Please be patient as the page loads
|
GNL3_HUMAN
|
||||||
| SwissProt Accessions | Q9BVP2, Q96SV6, Q96SV7, Q9UJY0 | Gene names | GNL3, E2IG3, NS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein-like 3 (Nucleolar GTP-binding protein 3) (Nucleostemin) (E2-induced gene 3-protein) (Novel nucleolar protein 47) (NNP47). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
GNL3_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q9BVP2, Q96SV6, Q96SV7, Q9UJY0 | Gene names | GNL3, E2IG3, NS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein-like 3 (Nucleolar GTP-binding protein 3) (Nucleostemin) (E2-induced gene 3-protein) (Novel nucleolar protein 47) (NNP47). | |||||
|
GNL3_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.959770 (rank : 2) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8CI11, Q811R9, Q811S8, Q8BK21 | Gene names | Gnl3, Ns | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein-like 3 (Nucleolar GTP-binding protein 3) (Nucleostemin). | |||||
|
NOG2_MOUSE
|
||||||
| θ value | 1.16164e-39 (rank : 3) | NC score | 0.777967 (rank : 3) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q99LH1, Q8BIF8 | Gene names | Gnl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar GTP-binding protein 2. | |||||
|
NOG2_HUMAN
|
||||||
| θ value | 1.98146e-39 (rank : 4) | NC score | 0.769681 (rank : 4) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q13823, Q9BWN7 | Gene names | GNL2, NGP1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar GTP-binding protein 2 (Autoantigen NGP-1). | |||||
|
GNL1_HUMAN
|
||||||
| θ value | 5.62301e-10 (rank : 5) | NC score | 0.508943 (rank : 5) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P36915 | Gene names | GNL1, HSR1 | |||
|
Domain Architecture |
|
|||||
| Description | Guanine nucleotide-binding protein-like 1 (GTP-binding protein HSR1). | |||||
|
GNL1_MOUSE
|
||||||
| θ value | 2.13673e-09 (rank : 6) | NC score | 0.504866 (rank : 6) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P36916 | Gene names | Gnl1, Gna-rs1, Mmr1 | |||
|
Domain Architecture |
|
|||||
| Description | Guanine nucleotide-binding protein-like 1 (GTP-binding protein MMR1). | |||||
|
CCD96_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 7) | NC score | 0.078258 (rank : 15) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9CR92 | Gene names | Ccdc96 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 96. | |||||
|
SMC1A_MOUSE
|
||||||
| θ value | 0.125558 (rank : 8) | NC score | 0.051580 (rank : 24) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1384 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9CU62, Q3V480, Q9CUX9, Q9D959, Q9WTU1 | Gene names | Smc1l1, Sb1.8, Smc1, Smcb | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Chromosome segregation protein SmcB) (Sb1.8). | |||||
|
ERAL_HUMAN
|
||||||
| θ value | 0.21417 (rank : 9) | NC score | 0.137479 (rank : 7) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O75616, O75617, Q8WUY4, Q96LE2, Q96TC0 | Gene names | ERAL1, HERA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GTP-binding protein era homolog (hERA) (ERA-W) (Conserved ERA-like GTPase) (CEGA). | |||||
|
KINH_MOUSE
|
||||||
| θ value | 0.21417 (rank : 10) | NC score | 0.032279 (rank : 49) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q61768, O08711, Q61580 | Gene names | Kif5b, Khcs, Kns1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain (Ubiquitous kinesin heavy chain) (UKHC). | |||||
|
CK5P2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 11) | NC score | 0.049993 (rank : 27) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q96SN8, Q7Z3L4, Q7Z3U1, Q7Z7I6, Q9BSW0, Q9H6J6, Q9HCD9, Q9NV90, Q9UIW9 | Gene names | CDK5RAP2, CEP215, KIAA1633 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48) (Centrosome-associated protein 215). | |||||
|
RBNS5_MOUSE
|
||||||
| θ value | 0.279714 (rank : 12) | NC score | 0.052990 (rank : 22) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q80Y56, Q8K0L6, Q9CTW0 | Gene names | Zfyve20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rabenosyn-5 (FYVE finger-containing Rab5 effector protein rabenosyn-5) (Zinc finger FYVE domain-containing protein 20). | |||||
|
SMC1A_HUMAN
|
||||||
| θ value | 0.279714 (rank : 13) | NC score | 0.052416 (rank : 23) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1346 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q14683, O14995, Q16351 | Gene names | SMC1L1, DXS423E, KIAA0178, SB1.8, SMC1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Sb1.8). | |||||
|
ERAL_MOUSE
|
||||||
| θ value | 0.365318 (rank : 14) | NC score | 0.131766 (rank : 8) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9CZU4, Q6NV78, Q925U1 | Gene names | Eral1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GTP-binding protein era homolog (ERA-W) (Conserved ERA-like GTPase) (CEGA). | |||||
|
TPR_HUMAN
|
||||||
| θ value | 0.365318 (rank : 15) | NC score | 0.041187 (rank : 35) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
TRIM7_HUMAN
|
||||||
| θ value | 0.365318 (rank : 16) | NC score | 0.015275 (rank : 77) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9C029, Q969F5, Q96F67, Q96J89, Q96J90 | Gene names | TRIM7, GNIP, RNF90 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 7 (Glycogenin-interacting protein) (RING finger protein 90). | |||||
|
VPS72_HUMAN
|
||||||
| θ value | 0.365318 (rank : 17) | NC score | 0.086823 (rank : 13) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q15906, Q53GJ2, Q5U0R4 | Gene names | VPS72, TCFL1, YL1 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 72 homolog (Transcription factor-like 1) (Protein YL-1). | |||||
|
VPS72_MOUSE
|
||||||
| θ value | 0.365318 (rank : 18) | NC score | 0.086403 (rank : 14) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q62481, Q3U2N4, Q810A9, Q99K81 | Gene names | Vps72, Tcfl1, Yl1 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 72 homolog (Transcription factor-like 1) (Protein YL-1). | |||||
|
DMD_MOUSE
|
||||||
| θ value | 0.47712 (rank : 19) | NC score | 0.042102 (rank : 34) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1111 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P11531, O35653, Q60703 | Gene names | Dmd | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
NOP14_HUMAN
|
||||||
| θ value | 0.47712 (rank : 20) | NC score | 0.060588 (rank : 20) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 573 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P78316, Q7LGI5, Q7Z6K0, Q8TBR6 | Gene names | NOP14, C4orf9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable nucleolar complex protein 14. | |||||
|
ANR18_HUMAN
|
||||||
| θ value | 0.62314 (rank : 21) | NC score | 0.029742 (rank : 52) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1106 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8IVF6, Q7Z468 | Gene names | ANKRD18A, KIAA2015 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 18A. | |||||
|
DMD_HUMAN
|
||||||
| θ value | 0.62314 (rank : 22) | NC score | 0.041183 (rank : 36) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P11532, Q02295, Q14169, Q14170, Q5JYU0, Q7KZ48 | Gene names | DMD | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
GOGA3_MOUSE
|
||||||
| θ value | 0.813845 (rank : 23) | NC score | 0.045111 (rank : 29) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1437 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P55937, Q80VF5, Q8CCK4, Q9QYT2, Q9QYT3 | Gene names | Golga3, Mea2 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Male-enhanced antigen 2) (MEA-2). | |||||
|
SEPT9_HUMAN
|
||||||
| θ value | 0.813845 (rank : 24) | NC score | 0.019848 (rank : 66) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
CD014_MOUSE
|
||||||
| θ value | 1.06291 (rank : 25) | NC score | 0.040296 (rank : 37) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JJG9, Q3T996, Q3U044, Q99JU4 | Gene names | ames=MNCb-4931 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C4orf14 homolog. | |||||
|
MYO5A_MOUSE
|
||||||
| θ value | 1.06291 (rank : 26) | NC score | 0.031821 (rank : 50) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1117 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q99104 | Gene names | Myo5a, Dilute | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5A (Myosin Va) (Dilute myosin heavy chain, non-muscle). | |||||
|
SH3K1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 27) | NC score | 0.025601 (rank : 59) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8R550, Q8CAL8, Q8CEF6, Q8R545, Q8R546, Q8R547, Q8R548, Q8R549, Q8R551, Q9CTQ9, Q9DC14, Q9JKC3 | Gene names | Sh3kbp1, Ruk, Seta | |||
|
Domain Architecture |

