Please be patient as the page loads
|
HPS1_MOUSE
|
||||||
| SwissProt Accessions | O08983, O35155, O35725, O35950 | Gene names | Hps1, Ep, Hps | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hermansky-Pudlak syndrome 1 protein homolog. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
HPS1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.987650 (rank : 2) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q92902, O15402, O15502 | Gene names | HPS1, HPS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hermansky-Pudlak syndrome 1 protein. | |||||
|
HPS1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O08983, O35155, O35725, O35950 | Gene names | Hps1, Ep, Hps | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hermansky-Pudlak syndrome 1 protein homolog. | |||||
|
PF21B_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 3) | NC score | 0.070727 (rank : 3) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96EK2, Q5TFL2, Q6ICC0 | Gene names | PHF21B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
GSCR1_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 4) | NC score | 0.062654 (rank : 4) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
ERCC6_HUMAN
|
||||||
| θ value | 1.06291 (rank : 5) | NC score | 0.018445 (rank : 12) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q03468 | Gene names | ERCC6, CSB | |||
|
Domain Architecture |

|
|||||
| Description | DNA excision repair protein ERCC-6 (EC 3.6.1.-) (ATP-dependent helicase ERCC6) (Cockayne syndrome protein CSB). | |||||
|
BPA1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 6) | NC score | 0.010700 (rank : 23) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q91ZU6, Q91ZU7 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 1/2/3/4 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
SALL3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 7) | NC score | 0.008248 (rank : 26) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 1066 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BXA9, Q9UGH1 | Gene names | SALL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 3 (Zinc finger protein SALL3) (hSALL3). | |||||
|
CFTR_MOUSE
|
||||||
| θ value | 2.36792 (rank : 8) | NC score | 0.013282 (rank : 20) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P26361, Q63893, Q63894, Q9JKQ6 | Gene names | Cftr, Abcc7 | |||
|
Domain Architecture |

|
|||||
| Description | Cystic fibrosis transmembrane conductance regulator (CFTR) (cAMP- dependent chloride channel) (ATP-binding cassette transporter sub- family C member 7). | |||||
|
TCEA1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 9) | NC score | 0.021678 (rank : 10) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P23193, Q15563, Q6FG87 | Gene names | TCEA1, TFIIS | |||
|
Domain Architecture |
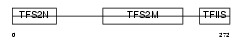
|
|||||
| Description | Transcription elongation factor A protein 1 (Transcription elongation factor S-II protein 1) (Transcription elongation factor TFIIS.o). | |||||
|
ATN1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 10) | NC score | 0.024730 (rank : 5) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
ELMO2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 11) | NC score | 0.023444 (rank : 9) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96JJ3, Q5JVZ6, Q96CJ2, Q96ME5, Q96PA9, Q9H938, Q9H9L5, Q9HAH0, Q9NQQ6 | Gene names | ELMO2, CED12A, KIAA1834 | |||
|
Domain Architecture |

|
|||||
| Description | Engulfment and cell motility protein 2 (CED-12 homolog A) (hCED-12A). | |||||
|
ELMO2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 12) | NC score | 0.023477 (rank : 8) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BHL5, Q8BHL9, Q8BQG1, Q8CBM8, Q8CC50, Q8CH98, Q91ZU2, Q9CT75 | Gene names | Elmo2, Kiaa1834 | |||
|
Domain Architecture |

