Please be patient as the page loads
|
GNPAT_HUMAN
|
||||||
| SwissProt Accessions | O15228 | Gene names | GNPAT, DAPAT, DHAPAT | |||
|
Domain Architecture |
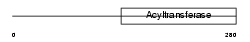
|
|||||
| Description | Dihydroxyacetone phosphate acyltransferase (EC 2.3.1.42) (DHAP-AT) (DAP-AT) (Glycerone-phosphate O-acyltransferase) (Acyl- CoA:dihydroxyacetonephosphateacyltransferase). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
GNPAT_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O15228 | Gene names | GNPAT, DAPAT, DHAPAT | |||
|
Domain Architecture |

|
|||||
| Description | Dihydroxyacetone phosphate acyltransferase (EC 2.3.1.42) (DHAP-AT) (DAP-AT) (Glycerone-phosphate O-acyltransferase) (Acyl- CoA:dihydroxyacetonephosphateacyltransferase). | |||||
|
GNPAT_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.997123 (rank : 2) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P98192, Q9WUT6 | Gene names | Gnpat, Dhapat | |||
|
Domain Architecture |
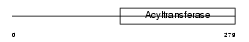
|
|||||
| Description | Dihydroxyacetone phosphate acyltransferase (EC 2.3.1.42) (DHAP-AT) (DAP-AT) (Glycerone-phosphate O-acyltransferase) (Acyl- CoA:dihydroxyacetonephosphateacyltransferase). | |||||
|
PLSB_HUMAN
|
||||||
| θ value | 1.16434e-31 (rank : 3) | NC score | 0.696843 (rank : 4) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HCL2, Q5VW51 | Gene names | GPAM, KIAA1560 | |||
|
Domain Architecture |
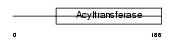
|
|||||
| Description | Glycerol-3-phosphate acyltransferase, mitochondrial precursor (EC 2.3.1.15) (GPAT). | |||||
|
PLSB_MOUSE
|
||||||
| θ value | 1.16434e-31 (rank : 4) | NC score | 0.697899 (rank : 3) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61586 | Gene names | Gpam | |||
|
Domain Architecture |

|
|||||
| Description | Glycerol-3-phosphate acyltransferase, mitochondrial precursor (EC 2.3.1.15) (GPAT) (P90). | |||||
|
MICA1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 5) | NC score | 0.022574 (rank : 7) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VDP3 | Gene names | Mical1, Mical, Nical | |||
|
Domain Architecture |