|
|||||
| Description | SH3-domain kinase-binding protein 1 (SH3-containing, expressed in tumorigenic astrocytes) (Regulator of ubiquitous kinase) (Ruk). | |||||
|
BLNK_MOUSE
|
||||||
| θ value | 1.38821 (rank : 28) | NC score | 0.028771 (rank : 54) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9QUN3, O88504 | Gene names | Blnk, Bash, Ly57, Slp65 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell linker protein (Cytoplasmic adapter protein) (B-cell adapter containing SH2 domain protein) (B-cell adapter containing Src homology 2 domain protein) (Src homology 2 domain-containing leukocyte protein of 65 kDa) (Slp-65) (Lymphocyte antigen 57). | |||||
|
KTN1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 29) | NC score | 0.043639 (rank : 31) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1324 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q61595, Q8BG49, Q8BHF4, Q8BHM8, Q8C9Y5, Q8CG51, Q8CG52, Q8CG53, Q8CG54, Q8CG55, Q8CG56, Q8CG57, Q8CG58, Q8CG59, Q8CG60, Q8CG61, Q8CG62, Q8CG63 | Gene names | Ktn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin. | |||||
|
NOG1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 30) | NC score | 0.093910 (rank : 9) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9BZE4, O95446, Q9NVJ8 | Gene names | GTPBP4, CRFG, NOG1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar GTP-binding protein 1 (Chronic renal failure gene protein) (GTP-binding protein NGB). | |||||
|
NOG1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 31) | NC score | 0.093367 (rank : 10) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q99ME9, Q99K16, Q99P78, Q9CT02 | Gene names | Gtpbp4, Crfg, Nog1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar GTP-binding protein 1 (Chronic renal failure gene protein) (GTP-binding protein NGB). | |||||
|
TAOK1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 32) | NC score | 0.020777 (rank : 65) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1734 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q7L7X3, Q96L75, Q9H2K7, Q9H7S5, Q9P2I6 | Gene names | TAOK1, KIAA1361, MAP3K16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase TAO1 (EC 2.7.11.1) (Thousand and one amino acid protein 1) (STE20-like kinase PSK2) (Kinase from chicken homolog B) (hKFC-B). | |||||
|
TAOK1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 33) | NC score | 0.020973 (rank : 64) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1720 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q5F2E8, Q69ZL2, Q8JZX2, Q91VG7 | Gene names | Taok1, Kiaa1361 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase TAO1 (EC 2.7.11.1) (Thousand and one amino acid protein 1). | |||||
|
TULP2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 34) | NC score | 0.025725 (rank : 58) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O00295 | Gene names | TULP2 | |||
|
Domain Architecture |

|
|||||
| Description | Tubby-related protein 2 (Tubby-like protein 2). | |||||
|
ZN318_MOUSE
|
||||||
| θ value | 1.38821 (rank : 35) | NC score | 0.050159 (rank : 26) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 642 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q99PP2, Q3TZL5, Q9JJ01 | Gene names | Znf318, Tzf, Zfp318 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 318 (Testicular zinc finger protein). | |||||
|
GNPAT_HUMAN
|
||||||
| θ value | 1.81305 (rank : 36) | NC score | 0.029664 (rank : 53) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15228 | Gene names | GNPAT, DAPAT, DHAPAT | |||
|
Domain Architecture |
|
|||||
| Description | Dihydroxyacetone phosphate acyltransferase (EC 2.3.1.42) (DHAP-AT) (DAP-AT) (Glycerone-phosphate O-acyltransferase) (Acyl- CoA:dihydroxyacetonephosphateacyltransferase). | |||||
|
PTD4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 37) | NC score | 0.065300 (rank : 17) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NTK5, Q8NCI8, Q96CU1, Q96SV2, Q9P1D3, Q9UNY9, Q9Y6G4 | Gene names | ames=PTD004, PRO2455 | |||
|
Domain Architecture |

|
|||||
| Description | Putative GTP-binding protein PTD004. | |||||
|
PTD4_MOUSE
|
||||||
| θ value | 1.81305 (rank : 38) | NC score | 0.065332 (rank : 16) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9CZ30, Q9CWE2, Q9CX91, Q9CYC9, Q9CZ56 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Putative GTP-binding protein PTD004 homolog. | |||||
|
STMN1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 39) | NC score | 0.033611 (rank : 46) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P54227 | Gene names | Stmn1, Lag, Lap18, Pr22 | |||
|
Domain Architecture |

