Please be patient as the page loads
|
LARGE_MOUSE
|
||||||
| SwissProt Accessions | Q9Z1M7, Q497S9, Q6P7U2, Q80TW0 | Gene names | Large, Kiaa0609, Large1 | |||
|
Domain Architecture |

|
|||||
| Description | Glycosyltransferase-like protein LARGE1 (EC 2.4.-.-) (Acetylglucosaminyltransferase-like 1A). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
LARG2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.992655 (rank : 3) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8N3Y3, Q8N8Y6, Q8NAK3, Q8WY62 | Gene names | GYLTL1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glycosyltransferase-like protein LARGE2 (EC 2.4.-.-) (Glycosyltransferase-like 1B). | |||||
|
LARG2_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.992108 (rank : 4) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5XPT3, Q5XPT2, Q8BJZ8, Q8K253 | Gene names | Gyltl1b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glycosyltransferase-like protein LARGE2 (EC 2.4.-.-) (Glycosyltransferase-like 1B). | |||||
|
LARGE_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.999354 (rank : 2) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O95461, O60348, Q9UGD1, Q9UGE7, Q9UGG3, Q9UGZ8, Q9UH22 | Gene names | LARGE, KIAA0609, LARGE1 | |||
|
Domain Architecture |

|
|||||
| Description | Glycosyltransferase-like protein LARGE1 (EC 2.4.-.-) (Acetylglucosaminyltransferase-like 1A). | |||||
|
LARGE_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Z1M7, Q497S9, Q6P7U2, Q80TW0 | Gene names | Large, Kiaa0609, Large1 | |||
|
Domain Architecture |

|
|||||
| Description | Glycosyltransferase-like protein LARGE1 (EC 2.4.-.-) (Acetylglucosaminyltransferase-like 1A). | |||||
|
B3GN6_MOUSE
|
||||||
| θ value | 1.02001e-27 (rank : 5) | NC score | 0.714798 (rank : 5) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BWP8, Q3TY43, Q8BJH9, Q99LW7 | Gene names | B3gnt6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase (EC 2.4.1.149) (Poly-N-acetyllactosamine extension enzyme) (I-beta- 1,3-N-acetylglucosaminyltransferase) (iGnT) (UDP-GlcNAc:betaGal beta- 1,3-N-acetylglucosaminyltransferase 6). | |||||
|
B3GN6_HUMAN
|
||||||
| θ value | 1.92365e-26 (rank : 6) | NC score | 0.703910 (rank : 6) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43505, Q4TTN0 | Gene names | B3GNT6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase (EC 2.4.1.149) (Poly-N-acetyllactosamine extension enzyme) (I-beta- 1,3-N-acetylglucosaminyltransferase) (iGnT) (UDP-GlcNAc:betaGal beta- 1,3-N-acetylglucosaminyltransferase 6). | |||||
|
CC021_HUMAN
|
||||||
| θ value | 9.26847e-13 (rank : 7) | NC score | 0.541273 (rank : 7) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NBI6, Q8NAL3, Q8WV03, Q96ME0 | Gene names | C3orf21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C3orf21. | |||||
|
CC021_MOUSE
|
||||||
| θ value | 3.52202e-12 (rank : 8) | NC score | 0.535783 (rank : 8) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q3U4G3, Q3TLX5, Q8K2I0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C3orf21 homolog. | |||||
|
RTN2_MOUSE
|
||||||
| θ value | 0.21417 (rank : 9) | NC score | 0.049594 (rank : 9) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O70622, O70620 | Gene names | Rtn2, Nspl1 | |||
|
Domain Architecture |

|
|||||
| Description | Reticulon-2 (Neuroendocrine-specific protein-like 1) (NSP-like protein 1) (NSPLI). | |||||
|
ZN644_HUMAN
|
||||||
| θ value | 0.62314 (rank : 10) | NC score | 0.013737 (rank : 16) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H582, Q5TCC0, Q6BEP7, Q6P446, Q6PI06, Q7LG67, Q9ULJ9 | Gene names | ZNF644, KIAA1221, ZEP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 644 (Zinc finger motif enhancer-binding protein 2) (Zep-2). | |||||
|
MYOC_HUMAN
|
||||||
| θ value | 0.813845 (rank : 11) | NC score | 0.025702 (rank : 14) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 489 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99972, O00620, Q7Z6Q9 | Gene names | MYOC, GLC1A, TIGR | |||
|
Domain Architecture |

