Please be patient as the page loads
|
GABR2_HUMAN
|
||||||
| SwissProt Accessions | O75899, O75974, O75975, Q5VXZ2, Q8WX04, Q9P1R2, Q9UNR1, Q9UNS9 | Gene names | GABBR2, GPR51, GPRC3B | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric acid type B receptor, subunit 2 precursor (GABA-B receptor 2) (GABA-B-R2) (Gb2) (GABABR2) (G-protein coupled receptor 51) (HG20). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
GABR2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 101 | |
| SwissProt Accessions | O75899, O75974, O75975, Q5VXZ2, Q8WX04, Q9P1R2, Q9UNR1, Q9UNS9 | Gene names | GABBR2, GPR51, GPRC3B | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric acid type B receptor, subunit 2 precursor (GABA-B receptor 2) (GABA-B-R2) (Gb2) (GABABR2) (G-protein coupled receptor 51) (HG20). | |||||
|
GABR1_MOUSE
|
||||||
| θ value | 2.69525e-129 (rank : 2) | NC score | 0.841151 (rank : 3) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9WV18, Q9WU48, Q9WV15, Q9WV16, Q9WV17 | Gene names | Gabbr1 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric acid type B receptor, subunit 1 precursor (GABA-B receptor 1) (GABA-B-R1) (Gb1). | |||||
|
GABR1_HUMAN
|
||||||
| θ value | 1.02419e-128 (rank : 3) | NC score | 0.841848 (rank : 2) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9UBS5, O95375, O95468, O95975, O96022, Q86W60, Q9UQQ0 | Gene names | GABBR1, GPRC3A | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric acid type B receptor, subunit 1 precursor (GABA-B receptor 1) (GABA-B-R1) (Gb1). | |||||
|
GP156_HUMAN
|
||||||
| θ value | 4.89182e-30 (rank : 4) | NC score | 0.593869 (rank : 4) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8NFN8, Q86SN6 | Gene names | GPR156, GABABL, PGR28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 156 (GABAB-related G-protein coupled receptor) (G-protein coupled receptor PGR28). | |||||
|
GP156_MOUSE
|
||||||
| θ value | 2.05525e-28 (rank : 5) | NC score | 0.564731 (rank : 5) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6PCP7, Q8K452 | Gene names | Gpr156, Gababl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 156 (GABAB-related G-protein coupled receptor). | |||||
|
MGR8_MOUSE
|
||||||
| θ value | 1.02475e-11 (rank : 6) | NC score | 0.548521 (rank : 9) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P47743, Q6B964 | Gene names | Grm8, Gprc1h, Mglur8 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 8 precursor (mGluR8). | |||||
|
MGR8_HUMAN
|
||||||
| θ value | 1.74796e-11 (rank : 7) | NC score | 0.547714 (rank : 10) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O00222, O15493, O95945, O95946, Q3MIV9, Q52M02, Q6J165 | Gene names | GRM8, GPRC1H, MGLUR8 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 8 precursor (mGluR8). | |||||
|
MGR7_HUMAN
|
||||||
| θ value | 1.9326e-10 (rank : 8) | NC score | 0.550485 (rank : 7) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q14831, Q8NFS2, Q8NFS3, Q8NFS4 | Gene names | GRM7, GPRC1G, MGLUR7 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 7 precursor (mGluR7). | |||||
|
MGR7_MOUSE
|
||||||
| θ value | 1.9326e-10 (rank : 9) | NC score | 0.550229 (rank : 8) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q68ED2 | Gene names | Grm7, Gprc1g, Mglur7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metabotropic glutamate receptor 7 precursor (mGluR7). | |||||
|
MGR5_HUMAN
|
||||||
| θ value | 7.34386e-10 (rank : 10) | NC score | 0.539900 (rank : 11) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P41594 | Gene names | GRM5, GPRC1E, MGLUR5 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 5 precursor (mGluR5). | |||||
|
TS1R3_MOUSE
|
||||||
| θ value | 2.79066e-09 (rank : 11) | NC score | 0.534080 (rank : 16) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q925D8, Q91VA4, Q923K0, Q925A4, Q925D9 | Gene names | Tas1r3, Sac, T1r3, Tr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 1 member 3 precursor (Sweet taste receptor T1R3) (Saccharin preference protein). | |||||
|
MGR3_MOUSE
|
||||||
| θ value | 3.64472e-09 (rank : 12) | NC score | 0.537325 (rank : 14) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9QYS2 | Gene names | Grm3, Gprc1c, Mglur3 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 3 precursor (mGluR3). | |||||
|
CASR_HUMAN
|
||||||
| θ value | 2.36244e-08 (rank : 13) | NC score | 0.534264 (rank : 15) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P41180, Q13912, Q16108, Q16109, Q16110, Q16379, Q4PJ19 | Gene names | CASR, GPRC2A, PCAR1 | |||
|
Domain Architecture |

|
|||||
| Description | Extracellular calcium-sensing receptor precursor (CaSR) (Parathyroid Cell calcium-sensing receptor). | |||||
|
CASR_MOUSE
|
||||||
| θ value | 2.36244e-08 (rank : 14) | NC score | 0.533174 (rank : 18) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9QY96, O08968, O88519, Q9QY95, Q9QZU8, Q9R1D6, Q9R1Y2 | Gene names | Casr, Gprc2a | |||
|
Domain Architecture |

|
|||||
| Description | Extracellular calcium-sensing receptor precursor (CaSR) (Parathyroid Cell calcium-sensing receptor). | |||||
|
MGR3_HUMAN
|
||||||
| θ value | 4.0297e-08 (rank : 15) | NC score | 0.531497 (rank : 21) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q14832, Q75MV4, Q75N17, Q86YG6, Q8TBH9 | Gene names | GRM3, GPRC1C, MGLUR3 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 3 precursor (mGluR3). | |||||
|
TS1R3_HUMAN
|
||||||
| θ value | 8.97725e-08 (rank : 16) | NC score | 0.539687 (rank : 12) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q7RTX0, Q5TA49, Q8NGW9 | Gene names | TAS1R3, T1R3, TR3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 1 member 3 precursor (Sweet taste receptor T1R3). | |||||