|
|||||
| Description | Engulfment and cell motility protein 2 (CED-12 homolog A). | |||||
|
HB2Q_HUMAN
|
||||||
| θ value | 4.03905 (rank : 13) | NC score | 0.006044 (rank : 29) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P13763 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | HLA class II histocompatibility antigen, DP(W2) beta chain precursor (SB-2-beta). | |||||
|
MDN1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 14) | NC score | 0.024317 (rank : 6) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
ZN236_HUMAN
|
||||||
| θ value | 4.03905 (rank : 15) | NC score | 0.002741 (rank : 34) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 844 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UL36, Q9UL37 | Gene names | ZNF236 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 236. | |||||
|
MAVS_HUMAN
|
||||||
| θ value | 5.27518 (rank : 16) | NC score | 0.023978 (rank : 7) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7Z434, Q3I0Y2, Q5T7I6, Q86VY7, Q9H1H3, Q9H4Y1, Q9H8D3, Q9ULE9 | Gene names | MAVS, IPS1, KIAA1271, VISA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial antiviral signaling protein (Interferon-beta promoter stimulator protein 1) (IPS-1) (Virus-induced signaling adapter) (CARD adapter inducing interferon-beta) (Cardif) (Putative NF-kappa-B- activating protein 031N). | |||||
|
SYP2L_HUMAN
|
||||||
| θ value | 5.27518 (rank : 17) | NC score | 0.012927 (rank : 21) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H987 | Gene names | SYNPO2L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin 2-like protein. | |||||
|
ZN447_HUMAN
|
||||||
| θ value | 5.27518 (rank : 18) | NC score | 0.005191 (rank : 31) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 899 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8TBC5, Q9BRK7, Q9H9A0 | Gene names | ZNF447 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 447. | |||||
|
AP1G1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 19) | NC score | 0.014621 (rank : 18) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43747, O75709, O75842, Q9UG09, Q9Y3U4 | Gene names | AP1G1, ADTG, CLAPG1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-1 (Adapter-related protein complex 1 gamma- 1 subunit) (Gamma-adaptin) (Adaptor protein complex AP-1 gamma-1 subunit) (Golgi adaptor HA1/AP1 adaptin subunit gamma-1) (Clathrin assembly protein complex 1 gamma-1 large chain). | |||||
|
AP1G1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 20) | NC score | 0.014657 (rank : 17) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P22892 | Gene names | Ap1g1, Adtg, Clapg1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-1 (Adapter-related protein complex 1 gamma- 1 subunit) (Gamma-adaptin) (Adaptor protein complex AP-1 gamma-1 subunit) (Golgi adaptor HA1/AP1 adaptin subunit gamma-1) (Clathrin assembly protein complex 1 gamma-1 large chain). | |||||
|
FOXL1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 21) | NC score | 0.006582 (rank : 28) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q64731 | Gene names | Foxl1, Fkh6, Fkhl11 | |||
|
Domain Architecture |
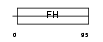
|
|||||
| Description | Forkhead box protein L1 (Forkhead-related protein FKHL11) (Transcription factor FKH-6). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 22) | NC score | 0.015740 (rank : 16) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 23) | NC score | 0.015996 (rank : 14) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
NALP5_MOUSE
|
||||||
| θ value | 6.88961 (rank : 24) | NC score | 0.009543 (rank : 24) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9R1M5, Q9JLR2 | Gene names | Nalp5, Mater | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Maternal antigen that embryos require) (Mater protein) (Ooplasm-specific protein 1) (OP1). | |||||
|
SCN4A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 25) | NC score | 0.005956 (rank : 30) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P35499, Q15478, Q16447, Q7Z6B1 | Gene names | SCN4A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 4 subunit alpha (Sodium channel protein type IV subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.4) (Sodium channel protein, skeletal muscle subunit alpha) (SkM1). | |||||
|
TPO_MOUSE
|
||||||
| θ value | 6.88961 (rank : 26) | NC score | 0.019829 (rank : 11) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P40226 | Gene names | Thpo | |||
|
Domain Architecture |

|
|||||
| Description | Thrombopoietin precursor (Megakaryocyte colony-stimulating factor) (Myeloproliferative leukemia virus oncogene ligand) (C-mpl ligand) (ML) (Megakaryocyte growth and development factor) (MGDF). | |||||
|
TPR_HUMAN
|
||||||
| θ value | 6.88961 (rank : 27) | NC score | 0.004647 (rank : 33) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
BRSK1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 28) | NC score | 0.000099 (rank : 35) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 942 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TDC3, Q5J5B5, Q8NDD0, Q8NDR4, Q8TDC2, Q96AV4, Q96JL4 | Gene names | BRSK1, KIAA1811, SAD1 | |||
|
Domain Architecture |