|
|||||
| Description | Myocilin precursor (Trabecular meshwork-induced glucocorticoid response protein). | |||||
|
RF1M_HUMAN
|
||||||
| θ value | 1.38821 (rank : 12) | NC score | 0.046605 (rank : 10) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75570, Q5T6Y5, Q8IUQ6 | Gene names | MTRF1 | |||
|
Domain Architecture |
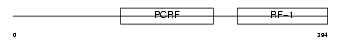
|
|||||
| Description | Peptide chain release factor 1, mitochondrial precursor (MRF-1). | |||||
|
PERI_HUMAN
|
||||||
| θ value | 1.81305 (rank : 13) | NC score | 0.007727 (rank : 18) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P41219, Q8N577 | Gene names | PRPH | |||
|
Domain Architecture |
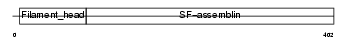
|
|||||
| Description | Peripherin. | |||||
|
PERI_MOUSE
|
||||||
| θ value | 1.81305 (rank : 14) | NC score | 0.007678 (rank : 19) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P15331, O35688, O35689 | Gene names | Prph, Prph1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripherin. | |||||
|
UGGG1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 15) | NC score | 0.030873 (rank : 12) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6P5E4, Q6NV70 | Gene names | Ugcgl1, Gt, Uggt, Ugt1, Ugtr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UDP-glucose:glycoprotein glucosyltransferase 1 precursor (EC 2.4.1.-) (UDP-glucose ceramide glucosyltransferase-like 1) (UDP-- Glc:glycoprotein glucosyltransferase). | |||||
|
RF1M_MOUSE
|
||||||
| θ value | 3.0926 (rank : 16) | NC score | 0.040148 (rank : 11) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K126 | Gene names | Mtrf1 | |||
|
Domain Architecture |
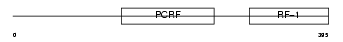
|
|||||
| Description | Peptide chain release factor 1, mitochondrial precursor (MRF-1). | |||||
|
CK5P2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 17) | NC score | 0.004634 (rank : 21) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96SN8, Q7Z3L4, Q7Z3U1, Q7Z7I6, Q9BSW0, Q9H6J6, Q9HCD9, Q9NV90, Q9UIW9 | Gene names | CDK5RAP2, CEP215, KIAA1633 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48) (Centrosome-associated protein 215). | |||||
|
FPRP_MOUSE
|
||||||
| θ value | 4.03905 (rank : 18) | NC score | 0.018001 (rank : 15) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9WV91 | Gene names | Ptgfrn, Fprp | |||
|
Domain Architecture |

|
|||||
| Description | Prostaglandin F2 receptor negative regulator precursor (Prostaglandin F2-alpha receptor regulatory protein) (Prostaglandin F2-alpha receptor-associated protein) (CD315 antigen). | |||||
|
UGGG1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 19) | NC score | 0.028444 (rank : 13) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NYU2, Q8IW30, Q9H8I4 | Gene names | UGCGL1, GT, UGGT, UGT1, UGTR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UDP-glucose:glycoprotein glucosyltransferase 1 precursor (EC 2.4.1.-) (UDP-glucose ceramide glucosyltransferase-like 1) (UDP-- Glc:glycoprotein glucosyltransferase) (HUGT1). | |||||
|
BIRC6_HUMAN
|
||||||
| θ value | 5.27518 (rank : 20) | NC score | 0.011970 (rank : 17) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NR09, Q9ULD1 | Gene names | BIRC6, KIAA1289 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 6 (Ubiquitin-conjugating BIR-domain enzyme apollon). | |||||
|
EVL_MOUSE
|
||||||
| θ value | 8.99809 (rank : 21) | NC score | 0.006702 (rank : 20) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P70429, Q9ERU8 | Gene names | Evl | |||
|
Domain Architecture |