|
MGR4_HUMAN
|
||||||
| θ value | 1.53129e-07 (rank : 17) | NC score | 0.552022 (rank : 6) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q14833, Q5SZ84 | Gene names | GRM4, GPRC1D, MGLUR4 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 4 precursor (mGluR4). | |||||
|
MGR6_HUMAN
|
||||||
| θ value | 2.61198e-07 (rank : 18) | NC score | 0.538123 (rank : 13) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O15303 | Gene names | GRM6, GPRC1F, MGLUR6 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 6 precursor (mGluR6). | |||||
|
MGR6_MOUSE
|
||||||
| θ value | 2.61198e-07 (rank : 19) | NC score | 0.533495 (rank : 17) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q5NCH9 | Gene names | Grm6, Gprc1f, Mglur6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metabotropic glutamate receptor 6 precursor (mGluR6). | |||||
|
GP179_HUMAN
|
||||||
| θ value | 4.45536e-07 (rank : 20) | NC score | 0.218523 (rank : 35) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6PRD1 | Gene names | GPR179, GPR158L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 179 precursor (Probable G-protein coupled receptor 158-like 1). | |||||
|
GUC2F_HUMAN
|
||||||
| θ value | 4.45536e-07 (rank : 21) | NC score | 0.070286 (rank : 42) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 740 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P51841, Q9UJF1 | Gene names | GUCY2F, GUC2F, RETGC2 | |||
|
Domain Architecture |

|
|||||
| Description | Retinal guanylyl cyclase 2 precursor (EC 4.6.1.2) (Guanylate cyclase 2F, retinal) (RETGC-2) (Rod outer segment membrane guanylate cyclase 2) (ROS-GC2) (Guanylate cyclase F) (GC-F). | |||||
|
GPC6A_MOUSE
|
||||||
| θ value | 7.59969e-07 (rank : 22) | NC score | 0.519180 (rank : 25) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8K4Z6, Q5DK51, Q5DK52 | Gene names | Gprc6a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor family C group 6 member A precursor. | |||||
|
MGR2_HUMAN
|
||||||
| θ value | 7.59969e-07 (rank : 23) | NC score | 0.531732 (rank : 20) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q14416, Q52MC6, Q9H3N6 | Gene names | GRM2, GPRC1B, MGLUR2 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 2 precursor (mGluR2). | |||||
|
TS1R1_HUMAN
|
||||||
| θ value | 7.59969e-07 (rank : 24) | NC score | 0.526939 (rank : 23) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q7RTX1, Q8NGZ7, Q8TDJ7, Q8TDJ8, Q8TDJ9, Q8TDK0 | Gene names | TAS1R1, GPR70, T1R1, TR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 1 member 1 precursor (G-protein coupled receptor 70) (Gm148). | |||||
|
MGR1_HUMAN
|
||||||
| θ value | 1.29631e-06 (rank : 25) | NC score | 0.531899 (rank : 19) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q13255, Q13256, Q14757, Q14758, Q5VTF7, Q5VTF8, Q9NU10, Q9UGS9, Q9UGT0 | Gene names | GRM1, GPRC1A, MGLUR1 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 1 precursor (mGluR1). | |||||
|
MGR1_MOUSE
|
||||||
| θ value | 1.29631e-06 (rank : 26) | NC score | 0.531294 (rank : 22) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P97772, Q6AXG4, Q9EPV6 | Gene names | Grm1, Gprc1a, Mglur1 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 1 precursor (mGluR1). | |||||
|
GPC6A_HUMAN
|
||||||
| θ value | 2.21117e-06 (rank : 27) | NC score | 0.515188 (rank : 27) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q5T6X5, Q6JK43, Q6JK44, Q8NGU8, Q8NHZ9, Q8TDT6 | Gene names | GPRC6A, GPCR33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor family C group 6 member A precursor (hGPRC6A) (G-protein coupled receptor 33) (hGPCR33). | |||||
|
TS1R2_MOUSE
|
||||||
| θ value | 3.77169e-06 (rank : 28) | NC score | 0.518495 (rank : 26) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q925I4, Q923J8 | Gene names | Tas1r2, Gpr71, T1r2, Tr2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 1 member 2 precursor (G-protein coupled receptor 71) (Sweet taste receptor T1R2). | |||||
|
V2RX_MOUSE
|
||||||
| θ value | 4.92598e-06 (rank : 29) | NC score | 0.524592 (rank : 24) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O70410 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vomeronasal type-2 receptor X precursor (Putative pheromone receptor V2RX). | |||||
|
TS1R2_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 30) | NC score | 0.507661 (rank : 29) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8TE23, Q5TZ19 | Gene names | TAS1R2, GPR71, T1R2, TR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 1 member 2 precursor (G-protein coupled receptor 71) (Sweet taste receptor T1R2). | |||||
|
ANPRC_MOUSE
|
||||||
| θ value | 3.19293e-05 (rank : 31) | NC score | 0.247306 (rank : 32) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P70180, P97804, Q9R025, Q9R027, Q9R028 | Gene names | Npr3 | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic peptide clearance receptor precursor (ANP-C) (ANPRC) (NPR-C) (Atrial natriuretic peptide C-type receptor) (EF-2). | |||||
|
ANPRC_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 32) | NC score | 0.239653 (rank : 34) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P17342 | Gene names | NPR3, ANPRC | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic peptide clearance receptor precursor (ANP-C) (ANPRC) (NPR-C) (Atrial natriuretic peptide C-type receptor). | |||||
|
GP158_MOUSE
|
||||||
| θ value | 0.000158464 (rank : 33) | NC score | 0.239797 (rank : 33) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8C419, Q8CHB0 | Gene names | Gpr158, Kiaa1136 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 158 precursor. | |||||