|
|||||
| Description | Stathmin (Phosphoprotein p19) (pp19) (Oncoprotein 18) (Op18) (Leukemia-associated phosphoprotein p18) (pp17) (Prosolin) (Metablastin) (Pr22 protein) (Leukemia-associated gene protein). | |||||
|
STOX1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 40) | NC score | 0.042858 (rank : 33) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6ZVD7, Q4F8Q6, Q5I946, Q5I947, Q5I948, Q5VX38, Q5VX39, Q6ZRY3, Q96LR3, Q96LS0 | Gene names | STOX1, C10orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Storkhead-box protein 1 (Winged helix domain-containing protein). | |||||
|
STK10_MOUSE
|
||||||
| θ value | 2.36792 (rank : 41) | NC score | 0.019600 (rank : 69) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 2132 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O55098 | Gene names | Stk10, Lok | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
CK5P2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 42) | NC score | 0.045157 (rank : 28) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1151 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8K389, Q6PCN1, Q6ZPL0 | Gene names | Cdk5rap2, Kiaa1633 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48). | |||||
|
CLSPN_HUMAN
|
||||||
| θ value | 3.0926 (rank : 43) | NC score | 0.036733 (rank : 41) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9HAW4, Q5VYG0, Q6P6H5, Q8IWI1 | Gene names | CLSPN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Claspin (hClaspin) (Hu-Claspin). | |||||
|
DRG1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 44) | NC score | 0.092542 (rank : 11) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Y295, Q6FGP8, Q8WW69, Q9UGF2 | Gene names | DRG1 | |||
|
Domain Architecture |
|
|||||
| Description | Developmentally-regulated GTP-binding protein 1 (DRG 1). | |||||
|
DRG1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 45) | NC score | 0.092502 (rank : 12) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P32233 | Gene names | Drg1, Drg, Nedd-3, Nedd3 | |||
|
Domain Architecture |
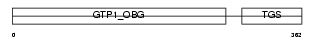
|
|||||
| Description | Developmentally-regulated GTP-binding protein 1 (DRG 1) (Protein NEDD3) (Neural precursor cell expressed developmentally down-regulated protein 3). | |||||
|
RUFY2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 46) | NC score | 0.031258 (rank : 51) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1208 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8WXA3, Q96P51, Q9P1Z1 | Gene names | RUFY2, KIAA1537, RABIP4R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 2 (Rab4-interacting protein related). | |||||
|
STMN1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 47) | NC score | 0.032703 (rank : 47) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 554 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P16949 | Gene names | STMN1, LAP18, OP18 | |||
|
Domain Architecture |

|
|||||
| Description | Stathmin (Phosphoprotein p19) (pp19) (Oncoprotein 18) (Op18) (Leukemia-associated phosphoprotein p18) (pp17) (Prosolin) (Metablastin) (Protein Pr22). | |||||
|
SVIL_MOUSE
|
||||||
| θ value | 3.0926 (rank : 48) | NC score | 0.016267 (rank : 74) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8K4L3 | Gene names | Svil | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Supervillin (Archvillin) (p205/p250). | |||||
|
SYCP3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 49) | NC score | 0.025816 (rank : 57) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8IZU3 | Gene names | SYCP3, SCP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptonemal complex protein 3 (SCP-3). | |||||
|
AKAP9_HUMAN
|
||||||
| θ value | 4.03905 (rank : 50) | NC score | 0.039998 (rank : 38) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
CCD96_HUMAN
|
||||||
| θ value | 4.03905 (rank : 51) | NC score | 0.050989 (rank : 25) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 595 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q2M329, Q8N2I7 | Gene names | CCDC96 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 96. | |||||
|
CDC37_HUMAN
|
||||||
| θ value | 4.03905 (rank : 52) | NC score | 0.034292 (rank : 45) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q16543 | Gene names | CDC37 | |||
|
Domain Architecture |

|
|||||
| Description | Hsp90 co-chaperone Cdc37 (Hsp90 chaperone protein kinase-targeting subunit) (p50Cdc37). | |||||
|
CI078_HUMAN
|
||||||
| θ value | 4.03905 (rank : 53) | NC score | 0.035700 (rank : 44) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NZ63, Q8WVU6, Q9NT39 | Gene names | C9orf78 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf78 (Hepatocellular carcinoma-associated antigen 59). | |||||
|
CING_HUMAN
|
||||||
| θ value | 4.03905 (rank : 54) | NC score | 0.037894 (rank : 39) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9P2M7, Q5T386, Q9NR25 | Gene names | CGN, KIAA1319 | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
CP250_HUMAN
|
||||||
| θ value | 4.03905 (rank : 55) | NC score | 0.035934 (rank : 43) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
RBBP6_MOUSE
|
||||||
| θ value | 4.03905 (rank : 56) | NC score | 0.026502 (rank : 56) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
RBCC1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 57) | NC score | 0.044517 (rank : 30) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1048 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9ESK9, Q3TXX2, Q61384, Q8BRY9, Q9JK14 | Gene names | Rb1cc1, Cc1, Kiaa0203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RB1-inducible coiled-coil protein 1 (Coiled-coil-forming protein 1) (LaXp180 protein). | |||||
|
TCRG1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 58) | NC score | 0.061212 (rank : 19) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O14776 | Gene names | TCERG1, CA150, TAF2S | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150). | |||||
|
TCRG1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 59) | NC score | 0.061293 (rank : 18) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8CGF7, Q61051, Q8C490, Q8CHT8, Q9R0R5 | Gene names | Tcerg1, Taf2s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150) (p144) (Formin- binding protein 28) (FBP 28). | |||||
|
CUTL2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 60) | NC score | 0.017934 (rank : 72) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O14529 | Gene names | CUTL2, KIAA0293 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 2 (Homeobox protein Cux-2) (Cut-like 2). | |||||
|
IMA2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 61) | NC score | 0.010318 (rank : 81) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P52293, Q64292 | Gene names | Kpna2, Rch1 | |||
|
Domain Architecture |
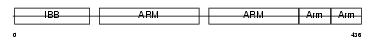
|
|||||
| Description | Importin alpha-2 subunit (Karyopherin alpha-2 subunit) (SRP1-alpha) (RAG cohort protein 1) (Pendulin) (Pore targeting complex 58 kDa subunit) (PTAC58) (Importin alpha P1). | |||||
|
LIPA2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 62) | NC score | 0.043110 (rank : 32) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1282 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8BSS9, Q8BN73 | Gene names | Ppfia2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-2) (PTPRF-interacting protein alpha-2). | |||||
|
RBM6_HUMAN
|
||||||
| θ value | 5.27518 (rank : 63) | NC score | 0.018714 (rank : 70) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P78332, O60549, O75524 | Gene names | RBM6, DEF3 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 6 (RNA-binding motif protein 6) (RNA-binding protein DEF-3) (Lung cancer antigen NY-LU-12) (Protein G16). | |||||
|
B4GT7_MOUSE
|
||||||
| θ value | 6.88961 (rank : 64) | NC score | 0.011571 (rank : 79) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8R087, Q7TNU3 | Gene names | B4galt7, Xgalt1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,4-galactosyltransferase 7 (EC 2.4.1.-) (Beta-1,4-GalTase 7) (Beta4Gal-T7) (b4Gal-T7) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 7) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 7) [Includes: Xylosylprotein 4-beta- galactosyltransferase (EC 2.4.1.133) (UDP-galactose:beta-xylose beta- 1,4-galactosyltransferase) (Xylosylprotein beta-1,4- galactosyltransferase) (XGPT) (Proteoglycan UDP-galactose:beta-xylose beta1,4-galactosyltransferase I) (XGalT-1)]. | |||||
|
C8AP2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 65) | NC score | 0.016159 (rank : 75) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9WUF3, Q9CSW2 | Gene names | Casp8ap2, Flash | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CASP8-associated protein 2 (FLICE-associated huge protein). | |||||
|
CI078_MOUSE
|
||||||
| θ value | 6.88961 (rank : 66) | NC score | 0.032397 (rank : 48) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q3TQI7 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf78 homolog. | |||||
|
CR1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 67) | NC score | 0.007316 (rank : 82) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P17927, Q16745, Q9UQV2 | Gene names | CR1, C3BR | |||
|
Domain Architecture |

