 (rank : 45) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q7KYR7, O00475, P78408, Q7KYQ7, Q7Z386, Q9NU62 | Gene names | BTN2A1, BT2.1, BTF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Butyrophilin subfamily 2 member A1 precursor. | |||||
|
JPH2_HUMAN
|
||||||
| NC score | 0.013233 (rank : 90) | θ value | 0.813845 (rank : 66) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
EXOC8_HUMAN
|
||||||
| NC score | 0.011360 (rank : 91) | θ value | 5.27518 (rank : 89) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IYI6, Q5TE82 | Gene names | EXOC8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Exocyst complex component 8 (Exocyst complex 84 kDa subunit). | |||||
|
EPN3_HUMAN
|
||||||
| NC score | 0.011045 (rank : 92) | θ value | 0.813845 (rank : 65) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H201, Q9BVN6, Q9NWK2 | Gene names | EPN3 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-3 (EPS-15-interacting protein 3). | |||||
|
RIN3_HUMAN
|
||||||
| NC score | 0.011041 (rank : 93) | θ value | 2.36792 (rank : 75) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8TB24, Q8NF30, Q8TEE8, Q8WYP4, Q9H6A5, Q9HAG1 | Gene names | RIN3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras and Rab interactor 3 (Ras interaction/interference protein 3). | |||||
|
CCNK_MOUSE
|
||||||
| NC score | 0.010939 (rank : 94) | θ value | 5.27518 (rank : 87) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O88874, Q8R068 | Gene names | Ccnk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-K. | |||||
|
UBP43_MOUSE
|
||||||
| NC score | 0.009805 (rank : 95) | θ value | 2.36792 (rank : 77) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BUM9, Q8VDP5 | Gene names | Usp43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 43 (EC 3.1.2.15) (Ubiquitin thioesterase 43) (Ubiquitin-specific-processing protease 43) (Deubiquitinating enzyme 43). | |||||
|
PJA1_MOUSE
|
||||||
| NC score | 0.009619 (rank : 96) | θ value | 1.81305 (rank : 71) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O55176, Q8CFU2, Q99MJ1, Q99MJ2, Q99MJ3, Q9DB04 | Gene names | Pja1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin protein ligase Praja1 (EC 6.3.2.-). | |||||
|
LRC24_HUMAN
|
||||||
| NC score | 0.008938 (rank : 97) | θ value | 6.88961 (rank : 97) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 404 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q50LG9 | Gene names | LRRC24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 24 precursor. | |||||
|
RTN4_MOUSE
|
||||||
| NC score | 0.008777 (rank : 98) | θ value | 4.03905 (rank : 86) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99P72, Q5DTK9, Q7TNB7, Q80W95, Q8BGK7, Q8BGM9, Q8K290, Q8K3G8, Q9CTE3 | Gene names | Rtn4, Kiaa0886, Nogo | |||
|
Domain Architecture |

|
|||||
| Description | Reticulon-4 (Neurite outgrowth inhibitor) (Nogo protein). | |||||
|
PFTA_MOUSE
|
||||||
| NC score | 0.008757 (rank : 99) | θ value | 5.27518 (rank : 92) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61239, Q921F7 | Gene names | Fnta | |||
|
Domain Architecture |

|
|||||
| Description | Protein farnesyltransferase/geranylgeranyltransferase type I alpha subunit (EC 2.5.1.58) (EC 2.5.1.59) (CAAX farnesyltransferase alpha subunit) (Ras proteins prenyltransferase alpha) (FTase-alpha) (Type I protein geranyl-geranyltransferase alpha subunit) (GGTase-I-alpha). | |||||
|
PTN22_MOUSE
|
||||||
| NC score | 0.008733 (rank : 100) | θ value | 1.81305 (rank : 72) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P29352, Q7TMP9 | Gene names | Ptpn22, Ptpn8 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 22 (EC 3.1.3.48) (Hematopoietic cell protein-tyrosine phosphatase 70Z-PEP). | |||||
|
PTPRU_HUMAN
|
||||||