|
TS1R1_MOUSE
|
||||||
| θ value | 0.000158464 (rank : 34) | NC score | 0.511171 (rank : 28) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q99PG6, Q6NS58, Q923J9, Q925I5, Q99PG5 | Gene names | Tas1r1, Gpr70, T1r1, Tr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 1 member 1 precursor (G-protein coupled receptor 70). | |||||
|
GP158_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 35) | NC score | 0.248411 (rank : 31) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q5T848, Q6QR81, Q9ULT3 | Gene names | GPR158, KIAA1136 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 158 precursor. | |||||
|
NMDZ1_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 36) | NC score | 0.145981 (rank : 39) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q05586, P35437, Q12867, Q12868 | Gene names | GRIN1, NMDAR1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit zeta 1 precursor (N-methyl-D- aspartate receptor subunit NR1). | |||||
|
NMDZ1_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 37) | NC score | 0.146142 (rank : 38) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P35438, Q8CFS4 | Gene names | Grin1, Glurz1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit zeta 1 precursor (N-methyl-D- aspartate receptor subunit NR1). | |||||
|
GPC5B_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 38) | NC score | 0.179566 (rank : 37) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9NZH0, O75205, Q8NBZ8 | Gene names | GPRC5B, RAIG2 | |||
|
Domain Architecture |

|
|||||
| Description | G-protein coupled receptor family C group 5 member B precursor (Retinoic acid-induced gene 2 protein) (RAIG-2) (A-69G12.1). | |||||
|
GUC2G_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 39) | NC score | 0.062583 (rank : 46) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6TL19, Q8BWU7 | Gene names | Gucy2g | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanylate cyclase 2G precursor (EC 4.6.1.2) (Guanylyl cyclase receptor G) (mGC-G). | |||||
|
K1C10_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 40) | NC score | 0.049703 (rank : 51) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 466 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P02535, P08731, Q8BUX3, Q8BV09, Q8BVU3, Q9CXH6, Q9JKB4 | Gene names | Krt10, Krt1-10 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 10 (Cytokeratin-10) (CK-10) (Keratin-10) (K10) (56 kDa cytokeratin) (Keratin, type I cytoskeletal 59 kDa). | |||||
|
V2R1B_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 41) | NC score | 0.458942 (rank : 30) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6TAC4, O70409 | Gene names | V2r1b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vomeronasal type-2 receptor 1b precursor (Putative pheromone receptor V2R1b). | |||||
|
K1C10_HUMAN
|
||||||
| θ value | 0.125558 (rank : 42) | NC score | 0.049334 (rank : 52) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 526 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P13645, Q14664, Q8N175 | Gene names | KRT10 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 10 (Cytokeratin-10) (CK-10) (Keratin-10) (K10). | |||||
|
SYTL1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 43) | NC score | 0.022657 (rank : 94) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IYJ3, Q96BB6, Q96GU6, Q96S89, Q96SI0 | Gene names | SYTL1, SLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 1 (Exophilin-7) (Protein JFC1) (SB146). | |||||
|
GPC5B_MOUSE
|
||||||
| θ value | 0.279714 (rank : 44) | NC score | 0.182346 (rank : 36) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q923Z0, Q8CCV3 | Gene names | Gprc5b, Raig2 | |||
|
Domain Architecture |

|
|||||
| Description | G-protein coupled receptor family C group 5 member B precursor (Retinoic acid-induced gene 2 protein) (RAIG-2). | |||||
|
VDP_HUMAN
|
||||||
| θ value | 0.279714 (rank : 45) | NC score | 0.045134 (rank : 64) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O60763 | Gene names | VDP | |||
|
Domain Architecture |

|
|||||
| Description | General vesicular transport factor p115 (Transcytosis-associated protein) (TAP) (Vesicle docking protein). | |||||
|
NMDE2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 46) | NC score | 0.045638 (rank : 61) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13224, Q12919, Q13220, Q13225, Q9UM56 | Gene names | GRIN2B | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 2 precursor (N-methyl D- aspartate receptor subtype 2B) (NR2B) (NMDAR2B) (N-methyl-D-aspartate receptor subunit 3) (NR3) (hNR3). | |||||
|
NMDE2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 47) | NC score | 0.048531 (rank : 53) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q01097, Q9DCB2 | Gene names | Grin2b | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 2 precursor (N-methyl D- aspartate receptor subtype 2B) (NR2B) (NMDAR2B). | |||||
|
K1C15_MOUSE
|
||||||
| θ value | 0.813845 (rank : 48) | NC score | 0.047350 (rank : 56) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 429 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q61414, Q6PEQ0 | Gene names | Krt15, Krt1-15 | |||
|
Domain Architecture |
|
|||||
| Description | Keratin, type I cytoskeletal 15 (Cytokeratin-15) (CK-15) (Keratin-15) (K15). | |||||
|
K1C13_HUMAN
|
||||||
| θ value | 1.06291 (rank : 49) | NC score | 0.045943 (rank : 60) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 386 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P13646, Q53G54, Q6AZK5, Q8N240 | Gene names | KRT13 | |||
|
Domain Architecture |
|
|||||