|
|||||
| Description | Complement receptor type 1 precursor (C3b/C4b receptor) (CD35 antigen). | |||||
|
CRCM_HUMAN
|
||||||
| θ value | 6.88961 (rank : 68) | NC score | 0.037040 (rank : 40) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 520 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P23508 | Gene names | MCC | |||
|
Domain Architecture |
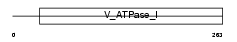
|
|||||
| Description | Colorectal mutant cancer protein (Protein MCC). | |||||
|
CTRO_MOUSE
|
||||||
| θ value | 6.88961 (rank : 69) | NC score | 0.019734 (rank : 68) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 2344 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P49025, O88528, O88937, O88938, Q3UM99, Q8CIJ1 | Gene names | Cit | |||
|
Domain Architecture |

|
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
MYO5C_HUMAN
|
||||||
| θ value | 6.88961 (rank : 70) | NC score | 0.028106 (rank : 55) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1206 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9NQX4 | Gene names | MYO5C | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5C (Myosin Vc). | |||||
|
PDCD2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 71) | NC score | 0.018000 (rank : 71) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q16342, Q9UH12 | Gene names | PDCD2, RP8, ZMYND7 | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death protein 2 (Zinc finger protein Rp-8) (Zinc finger MYND domain-containing protein 7). | |||||
|
ROCK2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 72) | NC score | 0.022095 (rank : 61) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 2073 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O75116, Q9UQN5 | Gene names | ROCK2, KIAA0619 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2) (Rho kinase 2). | |||||
|
SEPT9_MOUSE
|
||||||
| θ value | 6.88961 (rank : 73) | NC score | 0.015914 (rank : 76) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80UG5, Q3URP2, Q80TM7, Q9QYX9 | Gene names | Sept9, Kiaa0991, Sint1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (SL3-3 integration site 1 protein). | |||||
|
ZN452_HUMAN
|
||||||
| θ value | 6.88961 (rank : 74) | NC score | 0.006731 (rank : 84) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6R2W3, Q2NKL9, Q5SRJ3, Q8TCN2, Q96PW3 | Gene names | ZNF452, KIAA1925 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF452. | |||||
|
CCD16_HUMAN
|
||||||
| θ value | 8.99809 (rank : 75) | NC score | 0.021879 (rank : 62) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96NB3, Q96F60, Q96GZ5, Q9BU38 | Gene names | CCDC16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 16. | |||||
|
FSCN1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 76) | NC score | 0.010729 (rank : 80) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q16658, Q96IC5, Q9BRF1 | Gene names | FSCN1, FAN1, HSN, SNL | |||
|
Domain Architecture |
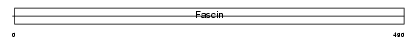
|
|||||
| Description | Fascin (Singed-like protein) (55 kDa actin-bundling protein) (p55). | |||||
|
GDS1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 77) | NC score | 0.019761 (rank : 67) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P52306, Q9NYM2, Q9NZA8 | Gene names | RAP1GDS1 | |||
|
Domain Architecture |