| NC score | 0.008612 (rank : 101) | θ value | 2.36792 (rank : 74) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q92729 | Gene names | PTPRU, PCP2 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase U precursor (EC 3.1.3.48) (R-PTP-U) (Protein-tyrosine phosphatase J) (PTP-J) (Pancreatic carcinoma phosphatase 2) (PCP-2). | |||||
|
MINT_MOUSE
|
||||||
| NC score | 0.008564 (rank : 102) | θ value | 6.88961 (rank : 102) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
PI5PA_MOUSE
|
||||||
| NC score | 0.007332 (rank : 103) | θ value | 5.27518 (rank : 93) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P59644 | Gene names | Pib5pa | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
RIMS2_HUMAN
|
||||||
| NC score | 0.006076 (rank : 104) | θ value | 6.88961 (rank : 105) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UQ26, O43413, Q86XL9, Q8IWV9, Q8IWW1 | Gene names | RIMS2, KIAA0751, RIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 2 (Rab3-interacting molecule 2) (RIM 2). | |||||
|
RIMS2_MOUSE
|
||||||
| NC score | 0.005908 (rank : 105) | θ value | 6.88961 (rank : 106) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9EQZ7, Q8C433, Q8CCK2 | Gene names | Rims2, Rab3ip2, Rim2 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 2 (Rab3-interacting molecule 2) (RIM 2) (Rab3-interacting protein 2). | |||||
|
LR37B_HUMAN
|
||||||
| NC score | 0.005652 (rank : 106) | θ value | 3.0926 (rank : 79) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96QE4, Q5YKG6 | Gene names | LRRC37B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 37B precursor (C66 SLIT-like testicular protein). | |||||
|
OXR1_HUMAN
|
||||||
| NC score | 0.005637 (rank : 107) | θ value | 4.03905 (rank : 85) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N573, Q3LIB5, Q6ZVK9, Q8N8V0, Q9H266, Q9NWC7 | Gene names | OXR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oxidation resistance protein 1. | |||||
|
DOK7_HUMAN
|
||||||
| NC score | 0.005554 (rank : 108) | θ value | 8.99809 (rank : 110) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q18PE1, Q6P6A6, Q86XG5, Q8N2J3, Q8NBC1 | Gene names | DOK7, C4orf25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Dok-7 (Downstream of tyrosine kinase 7). | |||||
|
RBM6_HUMAN
|
||||||
| NC score | 0.005402 (rank : 109) | θ value | 8.99809 (rank : 116) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P78332, O60549, O75524 | Gene names | RBM6, DEF3 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 6 (RNA-binding motif protein 6) (RNA-binding protein DEF-3) (Lung cancer antigen NY-LU-12) (Protein G16). | |||||
|
ATBF1_HUMAN
|
||||||
| NC score | 0.004882 (rank : 110) | θ value | 0.62314 (rank : 60) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1547 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15911, O15101, Q13719 | Gene names | ATBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
CNTP4_HUMAN
|
||||||
| NC score | 0.004691 (rank : 111) | θ value | 6.88961 (rank : 94) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9C0A0 | Gene names | CNTNAP4, CASPR4, KIAA1763 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein-like 4 precursor (Cell recognition molecule Caspr4). | |||||
|
LRRC4_HUMAN
|
||||||
| NC score | 0.004628 (rank : 112) | θ value | 8.99809 (rank : 112) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9HBW1, Q6ZMI8, Q96A85 | Gene names | LRRC4, BAG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 4 precursor (Brain tumor- associated protein LRRC4) (NAG14). | |||||
|
LRRC4_MOUSE
|
||||||
| NC score | 0.004548 (rank : 113) | θ value | 6.88961 (rank : 98) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q99PH1, Q8VI35 | Gene names | Lrrc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing 4 protein precursor (Brain tumor- associated protein MBAG1). | |||||
|
DNL1_HUMAN
|
||||||
| NC score | 0.004044 (rank : 114) | θ value | 8.99809 (rank : 109) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P18858 | Gene names | LIG1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA ligase 1 (EC 6.5.1.1) (DNA ligase I) (Polydeoxyribonucleotide synthase [ATP] 1). | |||||