| Description | Keratin, type I cytoskeletal 13 (Cytokeratin-13) (CK-13) (Keratin-13) (K13). | |||||
|
K1C15_HUMAN
|
||||||
| θ value | 1.06291 (rank : 50) | NC score | 0.045997 (rank : 59) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 367 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P19012, Q53XV8, Q9BUG4 | Gene names | KRT15, KRTB | |||
|
Domain Architecture |
|
|||||
| Description | Keratin, type I cytoskeletal 15 (Cytokeratin-15) (CK-15) (Keratin-15) (K15). | |||||
|
MYH8_MOUSE
|
||||||
| θ value | 1.06291 (rank : 51) | NC score | 0.028576 (rank : 77) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1600 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P13542, Q5SX36 | Gene names | Myh8, Myhsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
MYH9_MOUSE
|
||||||
| θ value | 1.06291 (rank : 52) | NC score | 0.028655 (rank : 76) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
GCC2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 53) | NC score | 0.028849 (rank : 75) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
K1C12_HUMAN
|
||||||
| θ value | 1.38821 (rank : 54) | NC score | 0.045418 (rank : 62) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q99456 | Gene names | KRT12 | |||
|
Domain Architecture |
|
|||||
| Description | Keratin, type I cytoskeletal 12 (Cytokeratin-12) (CK-12) (Keratin-12) (K12). | |||||
|
K1C13_MOUSE
|
||||||
| θ value | 1.38821 (rank : 55) | NC score | 0.045287 (rank : 63) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 410 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P08730, Q3V176, Q6PAI1 | Gene names | Krt13, Krt1-13 | |||
|
Domain Architecture |
|
|||||
| Description | Keratin, type I cytoskeletal 13 (Cytokeratin-13) (CK-13) (Keratin-13) (K13) (47 kDa cytokeratin). | |||||
|
K1C17_HUMAN
|
||||||
| θ value | 1.38821 (rank : 56) | NC score | 0.046640 (rank : 57) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 463 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q04695 | Gene names | KRT17 | |||
|
Domain Architecture |
|
|||||
| Description | Keratin, type I cytoskeletal 17 (Cytokeratin-17) (CK-17) (Keratin-17) (K17) (39.1). | |||||
|
K1C17_MOUSE
|
||||||
| θ value | 1.38821 (rank : 57) | NC score | 0.046350 (rank : 58) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9QWL7, Q61783 | Gene names | Krt17, Krt1-17 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 17 (Cytokeratin-17) (CK-17) (Keratin-17) (K17). | |||||
|
KNTC2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 58) | NC score | 0.032150 (rank : 72) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 692 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9D0F1, Q3TQT6, Q3UWM5, Q99P70 | Gene names | Kntc2, Hec1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Hec1 (Kinetochore-associated protein 2). | |||||
|
ITB5_HUMAN
|
||||||
| θ value | 1.81305 (rank : 59) | NC score | 0.013588 (rank : 102) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P18084 | Gene names | ITGB5 | |||
|
Domain Architecture |
|
|||||
| Description | Integrin beta-5 precursor. | |||||
|
K1C14_HUMAN
|
||||||
| θ value | 1.81305 (rank : 60) | NC score | 0.047372 (rank : 55) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P02533, Q14715, Q53XY3, Q9BUE3, Q9UBN2, Q9UBN3, Q9UCY4 | Gene names | KRT14 | |||
|
Domain Architecture |
|
|||||
| Description | Keratin, type I cytoskeletal 14 (Cytokeratin-14) (CK-14) (Keratin-14) (K14). | |||||
|
KIF5A_MOUSE
|
||||||
| θ value | 1.81305 (rank : 61) | NC score | 0.020711 (rank : 96) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 859 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P33175, Q5DTP1, Q6PDY7, Q9Z2F9 | Gene names | Kif5a, Kiaa4086, Kif5, Nkhc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin heavy chain isoform 5A (Neuronal kinesin heavy chain) (NKHC) (Kinesin heavy chain neuron-specific 1). | |||||
|
NDE1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 62) | NC score | 0.037073 (rank : 68) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 602 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9CZA6, Q3UBS6, Q3UIC1, Q9ERR0 | Gene names | Nde1, Nude | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear distribution protein nudE homolog 1 (NudE) (mNudE). | |||||
|
ITB5_MOUSE
|
||||||
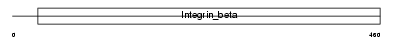
| θ value | 2.36792 (rank : 63) | NC score | 0.012785 (rank : 106) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O70309, O70308, O88347 | Gene names | Itgb5 | |||
|
Domain Architecture |
|
|||||
| Description | Integrin beta-5 precursor. | |||||
|
MYH9_HUMAN
|
||||||
| θ value | 2.36792 (rank : 64) | NC score | 0.027623 (rank : 80) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
HOOK3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 65) | NC score | 0.026225 (rank : 83) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8BUK6, Q8BV48, Q8BW57, Q8BY41 | Gene names | Hook3 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 3. | |||||
|
LAMA1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 66) | NC score | 0.008006 (rank : 108) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 804 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P25391 | Gene names | LAMA1, LAMA | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-1 chain precursor (Laminin A chain). | |||||
|
MYH4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 67) | NC score | 0.028035 (rank : 79) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
UACA_HUMAN
|
||||||