|
|||||
| Description | Rap1 GTPase-GDP dissociation stimulator 1 (SMG P21 stimulatory GDP/GTP exchange protein) (SMG GDS protein) (Exchange factor smgGDS). | |||||
|
MYO5B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 78) | NC score | 0.023453 (rank : 60) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 911 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9ULV0 | Gene names | MYO5B, KIAA1119 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5B (Myosin Vb). | |||||
|
ROCK2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 79) | NC score | 0.021325 (rank : 63) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 2072 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P70336, Q8BR64, Q8CBR0 | Gene names | Rock2 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2). | |||||
|
SDPR_MOUSE
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | 0.036434 (rank : 42) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q63918, Q3V1P6, Q78EC3, Q8CBT4 | Gene names | Sdpr, Sdr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serum deprivation-response protein (Phosphatidylserine-binding protein). | |||||
|
TAOK2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | 0.013845 (rank : 78) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1373 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6ZQ29, Q7TSS8 | Gene names | Taok2, Kiaa0881 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase TAO2 (EC 2.7.11.1) (Thousand and one amino acid protein 2). | |||||
|
TRPM8_HUMAN
|
||||||
| θ value | 8.99809 (rank : 82) | NC score | 0.007196 (rank : 83) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7Z2W7, Q6QNH9, Q8TAC3, Q8TDX8, Q9BVK1 | Gene names | TRPM8, LTRPC6, TRPP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 8 (Transient receptor potential-p8) (Trp-p8) (Long transient receptor potential channel 6) (LTrpC6). | |||||
|
UBF1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 83) | NC score | 0.017342 (rank : 73) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P25976 | Gene names | Ubtf, Tcfubf, Ubf-1, Ubf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1). | |||||
|
SM1L2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.053647 (rank : 21) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1019 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8NDV3, Q5TIC3, Q9Y3G5 | Gene names | SMC1L2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 2 protein (SMC1beta protein). | |||||
|
GNL3_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q9BVP2, Q96SV6, Q96SV7, Q9UJY0 | Gene names | GNL3, E2IG3, NS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein-like 3 (Nucleolar GTP-binding protein 3) (Nucleostemin) (E2-induced gene 3-protein) (Novel nucleolar protein 47) (NNP47). | |||||
|
GNL3_MOUSE
|
||||||
| NC score | 0.959770 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8CI11, Q811R9, Q811S8, Q8BK21 | Gene names | Gnl3, Ns | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein-like 3 (Nucleolar GTP-binding protein 3) (Nucleostemin). | |||||
|
NOG2_MOUSE
|
||||||
| NC score | 0.777967 (rank : 3) | θ value | 1.16164e-39 (rank : 3) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q99LH1, Q8BIF8 | Gene names | Gnl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar GTP-binding protein 2. | |||||
|
NOG2_HUMAN
|
||||||
| NC score | 0.769681 (rank : 4) | θ value | 1.98146e-39 (rank : 4) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q13823, Q9BWN7 | Gene names | GNL2, NGP1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar GTP-binding protein 2 (Autoantigen NGP-1). | |||||
|
GNL1_HUMAN
|
||||||
| NC score | 0.508943 (rank : 5) | θ value | 5.62301e-10 (rank : 5) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P36915 | Gene names | GNL1, HSR1 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-binding protein-like 1 (GTP-binding protein HSR1). | |||||
|
GNL1_MOUSE
|
||||||
| NC score | 0.504866 (rank : 6) | θ value | 2.13673e-09 (rank : 6) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P36916 | Gene names | Gnl1, Gna-rs1, Mmr1 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-binding protein-like 1 (GTP-binding protein MMR1). | |||||
|
ERAL_HUMAN
|
||||||
| NC score | 0.137479 (rank : 7) | θ value | 0.21417 (rank : 9) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O75616, O75617, Q8WUY4, Q96LE2, Q96TC0 | Gene names | ERAL1, HERA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GTP-binding protein era homolog (hERA) (ERA-W) (Conserved ERA-like GTPase) (CEGA). | |||||
|
ERAL_MOUSE
|
||||||
| NC score | 0.131766 (rank : 8) | θ value | 0.365318 (rank : 14) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9CZU4, Q6NV78, Q925U1 | Gene names | Eral1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GTP-binding protein era homolog (ERA-W) (Conserved ERA-like GTPase) (CEGA). | |||||
|
NOG1_HUMAN
|
||||||
| NC score | 0.093910 (rank : 9) | θ value | 1.38821 (rank : 30) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9BZE4, O95446, Q9NVJ8 | Gene names | GTPBP4, CRFG, NOG1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar GTP-binding protein 1 (Chronic renal failure gene protein) (GTP-binding protein NGB). | |||||
|
NOG1_MOUSE
|
||||||
| NC score | 0.093367 (rank : 10) | θ value | 1.38821 (rank : 31) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q99ME9, Q99K16, Q99P78, Q9CT02 | Gene names | Gtpbp4, Crfg, Nog1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar GTP-binding protein 1 (Chronic renal failure gene protein) (GTP-binding protein NGB). | |||||
|
DRG1_HUMAN
|
||||||
| NC score | 0.092542 (rank : 11) | θ value | 3.0926 (rank : 44) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Y295, Q6FGP8, Q8WW69, Q9UGF2 | Gene names | DRG1 | |||
|
Domain Architecture |

|
|||||
| Description | Developmentally-regulated GTP-binding protein 1 (DRG 1). | |||||
|
DRG1_MOUSE
|
||||||
| NC score | 0.092502 (rank : 12) | θ value | 3.0926 (rank : 45) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P32233 | Gene names | Drg1, Drg, Nedd-3, Nedd3 | |||
|
Domain Architecture |

|
|||||
| Description | Developmentally-regulated GTP-binding protein 1 (DRG 1) (Protein NEDD3) (Neural precursor cell expressed developmentally down-regulated protein 3). | |||||
|
VPS72_HUMAN
|
||||||
| NC score | 0.086823 (rank : 13) | θ value | 0.365318 (rank : 17) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q15906, Q53GJ2, Q5U0R4 | Gene names | VPS72, TCFL1, YL1 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 72 homolog (Transcription factor-like 1) (Protein YL-1). | |||||
|
VPS72_MOUSE
|
||||||
| NC score | 0.086403 (rank : 14) | θ value | 0.365318 (rank : 18) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q62481, Q3U2N4, Q810A9, Q99K81 | Gene names | Vps72, Tcfl1, Yl1 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 72 homolog (Transcription factor-like 1) (Protein YL-1). | |||||
|
CCD96_MOUSE
|
||||||
| NC score | 0.078258 (rank : 15) | θ value | 0.0330416 (rank : 7) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9CR92 | Gene names | Ccdc96 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 96. | |||||
|
PTD4_MOUSE
|
||||||
| NC score | 0.065332 (rank : 16) | θ value | 1.81305 (rank : 38) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9CZ30, Q9CWE2, Q9CX91, Q9CYC9, Q9CZ56 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Putative GTP-binding protein PTD004 homolog. | |||||
|
PTD4_HUMAN
|
||||||
| NC score | 0.065300 (rank : 17) | θ value | 1.81305 (rank : 37) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NTK5, Q8NCI8, Q96CU1, Q96SV2, Q9P1D3, Q9UNY9, Q9Y6G4 | Gene names | ames=PTD004, PRO2455 | |||
|
Domain Architecture |