|
|||||
| Description | BR serine/threonine-protein kinase 1 (EC 2.7.11.1) (SAD1 kinase) (SAD1A). | |||||
|
CIZ1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 29) | NC score | 0.018028 (rank : 13) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9ULV3, Q9NYM8, Q9UHK4, Q9Y3F9, Q9Y3G0 | Gene names | CIZ1, LSFR1, NP94 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cip1-interacting zinc finger protein (Nuclear protein NP94). | |||||
|
IF35_MOUSE
|
||||||
| θ value | 8.99809 (rank : 30) | NC score | 0.013431 (rank : 19) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DCH4 | Gene names | Eif3s5 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 5 (eIF-3 epsilon) (eIF3 p47 subunit) (eIF3f). | |||||
|
MYPT1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 31) | NC score | 0.005058 (rank : 32) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O14974, Q86WU3, Q8NFR6, Q9BYH0 | Gene names | PPP1R12A, MBS, MYPT1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12A (Myosin phosphatase- targeting subunit 1) (Myosin phosphatase target subunit 1) (Protein phosphatase myosin-binding subunit). | |||||
|
PAX8_HUMAN
|
||||||
| θ value | 8.99809 (rank : 32) | NC score | 0.007398 (rank : 27) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q06710, Q09155, Q16337, Q16338, Q16339, Q96J49 | Gene names | PAX8 | |||
|
Domain Architecture |
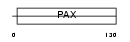
|
|||||
| Description | Paired box protein Pax-8. | |||||
|
RERE_MOUSE
|
||||||
| θ value | 8.99809 (rank : 33) | NC score | 0.015785 (rank : 15) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
TRIPC_HUMAN
|
||||||
| θ value | 8.99809 (rank : 34) | NC score | 0.009416 (rank : 25) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q14669, Q15644 | Gene names | TRIP12, KIAA0045 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid receptor-interacting protein 12 (TRIP12). | |||||
|
WIRE_HUMAN
|
||||||
| θ value | 8.99809 (rank : 35) | NC score | 0.012706 (rank : 22) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8TF74, Q658J8, Q71RE1, Q8TE44 | Gene names | WIRE, WICH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein) (WIP- and CR16-homologous protein). | |||||
|
HPS1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O08983, O35155, O35725, O35950 | Gene names | Hps1, Ep, Hps | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hermansky-Pudlak syndrome 1 protein homolog. | |||||
|
HPS1_HUMAN
|
||||||
| NC score | 0.987650 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q92902, O15402, O15502 | Gene names | HPS1, HPS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hermansky-Pudlak syndrome 1 protein. | |||||
|
PF21B_HUMAN
|
||||||
| NC score | 0.070727 (rank : 3) | θ value | 0.00509761 (rank : 3) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96EK2, Q5TFL2, Q6ICC0 | Gene names | PHF21B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
GSCR1_HUMAN
|
||||||
| NC score | 0.062654 (rank : 4) | θ value | 0.0330416 (rank : 4) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
ATN1_HUMAN
|
||||||
| NC score | 0.024730 (rank : 5) | θ value | 3.0926 (rank : 10) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
MDN1_HUMAN
|
||||||
| NC score | 0.024317 (rank : 6) | θ value | 4.03905 (rank : 14) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
MAVS_HUMAN
|
||||||
| NC score | 0.023978 (rank : 7) | θ value | 5.27518 (rank : 16) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7Z434, Q3I0Y2, Q5T7I6, Q86VY7, Q9H1H3, Q9H4Y1, Q9H8D3, Q9ULE9 | Gene names | MAVS, IPS1, KIAA1271, VISA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial antiviral signaling protein (Interferon-beta promoter stimulator protein 1) (IPS-1) (Virus-induced signaling adapter) (CARD adapter inducing interferon-beta) (Cardif) (Putative NF-kappa-B- activating protein 031N). | |||||
|
ELMO2_MOUSE
|
||||||
| NC score | 0.023477 (rank : 8) | θ value | 4.03905 (rank : 12) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BHL5, Q8BHL9, Q8BQG1, Q8CBM8, Q8CC50, Q8CH98, Q91ZU2, Q9CT75 | Gene names | Elmo2, Kiaa1834 | |||
|
Domain Architecture |

|
|||||
| Description | Engulfment and cell motility protein 2 (CED-12 homolog A). | |||||
|
ELMO2_HUMAN
|
||||||
| NC score | 0.023444 (rank : 9) | θ value | 4.03905 (rank : 11) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96JJ3, Q5JVZ6, Q96CJ2, Q96ME5, Q96PA9, Q9H938, Q9H9L5, Q9HAH0, Q9NQQ6 | Gene names | ELMO2, CED12A, KIAA1834 | |||
|
Domain Architecture |