|
|||||
| Description | NEDD9-interacting protein with calponin homology and LIM domains (Molecule interacting with CasL protein 1). | |||||
|
FHOD1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 6) | NC score | 0.039722 (rank : 5) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6P9Q4, Q8BMK2 | Gene names | Fhod1, Fhos1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FH1/FH2 domain-containing protein (Formin homolog overexpressed in spleen) (FHOS) (Formin homology 2 domain-containing protein 1). | |||||
|
GNL3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 7) | NC score | 0.029664 (rank : 6) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BVP2, Q96SV6, Q96SV7, Q9UJY0 | Gene names | GNL3, E2IG3, NS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein-like 3 (Nucleolar GTP-binding protein 3) (Nucleostemin) (E2-induced gene 3-protein) (Novel nucleolar protein 47) (NNP47). | |||||
|
UACA_MOUSE
|
||||||
| θ value | 2.36792 (rank : 8) | NC score | 0.005079 (rank : 14) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CGB3, Q69ZG3, Q7TN77, Q8BJC8 | Gene names | Uaca, Kiaa1561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uveal autoantigen with coiled-coil domains and ankyrin repeats protein (Nucling) (Nuclear membrane-binding protein). | |||||
|
LRSM1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 9) | NC score | 0.013358 (rank : 8) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 1078 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6UWE0, Q5VVV0, Q8NB40, Q96GT5, Q96MX5, Q96MZ7 | Gene names | LRSAM1, TAL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein LRSAM1 (EC 6.3.2.-) (Leucine-rich repeat and sterile alpha motif-containing protein 1) (Tsg101-associated ligase) (hTAL). | |||||
|
DDX3X_HUMAN
|
||||||
| θ value | 8.99809 (rank : 10) | NC score | 0.009544 (rank : 11) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O00571, O15536 | Gene names | DDX3X, DBX, DDX3 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3X (EC 3.6.1.-) (DEAD box protein 3, X- chromosomal) (Helicase-like protein 2) (HLP2) (DEAD box, X isoform). | |||||
|
DDX3X_MOUSE
|
||||||
| θ value | 8.99809 (rank : 11) | NC score | 0.009564 (rank : 10) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q62167, O09060, O09143 | Gene names | Ddx3x, D1Pas1-rs2, Ddx3, Dead3, Erh | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3X (EC 3.6.1.-) (DEAD box protein 3, X- chromosomal) (DEAD box RNA helicase DEAD3) (mDEAD3) (Embryonic RNA helicase) (D1Pas1-related sequence 2). | |||||
|
DDX3Y_HUMAN
|
||||||
| θ value | 8.99809 (rank : 12) | NC score | 0.009664 (rank : 9) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15523, Q8IYV7 | Gene names | DDX3Y, DBY | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3Y (EC 3.6.1.-) (DEAD box protein 3, Y- chromosomal). | |||||
|
PL10_MOUSE
|
||||||
| θ value | 8.99809 (rank : 13) | NC score | 0.009522 (rank : 12) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P16381 | Gene names | D1Pas1, Pl10 | |||
|
Domain Architecture |

|
|||||
| Description | Putative ATP-dependent RNA helicase Pl10 (EC 3.6.1.-). | |||||
|
UBR1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 14) | NC score | 0.009355 (rank : 13) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IWV7, O60708, O75492, Q68DN9, Q8IWY6, Q96JY4 | Gene names | UBR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase E3 component N-recognin-1 (EC 6.-.-.-) (Ubiquitin-protein ligase E3-alpha-1) (Ubiquitin-protein ligase E3- alpha-I). | |||||
|
GNPAT_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O15228 | Gene names | GNPAT, DAPAT, DHAPAT | |||
|
Domain Architecture |

|
|||||
| Description | Dihydroxyacetone phosphate acyltransferase (EC 2.3.1.42) (DHAP-AT) (DAP-AT) (Glycerone-phosphate O-acyltransferase) (Acyl- CoA:dihydroxyacetonephosphateacyltransferase). | |||||
|
GNPAT_MOUSE
|
||||||
| NC score | 0.997123 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P98192, Q9WUT6 | Gene names | Gnpat, Dhapat | |||
|
Domain Architecture |

|
|||||
| Description | Dihydroxyacetone phosphate acyltransferase (EC 2.3.1.42) (DHAP-AT) (DAP-AT) (Glycerone-phosphate O-acyltransferase) (Acyl- CoA:dihydroxyacetonephosphateacyltransferase). | |||||
|
PLSB_MOUSE
|
||||||
| NC score | 0.697899 (rank : 3) | θ value | 1.16434e-31 (rank : 4) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61586 | Gene names | Gpam | |||
|
Domain Architecture |

|
|||||
| Description | Glycerol-3-phosphate acyltransferase, mitochondrial precursor (EC 2.3.1.15) (GPAT) (P90). | |||||
|
PLSB_HUMAN
|
||||||
| NC score | 0.696843 (rank : 4) | θ value | 1.16434e-31 (rank : 3) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HCL2, Q5VW51 | Gene names | GPAM, KIAA1560 | |||
|
Domain Architecture |