|
|||||
| Description | Putative GTP-binding protein PTD004. | |||||
|
TCRG1_MOUSE
|
||||||
| NC score | 0.061293 (rank : 18) | θ value | 4.03905 (rank : 59) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8CGF7, Q61051, Q8C490, Q8CHT8, Q9R0R5 | Gene names | Tcerg1, Taf2s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150) (p144) (Formin- binding protein 28) (FBP 28). | |||||
|
TCRG1_HUMAN
|
||||||
| NC score | 0.061212 (rank : 19) | θ value | 4.03905 (rank : 58) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O14776 | Gene names | TCERG1, CA150, TAF2S | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150). | |||||
|
NOP14_HUMAN
|
||||||
| NC score | 0.060588 (rank : 20) | θ value | 0.47712 (rank : 20) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 573 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P78316, Q7LGI5, Q7Z6K0, Q8TBR6 | Gene names | NOP14, C4orf9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable nucleolar complex protein 14. | |||||
|
SM1L2_HUMAN
|
||||||
| NC score | 0.053647 (rank : 21) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1019 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8NDV3, Q5TIC3, Q9Y3G5 | Gene names | SMC1L2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 2 protein (SMC1beta protein). | |||||
|
RBNS5_MOUSE
|
||||||
| NC score | 0.052990 (rank : 22) | θ value | 0.279714 (rank : 12) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q80Y56, Q8K0L6, Q9CTW0 | Gene names | Zfyve20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rabenosyn-5 (FYVE finger-containing Rab5 effector protein rabenosyn-5) (Zinc finger FYVE domain-containing protein 20). | |||||
|
SMC1A_HUMAN
|
||||||
| NC score | 0.052416 (rank : 23) | θ value | 0.279714 (rank : 13) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1346 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q14683, O14995, Q16351 | Gene names | SMC1L1, DXS423E, KIAA0178, SB1.8, SMC1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Sb1.8). | |||||
|
SMC1A_MOUSE
|
||||||
| NC score | 0.051580 (rank : 24) | θ value | 0.125558 (rank : 8) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1384 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9CU62, Q3V480, Q9CUX9, Q9D959, Q9WTU1 | Gene names | Smc1l1, Sb1.8, Smc1, Smcb | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Chromosome segregation protein SmcB) (Sb1.8). | |||||
|
CCD96_HUMAN
|
||||||
| NC score | 0.050989 (rank : 25) | θ value | 4.03905 (rank : 51) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 595 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q2M329, Q8N2I7 | Gene names | CCDC96 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 96. | |||||
|
ZN318_MOUSE
|
||||||
| NC score | 0.050159 (rank : 26) | θ value | 1.38821 (rank : 35) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 642 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q99PP2, Q3TZL5, Q9JJ01 | Gene names | Znf318, Tzf, Zfp318 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 318 (Testicular zinc finger protein). | |||||
|
CK5P2_HUMAN
|
||||||
| NC score | 0.049993 (rank : 27) | θ value | 0.279714 (rank : 11) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q96SN8, Q7Z3L4, Q7Z3U1, Q7Z7I6, Q9BSW0, Q9H6J6, Q9HCD9, Q9NV90, Q9UIW9 | Gene names | CDK5RAP2, CEP215, KIAA1633 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48) (Centrosome-associated protein 215). | |||||
|
CK5P2_MOUSE
|
||||||
| NC score | 0.045157 (rank : 28) | θ value | 3.0926 (rank : 42) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1151 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8K389, Q6PCN1, Q6ZPL0 | Gene names | Cdk5rap2, Kiaa1633 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48). | |||||
|
GOGA3_MOUSE
|
||||||
| NC score | 0.045111 (rank : 29) | θ value | 0.813845 (rank : 23) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1437 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P55937, Q80VF5, Q8CCK4, Q9QYT2, Q9QYT3 | Gene names | Golga3, Mea2 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Male-enhanced antigen 2) (MEA-2). | |||||
|
RBCC1_MOUSE
|
||||||
| NC score | 0.044517 (rank : 30) | θ value | 4.03905 (rank : 57) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1048 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9ESK9, Q3TXX2, Q61384, Q8BRY9, Q9JK14 | Gene names | Rb1cc1, Cc1, Kiaa0203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RB1-inducible coiled-coil protein 1 (Coiled-coil-forming protein 1) (LaXp180 protein). | |||||
|
KTN1_MOUSE
|
||||||
| NC score | 0.043639 (rank : 31) | θ value | 1.38821 (rank : 29) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1324 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q61595, Q8BG49, Q8BHF4, Q8BHM8, Q8C9Y5, Q8CG51, Q8CG52, Q8CG53, Q8CG54, Q8CG55, Q8CG56, Q8CG57, Q8CG58, Q8CG59, Q8CG60, Q8CG61, Q8CG62, Q8CG63 | Gene names | Ktn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin. | |||||
|
LIPA2_MOUSE
|
||||||
| NC score | 0.043110 (rank : 32) | θ value | 5.27518 (rank : 62) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1282 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8BSS9, Q8BN73 | Gene names | Ppfia2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-2) (PTPRF-interacting protein alpha-2). | |||||
|
STOX1_HUMAN
|
||||||
| NC score | 0.042858 (rank : 33) | θ value | 1.81305 (rank : 40) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6ZVD7, Q4F8Q6, Q5I946, Q5I947, Q5I948, Q5VX38, Q5VX39, Q6ZRY3, Q96LR3, Q96LS0 | Gene names | STOX1, C10orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Storkhead-box protein 1 (Winged helix domain-containing protein). | |||||
|
DMD_MOUSE
|
||||||
| NC score | 0.042102 (rank : 34) | θ value | 0.47712 (rank : 19) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1111 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P11531, O35653, Q60703 | Gene names | Dmd | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
TPR_HUMAN
|
||||||
| NC score | 0.041187 (rank : 35) | θ value | 0.365318 (rank : 15) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
DMD_HUMAN
|
||||||
| NC score | 0.041183 (rank : 36) | θ value | 0.62314 (rank : 22) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P11532, Q02295, Q14169, Q14170, Q5JYU0, Q7KZ48 | Gene names | DMD | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
CD014_MOUSE
|
||||||
| NC score | 0.040296 (rank : 37) | θ value | 1.06291 (rank : 25) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JJG9, Q3T996, Q3U044, Q99JU4 | Gene names | ames=MNCb-4931 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C4orf14 homolog. | |||||
|
AKAP9_HUMAN
|
||||||
| NC score | 0.039998 (rank : 38) | θ value | 4.03905 (rank : 50) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
CING_HUMAN
|
||||||
| NC score | 0.037894 (rank : 39) | θ value | 4.03905 (rank : 54) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9P2M7, Q5T386, Q9NR25 | Gene names | CGN, KIAA1319 | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
CRCM_HUMAN
|
||||||
| NC score | 0.037040 (rank : 40) | θ value | 6.88961 (rank : 68) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 520 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P23508 | Gene names | MCC | |||
|
Domain Architecture |

|
|||||
| Description | Colorectal mutant cancer protein (Protein MCC). | |||||
|
CLSPN_HUMAN
|
||||||
| NC score | 0.036733 (rank : 41) | θ value | 3.0926 (rank : 43) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9HAW4, Q5VYG0, Q6P6H5, Q8IWI1 | Gene names | CLSPN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Claspin (hClaspin) (Hu-Claspin). | |||||
|
SDPR_MOUSE
|
||||||
| NC score | 0.036434 (rank : 42) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q63918, Q3V1P6, Q78EC3, Q8CBT4 | Gene names | Sdpr, Sdr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serum deprivation-response protein (Phosphatidylserine-binding protein). | |||||
|
CP250_HUMAN
|
||||||
| NC score | 0.035934 (rank : 43) | θ value | 4.03905 (rank : 55) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
CI078_HUMAN
|
||||||
| NC score | 0.035700 (rank : 44) | θ value | 4.03905 (rank : 53) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NZ63, Q8WVU6, Q9NT39 | Gene names | C9orf78 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf78 (Hepatocellular carcinoma-associated antigen 59). | |||||
|
CDC37_HUMAN
|
||||||
| NC score | 0.034292 (rank : 45) | θ value | 4.03905 (rank : 52) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q16543 | Gene names | CDC37 | |||
|
Domain Architecture |

|
|||||
| Description | Hsp90 co-chaperone Cdc37 (Hsp90 chaperone protein kinase-targeting subunit) (p50Cdc37). | |||||
|
STMN1_MOUSE
|
||||||
| NC score | 0.033611 (rank : 46) | θ value | 1.81305 (rank : 39) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P54227 | Gene names | Stmn1, Lag, Lap18, Pr22 | |||
|
Domain Architecture |