| θ value | 3.0926 (rank : 68) | NC score | 0.023838 (rank : 90) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1622 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9BZF9, Q8N3B8, Q9HCL1, Q9NWC6 | Gene names | UACA, KIAA1561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uveal autoantigen with coiled-coil domains and ankyrin repeats protein. | |||||
|
KIF5A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 69) | NC score | 0.018904 (rank : 98) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 843 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q12840, Q4LE26 | Gene names | KIF5A, NKHC1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain isoform 5A (Neuronal kinesin heavy chain) (NKHC) (Kinesin heavy chain neuron-specific 1). | |||||
|
KTN1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 70) | NC score | 0.025867 (rank : 84) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q86UP2, Q13999, Q14707, Q15387, Q86W57 | Gene names | KTN1, CG1, KIAA0004 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin (Kinesin receptor) (CG-1 antigen). | |||||
|
MYH2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 71) | NC score | 0.029072 (rank : 74) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1491 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9UKX2, Q14322, Q16229 | Gene names | MYH2, MYHSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, skeletal muscle, adult 2 (Myosin heavy chain IIa) (MyHC-IIa). | |||||
|
ROCK1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 72) | NC score | 0.010153 (rank : 107) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 2126 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P70335, Q8C3G4, Q8C7H0 | Gene names | Rock1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK). | |||||
|
WWTR1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 73) | NC score | 0.014648 (rank : 101) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9EPK5, Q99KI4 | Gene names | Wwtr1, Taz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing transcription regulator protein 1 (Transcriptional coactivator with PDZ-binding motif). | |||||
|
CE290_HUMAN
|
||||||
| θ value | 5.27518 (rank : 74) | NC score | 0.027328 (rank : 81) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
CK5P2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 75) | NC score | 0.029622 (rank : 73) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1151 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8K389, Q6PCN1, Q6ZPL0 | Gene names | Cdk5rap2, Kiaa1633 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48). | |||||
|
K1HB_HUMAN
|
||||||
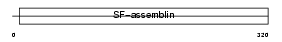
| θ value | 5.27518 (rank : 76) | NC score | 0.041368 (rank : 67) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 470 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q14525, O76010 | Gene names | KRTHA3B, HHA3-II, HKA3B | |||
|
Domain Architecture |
|
|||||
| Description | Keratin, type I cuticular Ha3-II (Hair keratin, type I Ha3-II). | |||||
|
MYH1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 77) | NC score | 0.028471 (rank : 78) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1461 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q5SX40 | Gene names | Myh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1). | |||||
|
NMDE1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 78) | NC score | 0.036046 (rank : 69) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q12879, O00669 | Gene names | GRIN2A | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 1 precursor (N-methyl D- aspartate receptor subtype 2A) (NR2A) (NMDAR2A) (hNR2A). | |||||
|
SPTB2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 79) | NC score | 0.017180 (rank : 100) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q01082, O60837, Q16057 | Gene names | SPTBN1, SPTB2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
SPTB2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 80) | NC score | 0.018010 (rank : 99) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q62261, Q5SQL8 | Gene names | Sptbn1, Spnb-2, Spnb2, Sptb2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
WASF2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 81) | NC score | 0.012991 (rank : 105) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y6W5, O60794, Q9UDY7 | Gene names | WASF2, WAVE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 2 (WASP-family protein member 2) (Protein WAVE-2) (Verprolin homology domain-containing protein 2). | |||||
|
ZNF8_HUMAN
|
||||||
| θ value | 5.27518 (rank : 82) | NC score | -0.003342 (rank : 109) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P17098, Q6PI99 | Gene names | ZNF8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 8 (Zinc finger protein HF.18). | |||||
|
AINX_HUMAN
|
||||||
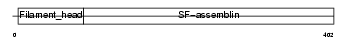
| θ value | 6.88961 (rank : 83) | NC score | 0.033804 (rank : 70) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 442 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q16352, Q9BRC5 | Gene names | INA | |||
|
Domain Architecture |
|
|||||
| Description | Alpha-internexin (Alpha-Inx) (66 kDa neurofilament protein) (Neurofilament-66) (NF-66). | |||||
|
CCD27_MOUSE
|
||||||
| θ value | 6.88961 (rank : 84) | NC score | 0.024338 (rank : 88) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q3V036 | Gene names | Ccdc27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 27. | |||||
|
CCD41_HUMAN
|
||||||
| θ value | 6.88961 (rank : 85) | NC score | 0.024695 (rank : 87) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1139 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9Y592 | Gene names | CCDC41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41 (NY-REN-58 antigen). | |||||