|
|||||
| Description | Engulfment and cell motility protein 2 (CED-12 homolog A) (hCED-12A). | |||||
|
TCEA1_HUMAN
|
||||||
| NC score | 0.021678 (rank : 10) | θ value | 2.36792 (rank : 9) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P23193, Q15563, Q6FG87 | Gene names | TCEA1, TFIIS | |||
|
Domain Architecture |

|
|||||
| Description | Transcription elongation factor A protein 1 (Transcription elongation factor S-II protein 1) (Transcription elongation factor TFIIS.o). | |||||
|
TPO_MOUSE
|
||||||
| NC score | 0.019829 (rank : 11) | θ value | 6.88961 (rank : 26) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P40226 | Gene names | Thpo | |||
|
Domain Architecture |

|
|||||
| Description | Thrombopoietin precursor (Megakaryocyte colony-stimulating factor) (Myeloproliferative leukemia virus oncogene ligand) (C-mpl ligand) (ML) (Megakaryocyte growth and development factor) (MGDF). | |||||
|
ERCC6_HUMAN
|
||||||
| NC score | 0.018445 (rank : 12) | θ value | 1.06291 (rank : 5) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q03468 | Gene names | ERCC6, CSB | |||
|
Domain Architecture |

|
|||||
| Description | DNA excision repair protein ERCC-6 (EC 3.6.1.-) (ATP-dependent helicase ERCC6) (Cockayne syndrome protein CSB). | |||||
|
CIZ1_HUMAN
|
||||||
| NC score | 0.018028 (rank : 13) | θ value | 8.99809 (rank : 29) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9ULV3, Q9NYM8, Q9UHK4, Q9Y3F9, Q9Y3G0 | Gene names | CIZ1, LSFR1, NP94 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cip1-interacting zinc finger protein (Nuclear protein NP94). | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.015996 (rank : 14) | θ value | 6.88961 (rank : 23) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
RERE_MOUSE
|
||||||
| NC score | 0.015785 (rank : 15) | θ value | 8.99809 (rank : 33) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.015740 (rank : 16) | θ value | 6.88961 (rank : 22) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
AP1G1_MOUSE
|
||||||
| NC score | 0.014657 (rank : 17) | θ value | 6.88961 (rank : 20) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P22892 | Gene names | Ap1g1, Adtg, Clapg1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-1 (Adapter-related protein complex 1 gamma- 1 subunit) (Gamma-adaptin) (Adaptor protein complex AP-1 gamma-1 subunit) (Golgi adaptor HA1/AP1 adaptin subunit gamma-1) (Clathrin assembly protein complex 1 gamma-1 large chain). | |||||
|
AP1G1_HUMAN
|
||||||
| NC score | 0.014621 (rank : 18) | θ value | 6.88961 (rank : 19) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43747, O75709, O75842, Q9UG09, Q9Y3U4 | Gene names | AP1G1, ADTG, CLAPG1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-1 (Adapter-related protein complex 1 gamma- 1 subunit) (Gamma-adaptin) (Adaptor protein complex AP-1 gamma-1 subunit) (Golgi adaptor HA1/AP1 adaptin subunit gamma-1) (Clathrin assembly protein complex 1 gamma-1 large chain). | |||||
|
IF35_MOUSE
|
||||||
| NC score | 0.013431 (rank : 19) | θ value | 8.99809 (rank : 30) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DCH4 | Gene names | Eif3s5 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 5 (eIF-3 epsilon) (eIF3 p47 subunit) (eIF3f). | |||||
|
CFTR_MOUSE
|
||||||
| NC score | 0.013282 (rank : 20) | θ value | 2.36792 (rank : 8) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P26361, Q63893, Q63894, Q9JKQ6 | Gene names | Cftr, Abcc7 | |||
|
Domain Architecture |