|
|||||
| Description | Ena/VASP-like protein (Ena/vasodilator-stimulated phosphoprotein- like). | |||||
|
LARGE_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Z1M7, Q497S9, Q6P7U2, Q80TW0 | Gene names | Large, Kiaa0609, Large1 | |||
|
Domain Architecture |

|
|||||
| Description | Glycosyltransferase-like protein LARGE1 (EC 2.4.-.-) (Acetylglucosaminyltransferase-like 1A). | |||||
|
LARGE_HUMAN
|
||||||
| NC score | 0.999354 (rank : 2) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O95461, O60348, Q9UGD1, Q9UGE7, Q9UGG3, Q9UGZ8, Q9UH22 | Gene names | LARGE, KIAA0609, LARGE1 | |||
|
Domain Architecture |

|
|||||
| Description | Glycosyltransferase-like protein LARGE1 (EC 2.4.-.-) (Acetylglucosaminyltransferase-like 1A). | |||||
|
LARG2_HUMAN
|
||||||
| NC score | 0.992655 (rank : 3) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8N3Y3, Q8N8Y6, Q8NAK3, Q8WY62 | Gene names | GYLTL1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glycosyltransferase-like protein LARGE2 (EC 2.4.-.-) (Glycosyltransferase-like 1B). | |||||
|
LARG2_MOUSE
|
||||||
| NC score | 0.992108 (rank : 4) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5XPT3, Q5XPT2, Q8BJZ8, Q8K253 | Gene names | Gyltl1b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glycosyltransferase-like protein LARGE2 (EC 2.4.-.-) (Glycosyltransferase-like 1B). | |||||
|
B3GN6_MOUSE
|
||||||
| NC score | 0.714798 (rank : 5) | θ value | 1.02001e-27 (rank : 5) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BWP8, Q3TY43, Q8BJH9, Q99LW7 | Gene names | B3gnt6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase (EC 2.4.1.149) (Poly-N-acetyllactosamine extension enzyme) (I-beta- 1,3-N-acetylglucosaminyltransferase) (iGnT) (UDP-GlcNAc:betaGal beta- 1,3-N-acetylglucosaminyltransferase 6). | |||||
|
B3GN6_HUMAN
|
||||||
| NC score | 0.703910 (rank : 6) | θ value | 1.92365e-26 (rank : 6) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43505, Q4TTN0 | Gene names | B3GNT6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase (EC 2.4.1.149) (Poly-N-acetyllactosamine extension enzyme) (I-beta- 1,3-N-acetylglucosaminyltransferase) (iGnT) (UDP-GlcNAc:betaGal beta- 1,3-N-acetylglucosaminyltransferase 6). | |||||
|
CC021_HUMAN
|
||||||
| NC score | 0.541273 (rank : 7) | θ value | 9.26847e-13 (rank : 7) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NBI6, Q8NAL3, Q8WV03, Q96ME0 | Gene names | C3orf21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C3orf21. | |||||
|
CC021_MOUSE
|
||||||
| NC score | 0.535783 (rank : 8) | θ value | 3.52202e-12 (rank : 8) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q3U4G3, Q3TLX5, Q8K2I0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C3orf21 homolog. | |||||
|
RTN2_MOUSE
|
||||||
| NC score | 0.049594 (rank : 9) | θ value | 0.21417 (rank : 9) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O70622, O70620 | Gene names | Rtn2, Nspl1 | |||
|
Domain Architecture |

|
|||||
| Description | Reticulon-2 (Neuroendocrine-specific protein-like 1) (NSP-like protein 1) (NSPLI). | |||||
|
RF1M_HUMAN
|
||||||
| NC score | 0.046605 (rank : 10) | θ value | 1.38821 (rank : 12) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75570, Q5T6Y5, Q8IUQ6 | Gene names | MTRF1 | |||
|
Domain Architecture |

|
|||||
| Description | Peptide chain release factor 1, mitochondrial precursor (MRF-1). | |||||
|
RF1M_MOUSE
|
||||||
| NC score | 0.040148 (rank : 11) | θ value | 3.0926 (rank : 16) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K126 | Gene names | Mtrf1 | |||
|
Domain Architecture |