|
EEA1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 86) | NC score | 0.023142 (rank : 91) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
HOOK3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 87) | NC score | 0.023839 (rank : 89) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1070 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q86VS8, Q8NBH0, Q9BY13 | Gene names | HOOK3 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 3 (hHK3). | |||||
|
K1C19_HUMAN
|
||||||
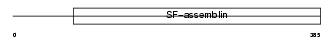
| θ value | 6.88961 (rank : 88) | NC score | 0.043079 (rank : 65) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 320 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P08727, Q5XG83, Q6NW33, Q7L5M9, Q96A53, Q96FV1, Q9BYF9, Q9P1Y4 | Gene names | KRT19 | |||
|
Domain Architecture |
|
|||||
| Description | Keratin, type I cytoskeletal 19 (Cytokeratin-19) (CK-19) (Keratin-19) (K19). | |||||
|
NDE1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 89) | NC score | 0.026888 (rank : 82) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 823 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9NXR1, Q49AQ2 | Gene names | NDE1, NUDE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear distribution protein nudE homolog 1 (NudE). | |||||
|
AINX_MOUSE
|
||||||
| θ value | 8.99809 (rank : 90) | NC score | 0.033601 (rank : 71) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 448 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P46660, Q61958, Q8VCW5 | Gene names | Ina | |||
|
Domain Architecture |
|
|||||
| Description | Alpha-internexin (Alpha-Inx) (66 kDa neurofilament protein) (Neurofilament-66) (NF-66). | |||||
|
ANPRA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 91) | NC score | 0.057058 (rank : 49) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 612 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P16066, Q6P4Q3 | Gene names | NPR1, ANPRA | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic peptide receptor A precursor (ANP-A) (ANPRA) (GC-A) (Guanylate cyclase) (EC 4.6.1.2) (NPR-A) (Atrial natriuretic peptide A-type receptor). | |||||
|
ERC2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 92) | NC score | 0.022759 (rank : 93) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1156 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O15083, Q86TK4 | Gene names | ERC2, KIAA0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2. | |||||
|
ERC2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 93) | NC score | 0.022372 (rank : 95) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1166 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q6PH08, Q80U20, Q8CCP1 | Gene names | Erc2, Cast1, D14Ertd171e, Kiaa0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2 (CAZ-associated structural protein 1) (CAST1). | |||||
|
FLIP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 94) | NC score | 0.020408 (rank : 97) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1176 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9CS72 | Gene names | Filip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
GOGA4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 95) | NC score | 0.022889 (rank : 92) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
K1C14_MOUSE
|
||||||
| θ value | 8.99809 (rank : 96) | NC score | 0.047961 (rank : 54) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 501 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q61781, Q91VQ4 | Gene names | Krt14, Krt1-14 | |||
|
Domain Architecture |
|
|||||
| Description | Keratin, type I cytoskeletal 14 (Cytokeratin-14) (CK-14) (Keratin-14) (K14). | |||||
|
K1C19_MOUSE
|
||||||
| θ value | 8.99809 (rank : 97) | NC score | 0.042801 (rank : 66) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P19001 | Gene names | Krt19, Krt1-19 | |||
|
Domain Architecture |
|
|||||
| Description | Keratin, type I cytoskeletal 19 (Cytokeratin-19) (CK-19) (Keratin-19) (K19). | |||||
|
MYH14_MOUSE
|
||||||
| θ value | 8.99809 (rank : 98) | NC score | 0.025148 (rank : 86) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1225 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q6URW6, Q80V64, Q80ZE6 | Gene names | Myh14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
NIN_MOUSE
|
||||||
| θ value | 8.99809 (rank : 99) | NC score | 0.025591 (rank : 85) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q61043, Q6ZPM7 | Gene names | Nin, Kiaa1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein. | |||||
|
WASF3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 100) | NC score | 0.013330 (rank : 104) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UPY6, O94974 | Gene names | WASF3, KIAA0900, SCAR3, WAVE3 | |||
|
Domain Architecture |

|
|||||
| Description | Wiskott-Aldrich syndrome protein family member 3 (WASP-family protein member 3) (Protein WAVE-3) (Verprolin homology domain-containing protein 3). | |||||
|
WASF3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 101) | NC score | 0.013519 (rank : 103) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8VHI6 | Gene names | Wasf3, Wave3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 3 (WASP-family protein member 3) (Protein WAVE-3). | |||||
|
ANPRA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.058241 (rank : 47) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 600 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P18293 | Gene names | Npr1, Npra | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic peptide receptor A precursor (ANP-A) (ANPRA) (GC-A) (Guanylate cyclase) (EC 4.6.1.2) (NPR-A) (Atrial natriuretic peptide A-type receptor). | |||||