|
|||||
| Description | Stathmin (Phosphoprotein p19) (pp19) (Oncoprotein 18) (Op18) (Leukemia-associated phosphoprotein p18) (pp17) (Prosolin) (Metablastin) (Pr22 protein) (Leukemia-associated gene protein). | |||||
|
STMN1_HUMAN
|
||||||
| NC score | 0.032703 (rank : 47) | θ value | 3.0926 (rank : 47) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 554 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P16949 | Gene names | STMN1, LAP18, OP18 | |||
|
Domain Architecture |

|
|||||
| Description | Stathmin (Phosphoprotein p19) (pp19) (Oncoprotein 18) (Op18) (Leukemia-associated phosphoprotein p18) (pp17) (Prosolin) (Metablastin) (Protein Pr22). | |||||
|
CI078_MOUSE
|
||||||
| NC score | 0.032397 (rank : 48) | θ value | 6.88961 (rank : 66) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q3TQI7 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf78 homolog. | |||||
|
KINH_MOUSE
|
||||||
| NC score | 0.032279 (rank : 49) | θ value | 0.21417 (rank : 10) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q61768, O08711, Q61580 | Gene names | Kif5b, Khcs, Kns1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain (Ubiquitous kinesin heavy chain) (UKHC). | |||||
|
MYO5A_MOUSE
|
||||||
| NC score | 0.031821 (rank : 50) | θ value | 1.06291 (rank : 26) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1117 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q99104 | Gene names | Myo5a, Dilute | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5A (Myosin Va) (Dilute myosin heavy chain, non-muscle). | |||||
|
RUFY2_HUMAN
|
||||||
| NC score | 0.031258 (rank : 51) | θ value | 3.0926 (rank : 46) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1208 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8WXA3, Q96P51, Q9P1Z1 | Gene names | RUFY2, KIAA1537, RABIP4R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 2 (Rab4-interacting protein related). | |||||
|
ANR18_HUMAN
|
||||||
| NC score | 0.029742 (rank : 52) | θ value | 0.62314 (rank : 21) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1106 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8IVF6, Q7Z468 | Gene names | ANKRD18A, KIAA2015 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 18A. | |||||
|
GNPAT_HUMAN
|
||||||
| NC score | 0.029664 (rank : 53) | θ value | 1.81305 (rank : 36) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15228 | Gene names | GNPAT, DAPAT, DHAPAT | |||
|
Domain Architecture |

|
|||||
| Description | Dihydroxyacetone phosphate acyltransferase (EC 2.3.1.42) (DHAP-AT) (DAP-AT) (Glycerone-phosphate O-acyltransferase) (Acyl- CoA:dihydroxyacetonephosphateacyltransferase). | |||||
|
BLNK_MOUSE
|
||||||
| NC score | 0.028771 (rank : 54) | θ value | 1.38821 (rank : 28) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9QUN3, O88504 | Gene names | Blnk, Bash, Ly57, Slp65 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell linker protein (Cytoplasmic adapter protein) (B-cell adapter containing SH2 domain protein) (B-cell adapter containing Src homology 2 domain protein) (Src homology 2 domain-containing leukocyte protein of 65 kDa) (Slp-65) (Lymphocyte antigen 57). | |||||
|
MYO5C_HUMAN
|
||||||
| NC score | 0.028106 (rank : 55) | θ value | 6.88961 (rank : 70) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1206 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9NQX4 | Gene names | MYO5C | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5C (Myosin Vc). | |||||
|
RBBP6_MOUSE
|
||||||
| NC score | 0.026502 (rank : 56) | θ value | 4.03905 (rank : 56) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
SYCP3_HUMAN
|
||||||
| NC score | 0.025816 (rank : 57) | θ value | 3.0926 (rank : 49) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8IZU3 | Gene names | SYCP3, SCP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptonemal complex protein 3 (SCP-3). | |||||
|
TULP2_HUMAN
|
||||||
| NC score | 0.025725 (rank : 58) | θ value | 1.38821 (rank : 34) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O00295 | Gene names | TULP2 | |||
|
Domain Architecture |

|
|||||
| Description | Tubby-related protein 2 (Tubby-like protein 2). | |||||
|
SH3K1_MOUSE
|
||||||
| NC score | 0.025601 (rank : 59) | θ value | 1.06291 (rank : 27) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8R550, Q8CAL8, Q8CEF6, Q8R545, Q8R546, Q8R547, Q8R548, Q8R549, Q8R551, Q9CTQ9, Q9DC14, Q9JKC3 | Gene names | Sh3kbp1, Ruk, Seta | |||
|
Domain Architecture |

|
|||||
| Description | SH3-domain kinase-binding protein 1 (SH3-containing, expressed in tumorigenic astrocytes) (Regulator of ubiquitous kinase) (Ruk). | |||||
|
MYO5B_HUMAN
|
||||||
| NC score | 0.023453 (rank : 60) | θ value | 8.99809 (rank : 78) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 911 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9ULV0 | Gene names | MYO5B, KIAA1119 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5B (Myosin Vb). | |||||
|
ROCK2_HUMAN
|
||||||
| NC score | 0.022095 (rank : 61) | θ value | 6.88961 (rank : 72) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 2073 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O75116, Q9UQN5 | Gene names | ROCK2, KIAA0619 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2) (Rho kinase 2). | |||||
|
CCD16_HUMAN
|
||||||
| NC score | 0.021879 (rank : 62) | θ value | 8.99809 (rank : 75) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96NB3, Q96F60, Q96GZ5, Q9BU38 | Gene names | CCDC16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 16. | |||||
|
ROCK2_MOUSE
|
||||||
| NC score | 0.021325 (rank : 63) | θ value | 8.99809 (rank : 79) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 2072 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P70336, Q8BR64, Q8CBR0 | Gene names | Rock2 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2). | |||||
|
TAOK1_MOUSE
|
||||||
| NC score | 0.020973 (rank : 64) | θ value | 1.38821 (rank : 33) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1720 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q5F2E8, Q69ZL2, Q8JZX2, Q91VG7 | Gene names | Taok1, Kiaa1361 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase TAO1 (EC 2.7.11.1) (Thousand and one amino acid protein 1). | |||||
|
TAOK1_HUMAN
|
||||||
| NC score | 0.020777 (rank : 65) | θ value | 1.38821 (rank : 32) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1734 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q7L7X3, Q96L75, Q9H2K7, Q9H7S5, Q9P2I6 | Gene names | TAOK1, KIAA1361, MAP3K16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase TAO1 (EC 2.7.11.1) (Thousand and one amino acid protein 1) (STE20-like kinase PSK2) (Kinase from chicken homolog B) (hKFC-B). | |||||
|
SEPT9_HUMAN
|
||||||
| NC score | 0.019848 (rank : 66) | θ value | 0.813845 (rank : 24) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
GDS1_HUMAN
|
||||||
| NC score | 0.019761 (rank : 67) | θ value | 8.99809 (rank : 77) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P52306, Q9NYM2, Q9NZA8 | Gene names | RAP1GDS1 | |||
|
Domain Architecture |