|
|||||
| Description | Peptide chain release factor 1, mitochondrial precursor (MRF-1). | |||||
|
UGGG1_MOUSE
|
||||||
| NC score | 0.030873 (rank : 12) | θ value | 2.36792 (rank : 15) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6P5E4, Q6NV70 | Gene names | Ugcgl1, Gt, Uggt, Ugt1, Ugtr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UDP-glucose:glycoprotein glucosyltransferase 1 precursor (EC 2.4.1.-) (UDP-glucose ceramide glucosyltransferase-like 1) (UDP-- Glc:glycoprotein glucosyltransferase). | |||||
|
UGGG1_HUMAN
|
||||||
| NC score | 0.028444 (rank : 13) | θ value | 4.03905 (rank : 19) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NYU2, Q8IW30, Q9H8I4 | Gene names | UGCGL1, GT, UGGT, UGT1, UGTR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UDP-glucose:glycoprotein glucosyltransferase 1 precursor (EC 2.4.1.-) (UDP-glucose ceramide glucosyltransferase-like 1) (UDP-- Glc:glycoprotein glucosyltransferase) (HUGT1). | |||||
|
MYOC_HUMAN
|
||||||
| NC score | 0.025702 (rank : 14) | θ value | 0.813845 (rank : 11) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 489 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99972, O00620, Q7Z6Q9 | Gene names | MYOC, GLC1A, TIGR | |||
|
Domain Architecture |

|
|||||
| Description | Myocilin precursor (Trabecular meshwork-induced glucocorticoid response protein). | |||||
|
FPRP_MOUSE
|
||||||
| NC score | 0.018001 (rank : 15) | θ value | 4.03905 (rank : 18) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9WV91 | Gene names | Ptgfrn, Fprp | |||
|
Domain Architecture |

|
|||||
| Description | Prostaglandin F2 receptor negative regulator precursor (Prostaglandin F2-alpha receptor regulatory protein) (Prostaglandin F2-alpha receptor-associated protein) (CD315 antigen). | |||||
|
ZN644_HUMAN
|
||||||
| NC score | 0.013737 (rank : 16) | θ value | 0.62314 (rank : 10) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H582, Q5TCC0, Q6BEP7, Q6P446, Q6PI06, Q7LG67, Q9ULJ9 | Gene names | ZNF644, KIAA1221, ZEP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 644 (Zinc finger motif enhancer-binding protein 2) (Zep-2). | |||||
|
BIRC6_HUMAN
|
||||||
| NC score | 0.011970 (rank : 17) | θ value | 5.27518 (rank : 20) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NR09, Q9ULD1 | Gene names | BIRC6, KIAA1289 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 6 (Ubiquitin-conjugating BIR-domain enzyme apollon). | |||||
|
PERI_HUMAN
|
||||||
| NC score | 0.007727 (rank : 18) | θ value | 1.81305 (rank : 13) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P41219, Q8N577 | Gene names | PRPH | |||
|
Domain Architecture |

|
|||||
| Description | Peripherin. | |||||
|
PERI_MOUSE
|
||||||
| NC score | 0.007678 (rank : 19) | θ value | 1.81305 (rank : 14) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P15331, O35688, O35689 | Gene names | Prph, Prph1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripherin. | |||||
|
EVL_MOUSE
|
||||||
| NC score | 0.006702 (rank : 20) | θ value | 8.99809 (rank : 21) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P70429, Q9ERU8 | Gene names | Evl | |||
|
Domain Architecture |

|
|||||
| Description | Ena/VASP-like protein (Ena/vasodilator-stimulated phosphoprotein- like). | |||||
|
CK5P2_HUMAN
|
||||||
| NC score | 0.004634 (rank : 21) | θ value | 4.03905 (rank : 17) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96SN8, Q7Z3L4, Q7Z3U1, Q7Z7I6, Q9BSW0, Q9H6J6, Q9HCD9, Q9NV90, Q9UIW9 | Gene names | CDK5RAP2, CEP215, KIAA1633 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48) (Centrosome-associated protein 215). | |||||