|
ANPRB_HUMAN
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.063429 (rank : 44) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 646 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P20594, O60871, Q9UQ50 | Gene names | NPR2, ANPRB | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic peptide receptor B precursor (ANP-B) (ANPRB) (GC-B) (Guanylate cyclase B) (EC 4.6.1.2) (NPR-B) (Atrial natriuretic peptide B-type receptor). | |||||
|
ANPRB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.067282 (rank : 43) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6VVW5, Q6VVW3, Q6VVW4, Q8CGA9 | Gene names | Npr2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Atrial natriuretic peptide receptor B precursor (ANP-B) (ANPRB) (GC-B) (Guanylate cyclase B) (EC 4.6.1.2) (NPR-B) (Atrial natriuretic peptide B-type receptor). | |||||
|
DAF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.058183 (rank : 48) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P08174, P09679, P78361 | Gene names | CD55, CR, DAF | |||
|
Domain Architecture |
|
|||||
| Description | Complement decay-accelerating factor precursor (CD55 antigen). | |||||
|
GPC5C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.074536 (rank : 41) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NQ84, Q9NZG5 | Gene names | GPRC5C, RAIG3 | |||
|
Domain Architecture |
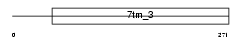
|
|||||
| Description | G-protein coupled receptor family C group 5 member C precursor (Retinoic acid-induced gene 3 protein) (RAIG-3). | |||||
|
GPC5C_MOUSE
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.082651 (rank : 40) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8K3J9, Q8K0H0 | Gene names | Gprc5c, Raig3 | |||
|
Domain Architecture |

|
|||||
| Description | G-protein coupled receptor family C group 5 member C precursor (Retinoic acid-induced gene 3 protein) (RAIG-3). | |||||
|
GPC5D_MOUSE
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.063023 (rank : 45) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9JIL6, Q8CJI0, Q8CJI1 | Gene names | Gprc5d | |||
|
Domain Architecture |
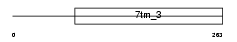
|
|||||
| Description | G-protein coupled receptor family C group 5 member D. | |||||
|
RAI3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.057019 (rank : 50) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BHL4, Q8K067, Q8K1G5 | Gene names | Gprc5a, Rai3, Raig1 | |||
|
Domain Architecture |
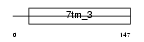
|
|||||
| Description | Retinoic acid-induced protein 3 (G-protein coupled receptor family C group 5 member A) (Retinoic acid-induced gene 1 protein) (RAIG-1). | |||||
|
GABR2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 101 | |
| SwissProt Accessions | O75899, O75974, O75975, Q5VXZ2, Q8WX04, Q9P1R2, Q9UNR1, Q9UNS9 | Gene names | GABBR2, GPR51, GPRC3B | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric acid type B receptor, subunit 2 precursor (GABA-B receptor 2) (GABA-B-R2) (Gb2) (GABABR2) (G-protein coupled receptor 51) (HG20). | |||||
|
GABR1_HUMAN
|
||||||
| NC score | 0.841848 (rank : 2) | θ value | 1.02419e-128 (rank : 3) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9UBS5, O95375, O95468, O95975, O96022, Q86W60, Q9UQQ0 | Gene names | GABBR1, GPRC3A | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric acid type B receptor, subunit 1 precursor (GABA-B receptor 1) (GABA-B-R1) (Gb1). | |||||
|
GABR1_MOUSE
|
||||||
| NC score | 0.841151 (rank : 3) | θ value | 2.69525e-129 (rank : 2) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9WV18, Q9WU48, Q9WV15, Q9WV16, Q9WV17 | Gene names | Gabbr1 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric acid type B receptor, subunit 1 precursor (GABA-B receptor 1) (GABA-B-R1) (Gb1). | |||||
|
GP156_HUMAN
|
||||||
| NC score | 0.593869 (rank : 4) | θ value | 4.89182e-30 (rank : 4) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8NFN8, Q86SN6 | Gene names | GPR156, GABABL, PGR28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 156 (GABAB-related G-protein coupled receptor) (G-protein coupled receptor PGR28). | |||||
|
GP156_MOUSE
|
||||||
| NC score | 0.564731 (rank : 5) | θ value | 2.05525e-28 (rank : 5) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6PCP7, Q8K452 | Gene names | Gpr156, Gababl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 156 (GABAB-related G-protein coupled receptor). | |||||
|
MGR4_HUMAN
|
||||||
| NC score | 0.552022 (rank : 6) | θ value | 1.53129e-07 (rank : 17) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q14833, Q5SZ84 | Gene names | GRM4, GPRC1D, MGLUR4 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 4 precursor (mGluR4). | |||||
|
MGR7_HUMAN
|
||||||
| NC score | 0.550485 (rank : 7) | θ value | 1.9326e-10 (rank : 8) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q14831, Q8NFS2, Q8NFS3, Q8NFS4 | Gene names | GRM7, GPRC1G, MGLUR7 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 7 precursor (mGluR7). | |||||
|
MGR7_MOUSE
|
||||||
| NC score | 0.550229 (rank : 8) | θ value | 1.9326e-10 (rank : 9) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q68ED2 | Gene names | Grm7, Gprc1g, Mglur7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metabotropic glutamate receptor 7 precursor (mGluR7). | |||||
|
MGR8_MOUSE
|
||||||
| NC score | 0.548521 (rank : 9) | θ value | 1.02475e-11 (rank : 6) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P47743, Q6B964 | Gene names | Grm8, Gprc1h, Mglur8 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 8 precursor (mGluR8). | |||||