|
|||||
| Description | Rap1 GTPase-GDP dissociation stimulator 1 (SMG P21 stimulatory GDP/GTP exchange protein) (SMG GDS protein) (Exchange factor smgGDS). | |||||
|
CTRO_MOUSE
|
||||||
| NC score | 0.019734 (rank : 68) | θ value | 6.88961 (rank : 69) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 2344 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P49025, O88528, O88937, O88938, Q3UM99, Q8CIJ1 | Gene names | Cit | |||
|
Domain Architecture |

|
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
STK10_MOUSE
|
||||||
| NC score | 0.019600 (rank : 69) | θ value | 2.36792 (rank : 41) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 2132 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O55098 | Gene names | Stk10, Lok | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
RBM6_HUMAN
|
||||||
| NC score | 0.018714 (rank : 70) | θ value | 5.27518 (rank : 63) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P78332, O60549, O75524 | Gene names | RBM6, DEF3 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 6 (RNA-binding motif protein 6) (RNA-binding protein DEF-3) (Lung cancer antigen NY-LU-12) (Protein G16). | |||||
|
PDCD2_HUMAN
|
||||||
| NC score | 0.018000 (rank : 71) | θ value | 6.88961 (rank : 71) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q16342, Q9UH12 | Gene names | PDCD2, RP8, ZMYND7 | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death protein 2 (Zinc finger protein Rp-8) (Zinc finger MYND domain-containing protein 7). | |||||
|
CUTL2_HUMAN
|
||||||
| NC score | 0.017934 (rank : 72) | θ value | 5.27518 (rank : 60) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O14529 | Gene names | CUTL2, KIAA0293 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 2 (Homeobox protein Cux-2) (Cut-like 2). | |||||
|
UBF1_MOUSE
|
||||||
| NC score | 0.017342 (rank : 73) | θ value | 8.99809 (rank : 83) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P25976 | Gene names | Ubtf, Tcfubf, Ubf-1, Ubf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1). | |||||
|
SVIL_MOUSE
|
||||||
| NC score | 0.016267 (rank : 74) | θ value | 3.0926 (rank : 48) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8K4L3 | Gene names | Svil | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Supervillin (Archvillin) (p205/p250). | |||||
|
C8AP2_MOUSE
|
||||||
| NC score | 0.016159 (rank : 75) | θ value | 6.88961 (rank : 65) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9WUF3, Q9CSW2 | Gene names | Casp8ap2, Flash | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CASP8-associated protein 2 (FLICE-associated huge protein). | |||||
|
SEPT9_MOUSE
|
||||||
| NC score | 0.015914 (rank : 76) | θ value | 6.88961 (rank : 73) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80UG5, Q3URP2, Q80TM7, Q9QYX9 | Gene names | Sept9, Kiaa0991, Sint1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (SL3-3 integration site 1 protein). | |||||
|
TRIM7_HUMAN
|
||||||
| NC score | 0.015275 (rank : 77) | θ value | 0.365318 (rank : 16) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9C029, Q969F5, Q96F67, Q96J89, Q96J90 | Gene names | TRIM7, GNIP, RNF90 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 7 (Glycogenin-interacting protein) (RING finger protein 90). | |||||
|
TAOK2_MOUSE
|
||||||
| NC score | 0.013845 (rank : 78) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1373 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6ZQ29, Q7TSS8 | Gene names | Taok2, Kiaa0881 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase TAO2 (EC 2.7.11.1) (Thousand and one amino acid protein 2). | |||||
|
B4GT7_MOUSE
|
||||||
| NC score | 0.011571 (rank : 79) | θ value | 6.88961 (rank : 64) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8R087, Q7TNU3 | Gene names | B4galt7, Xgalt1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,4-galactosyltransferase 7 (EC 2.4.1.-) (Beta-1,4-GalTase 7) (Beta4Gal-T7) (b4Gal-T7) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 7) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 7) [Includes: Xylosylprotein 4-beta- galactosyltransferase (EC 2.4.1.133) (UDP-galactose:beta-xylose beta- 1,4-galactosyltransferase) (Xylosylprotein beta-1,4- galactosyltransferase) (XGPT) (Proteoglycan UDP-galactose:beta-xylose beta1,4-galactosyltransferase I) (XGalT-1)]. | |||||
|
FSCN1_HUMAN
|
||||||
| NC score | 0.010729 (rank : 80) | θ value | 8.99809 (rank : 76) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q16658, Q96IC5, Q9BRF1 | Gene names | FSCN1, FAN1, HSN, SNL | |||
|
Domain Architecture |

|
|||||
| Description | Fascin (Singed-like protein) (55 kDa actin-bundling protein) (p55). | |||||
|
IMA2_MOUSE
|
||||||
| NC score | 0.010318 (rank : 81) | θ value | 5.27518 (rank : 61) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P52293, Q64292 | Gene names | Kpna2, Rch1 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-2 subunit (Karyopherin alpha-2 subunit) (SRP1-alpha) (RAG cohort protein 1) (Pendulin) (Pore targeting complex 58 kDa subunit) (PTAC58) (Importin alpha P1). | |||||
|
CR1_HUMAN
|
||||||
| NC score | 0.007316 (rank : 82) | θ value | 6.88961 (rank : 67) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P17927, Q16745, Q9UQV2 | Gene names | CR1, C3BR | |||
|
Domain Architecture |

|
|||||
| Description | Complement receptor type 1 precursor (C3b/C4b receptor) (CD35 antigen). | |||||
|
TRPM8_HUMAN
|
||||||
| NC score | 0.007196 (rank : 83) | θ value | 8.99809 (rank : 82) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7Z2W7, Q6QNH9, Q8TAC3, Q8TDX8, Q9BVK1 | Gene names | TRPM8, LTRPC6, TRPP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 8 (Transient receptor potential-p8) (Trp-p8) (Long transient receptor potential channel 6) (LTrpC6). | |||||
|
ZN452_HUMAN
|
||||||
| NC score | 0.006731 (rank : 84) | θ value | 6.88961 (rank : 74) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6R2W3, Q2NKL9, Q5SRJ3, Q8TCN2, Q96PW3 | Gene names | ZNF452, KIAA1925 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF452. | |||||