|
|||||
| Description | Glycerol-3-phosphate acyltransferase, mitochondrial precursor (EC 2.3.1.15) (GPAT). | |||||
|
FHOD1_MOUSE
|
||||||
| NC score | 0.039722 (rank : 5) | θ value | 1.81305 (rank : 6) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6P9Q4, Q8BMK2 | Gene names | Fhod1, Fhos1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FH1/FH2 domain-containing protein (Formin homolog overexpressed in spleen) (FHOS) (Formin homology 2 domain-containing protein 1). | |||||
|
GNL3_HUMAN
|
||||||
| NC score | 0.029664 (rank : 6) | θ value | 1.81305 (rank : 7) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BVP2, Q96SV6, Q96SV7, Q9UJY0 | Gene names | GNL3, E2IG3, NS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein-like 3 (Nucleolar GTP-binding protein 3) (Nucleostemin) (E2-induced gene 3-protein) (Novel nucleolar protein 47) (NNP47). | |||||
|
MICA1_MOUSE
|
||||||
| NC score | 0.022574 (rank : 7) | θ value | 0.62314 (rank : 5) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VDP3 | Gene names | Mical1, Mical, Nical | |||
|
Domain Architecture |

|
|||||
| Description | NEDD9-interacting protein with calponin homology and LIM domains (Molecule interacting with CasL protein 1). | |||||
|
LRSM1_HUMAN
|
||||||
| NC score | 0.013358 (rank : 8) | θ value | 5.27518 (rank : 9) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 1078 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6UWE0, Q5VVV0, Q8NB40, Q96GT5, Q96MX5, Q96MZ7 | Gene names | LRSAM1, TAL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein LRSAM1 (EC 6.3.2.-) (Leucine-rich repeat and sterile alpha motif-containing protein 1) (Tsg101-associated ligase) (hTAL). | |||||
|
DDX3Y_HUMAN
|
||||||
| NC score | 0.009664 (rank : 9) | θ value | 8.99809 (rank : 12) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15523, Q8IYV7 | Gene names | DDX3Y, DBY | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3Y (EC 3.6.1.-) (DEAD box protein 3, Y- chromosomal). | |||||
|
DDX3X_MOUSE
|
||||||
| NC score | 0.009564 (rank : 10) | θ value | 8.99809 (rank : 11) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q62167, O09060, O09143 | Gene names | Ddx3x, D1Pas1-rs2, Ddx3, Dead3, Erh | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3X (EC 3.6.1.-) (DEAD box protein 3, X- chromosomal) (DEAD box RNA helicase DEAD3) (mDEAD3) (Embryonic RNA helicase) (D1Pas1-related sequence 2). | |||||
|
DDX3X_HUMAN
|
||||||
| NC score | 0.009544 (rank : 11) | θ value | 8.99809 (rank : 10) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O00571, O15536 | Gene names | DDX3X, DBX, DDX3 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3X (EC 3.6.1.-) (DEAD box protein 3, X- chromosomal) (Helicase-like protein 2) (HLP2) (DEAD box, X isoform). | |||||
|
PL10_MOUSE
|
||||||
| NC score | 0.009522 (rank : 12) | θ value | 8.99809 (rank : 13) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P16381 | Gene names | D1Pas1, Pl10 | |||
|
Domain Architecture |

|
|||||
| Description | Putative ATP-dependent RNA helicase Pl10 (EC 3.6.1.-). | |||||
|
UBR1_HUMAN
|
||||||
| NC score | 0.009355 (rank : 13) | θ value | 8.99809 (rank : 14) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IWV7, O60708, O75492, Q68DN9, Q8IWY6, Q96JY4 | Gene names | UBR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase E3 component N-recognin-1 (EC 6.-.-.-) (Ubiquitin-protein ligase E3-alpha-1) (Ubiquitin-protein ligase E3- alpha-I). | |||||
|
UACA_MOUSE
|
||||||
| NC score | 0.005079 (rank : 14) | θ value | 2.36792 (rank : 8) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CGB3, Q69ZG3, Q7TN77, Q8BJC8 | Gene names | Uaca, Kiaa1561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uveal autoantigen with coiled-coil domains and ankyrin repeats protein (Nucling) (Nuclear membrane-binding protein). | |||||