|
ATBF1_MOUSE
|
||||||
| NC score | 0.004029 (rank : 115) | θ value | 0.62314 (rank : 61) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
PCDGF_HUMAN
|
||||||
| NC score | 0.003909 (rank : 116) | θ value | 0.365318 (rank : 58) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y5G1, Q9Y5C7 | Gene names | PCDHGB3 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma B3 precursor (PCDH-gamma-B3). | |||||
|
ATCAY_MOUSE
|
||||||
| NC score | 0.003613 (rank : 117) | θ value | 8.99809 (rank : 108) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BHE3, Q3TR94 | Gene names | Atcay | |||
|
Domain Architecture |

|
|||||
| Description | Caytaxin. | |||||
|
PKD1_HUMAN
|
||||||
| NC score | 0.003451 (rank : 118) | θ value | 6.88961 (rank : 103) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P98161, Q15140, Q15141 | Gene names | PKD1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-1 precursor (Autosomal dominant polycystic kidney disease protein 1). | |||||
|
SEM6C_HUMAN
|
||||||
| NC score | 0.003203 (rank : 119) | θ value | 2.36792 (rank : 76) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H3T2, Q5JR71, Q8WXT8, Q8WXT9, Q8WXU0, Q96JF8 | Gene names | SEMA6C, KIAA1869, SEMAY | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6C precursor (Semaphorin Y) (Sema Y). | |||||
|
MCAT_HUMAN
|
||||||
| NC score | 0.002126 (rank : 120) | θ value | 6.88961 (rank : 100) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O43772, Q9UIQ2 | Gene names | SLC25A20, CAC, CACT | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial carnitine/acylcarnitine carrier protein (Carnitine/acylcarnitine translocase) (CAC) (Solute carrier family 25 member 20). | |||||
|
SLAP1_MOUSE
|
||||||
| NC score | 0.001901 (rank : 121) | θ value | 6.88961 (rank : 107) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q60898, Q8C9Q8, Q8CAT0, Q8CBE9, Q8QZX8 | Gene names | Sla, Slap, Slap1 | |||
|
Domain Architecture |

|
|||||
| Description | SRC-like-adapter (Src-like-adapter protein 1) (mSLAP). | |||||
|
EP400_MOUSE
|
||||||
| NC score | 0.001408 (rank : 122) | θ value | 8.99809 (rank : 111) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
KIF12_MOUSE
|
||||||
| NC score | 0.000962 (rank : 123) | θ value | 6.88961 (rank : 96) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D2Z8, O35061 | Gene names | Kif12 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF12. | |||||
|
NFH_MOUSE
|
||||||
| NC score | -0.000339 (rank : 124) | θ value | 4.03905 (rank : 84) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
NR1H2_MOUSE
|
||||||
| NC score | -0.000369 (rank : 125) | θ value | 8.99809 (rank : 114) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q60644 | Gene names | Nr1h2, Lxrb, Rip15, Unr, Unr2 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterols receptor LXR-beta (Liver X receptor beta) (Nuclear orphan receptor LXR-beta) (Ubiquitously-expressed nuclear receptor) (Retinoid X receptor-interacting protein No.15). | |||||
|
RIMB1_MOUSE
|
||||||
| NC score | -0.002919 (rank : 126) | θ value | 6.88961 (rank : 104) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7TNF8, Q5NCP7, Q80TV9, Q8BIH5 | Gene names | Bzrap1, Kiaa0612, Rbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
M4K2_HUMAN
|
||||||
| NC score | -0.003790 (rank : 127) | θ value | 8.99809 (rank : 113) | |||
| Query Neighborhood Hits | 116 | Target Neighborhood Hits | 850 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q12851 | Gene names | MAP4K2, GCK, RAB8IP | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 2 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 2) (MEK kinase kinase 2) (MEKKK 2) (Germinal center kinase) (GC kinase) (Rab8-interacting protein) (B lymphocyte serine/threonine-protein kinase). | |||||