|
|||||
| Description | Cystic fibrosis transmembrane conductance regulator (CFTR) (cAMP- dependent chloride channel) (ATP-binding cassette transporter sub- family C member 7). | |||||
|
SYP2L_HUMAN
|
||||||
| NC score | 0.012927 (rank : 21) | θ value | 5.27518 (rank : 17) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H987 | Gene names | SYNPO2L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin 2-like protein. | |||||
|
WIRE_HUMAN
|
||||||
| NC score | 0.012706 (rank : 22) | θ value | 8.99809 (rank : 35) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8TF74, Q658J8, Q71RE1, Q8TE44 | Gene names | WIRE, WICH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein) (WIP- and CR16-homologous protein). | |||||
|
BPA1_MOUSE
|
||||||
| NC score | 0.010700 (rank : 23) | θ value | 1.38821 (rank : 6) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q91ZU6, Q91ZU7 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 1/2/3/4 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
NALP5_MOUSE
|
||||||
| NC score | 0.009543 (rank : 24) | θ value | 6.88961 (rank : 24) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9R1M5, Q9JLR2 | Gene names | Nalp5, Mater | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Maternal antigen that embryos require) (Mater protein) (Ooplasm-specific protein 1) (OP1). | |||||
|
TRIPC_HUMAN
|
||||||
| NC score | 0.009416 (rank : 25) | θ value | 8.99809 (rank : 34) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q14669, Q15644 | Gene names | TRIP12, KIAA0045 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid receptor-interacting protein 12 (TRIP12). | |||||
|
SALL3_HUMAN
|
||||||
| NC score | 0.008248 (rank : 26) | θ value | 1.38821 (rank : 7) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 1066 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BXA9, Q9UGH1 | Gene names | SALL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 3 (Zinc finger protein SALL3) (hSALL3). | |||||
|
PAX8_HUMAN
|
||||||
| NC score | 0.007398 (rank : 27) | θ value | 8.99809 (rank : 32) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q06710, Q09155, Q16337, Q16338, Q16339, Q96J49 | Gene names | PAX8 | |||
|
Domain Architecture |

|
|||||
| Description | Paired box protein Pax-8. | |||||
|
FOXL1_MOUSE
|
||||||
| NC score | 0.006582 (rank : 28) | θ value | 6.88961 (rank : 21) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q64731 | Gene names | Foxl1, Fkh6, Fkhl11 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein L1 (Forkhead-related protein FKHL11) (Transcription factor FKH-6). | |||||
|
HB2Q_HUMAN
|
||||||
| NC score | 0.006044 (rank : 29) | θ value | 4.03905 (rank : 13) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P13763 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | HLA class II histocompatibility antigen, DP(W2) beta chain precursor (SB-2-beta). | |||||
|
SCN4A_HUMAN
|
||||||
| NC score | 0.005956 (rank : 30) | θ value | 6.88961 (rank : 25) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P35499, Q15478, Q16447, Q7Z6B1 | Gene names | SCN4A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 4 subunit alpha (Sodium channel protein type IV subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.4) (Sodium channel protein, skeletal muscle subunit alpha) (SkM1). | |||||
|
ZN447_HUMAN
|
||||||
| NC score | 0.005191 (rank : 31) | θ value | 5.27518 (rank : 18) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 899 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8TBC5, Q9BRK7, Q9H9A0 | Gene names | ZNF447 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 447. | |||||
|
MYPT1_HUMAN
|
||||||
| NC score | 0.005058 (rank : 32) | θ value | 8.99809 (rank : 31) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O14974, Q86WU3, Q8NFR6, Q9BYH0 | Gene names | PPP1R12A, MBS, MYPT1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12A (Myosin phosphatase- targeting subunit 1) (Myosin phosphatase target subunit 1) (Protein phosphatase myosin-binding subunit). | |||||
|
TPR_HUMAN
|
||||||
| NC score | 0.004647 (rank : 33) | θ value | 6.88961 (rank : 27) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
ZN236_HUMAN
|
||||||
| NC score | 0.002741 (rank : 34) | θ value | 4.03905 (rank : 15) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 844 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UL36, Q9UL37 | Gene names | ZNF236 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 236. | |||||
|
BRSK1_HUMAN
|
||||||
| NC score | 0.000099 (rank : 35) | θ value | 8.99809 (rank : 28) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 942 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TDC3, Q5J5B5, Q8NDD0, Q8NDR4, Q8TDC2, Q96AV4, Q96JL4 | Gene names | BRSK1, KIAA1811, SAD1 | |||
|
Domain Architecture |

|
|||||
| Description | BR serine/threonine-protein kinase 1 (EC 2.7.11.1) (SAD1 kinase) (SAD1A). | |||||