|
MGR8_HUMAN
|
||||||
| NC score | 0.547714 (rank : 10) | θ value | 1.74796e-11 (rank : 7) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O00222, O15493, O95945, O95946, Q3MIV9, Q52M02, Q6J165 | Gene names | GRM8, GPRC1H, MGLUR8 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 8 precursor (mGluR8). | |||||
|
MGR5_HUMAN
|
||||||
| NC score | 0.539900 (rank : 11) | θ value | 7.34386e-10 (rank : 10) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P41594 | Gene names | GRM5, GPRC1E, MGLUR5 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 5 precursor (mGluR5). | |||||
|
TS1R3_HUMAN
|
||||||
| NC score | 0.539687 (rank : 12) | θ value | 8.97725e-08 (rank : 16) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q7RTX0, Q5TA49, Q8NGW9 | Gene names | TAS1R3, T1R3, TR3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 1 member 3 precursor (Sweet taste receptor T1R3). | |||||
|
MGR6_HUMAN
|
||||||
| NC score | 0.538123 (rank : 13) | θ value | 2.61198e-07 (rank : 18) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O15303 | Gene names | GRM6, GPRC1F, MGLUR6 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 6 precursor (mGluR6). | |||||
|
MGR3_MOUSE
|
||||||
| NC score | 0.537325 (rank : 14) | θ value | 3.64472e-09 (rank : 12) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9QYS2 | Gene names | Grm3, Gprc1c, Mglur3 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 3 precursor (mGluR3). | |||||
|
CASR_HUMAN
|
||||||
| NC score | 0.534264 (rank : 15) | θ value | 2.36244e-08 (rank : 13) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P41180, Q13912, Q16108, Q16109, Q16110, Q16379, Q4PJ19 | Gene names | CASR, GPRC2A, PCAR1 | |||
|
Domain Architecture |

|
|||||
| Description | Extracellular calcium-sensing receptor precursor (CaSR) (Parathyroid Cell calcium-sensing receptor). | |||||
|
TS1R3_MOUSE
|
||||||
| NC score | 0.534080 (rank : 16) | θ value | 2.79066e-09 (rank : 11) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q925D8, Q91VA4, Q923K0, Q925A4, Q925D9 | Gene names | Tas1r3, Sac, T1r3, Tr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 1 member 3 precursor (Sweet taste receptor T1R3) (Saccharin preference protein). | |||||
|
MGR6_MOUSE
|
||||||
| NC score | 0.533495 (rank : 17) | θ value | 2.61198e-07 (rank : 19) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q5NCH9 | Gene names | Grm6, Gprc1f, Mglur6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metabotropic glutamate receptor 6 precursor (mGluR6). | |||||
|
CASR_MOUSE
|
||||||
| NC score | 0.533174 (rank : 18) | θ value | 2.36244e-08 (rank : 14) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9QY96, O08968, O88519, Q9QY95, Q9QZU8, Q9R1D6, Q9R1Y2 | Gene names | Casr, Gprc2a | |||
|
Domain Architecture |

|
|||||
| Description | Extracellular calcium-sensing receptor precursor (CaSR) (Parathyroid Cell calcium-sensing receptor). | |||||
|
MGR1_HUMAN
|
||||||
| NC score | 0.531899 (rank : 19) | θ value | 1.29631e-06 (rank : 25) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q13255, Q13256, Q14757, Q14758, Q5VTF7, Q5VTF8, Q9NU10, Q9UGS9, Q9UGT0 | Gene names | GRM1, GPRC1A, MGLUR1 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 1 precursor (mGluR1). | |||||
|
MGR2_HUMAN
|
||||||
| NC score | 0.531732 (rank : 20) | θ value | 7.59969e-07 (rank : 23) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q14416, Q52MC6, Q9H3N6 | Gene names | GRM2, GPRC1B, MGLUR2 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 2 precursor (mGluR2). | |||||
|
MGR3_HUMAN
|
||||||
| NC score | 0.531497 (rank : 21) | θ value | 4.0297e-08 (rank : 15) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q14832, Q75MV4, Q75N17, Q86YG6, Q8TBH9 | Gene names | GRM3, GPRC1C, MGLUR3 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 3 precursor (mGluR3). | |||||
|
MGR1_MOUSE
|
||||||
| NC score | 0.531294 (rank : 22) | θ value | 1.29631e-06 (rank : 26) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P97772, Q6AXG4, Q9EPV6 | Gene names | Grm1, Gprc1a, Mglur1 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 1 precursor (mGluR1). | |||||
|
TS1R1_HUMAN
|
||||||
| NC score | 0.526939 (rank : 23) | θ value | 7.59969e-07 (rank : 24) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q7RTX1, Q8NGZ7, Q8TDJ7, Q8TDJ8, Q8TDJ9, Q8TDK0 | Gene names | TAS1R1, GPR70, T1R1, TR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 1 member 1 precursor (G-protein coupled receptor 70) (Gm148). | |||||
|
V2RX_MOUSE
|
||||||
| NC score | 0.524592 (rank : 24) | θ value | 4.92598e-06 (rank : 29) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O70410 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vomeronasal type-2 receptor X precursor (Putative pheromone receptor V2RX). | |||||
|
GPC6A_MOUSE
|
||||||
| NC score | 0.519180 (rank : 25) | θ value | 7.59969e-07 (rank : 22) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8K4Z6, Q5DK51, Q5DK52 | Gene names | Gprc6a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor family C group 6 member A precursor. | |||||
|
TS1R2_MOUSE
|
||||||
| NC score | 0.518495 (rank : 26) | θ value | 3.77169e-06 (rank : 28) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q925I4, Q923J8 | Gene names | Tas1r2, Gpr71, T1r2, Tr2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 1 member 2 precursor (G-protein coupled receptor 71) (Sweet taste receptor T1R2). | |||||
|
GPC6A_HUMAN
|
||||||
| NC score | 0.515188 (rank : 27) | θ value | 2.21117e-06 (rank : 27) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q5T6X5, Q6JK43, Q6JK44, Q8NGU8, Q8NHZ9, Q8TDT6 | Gene names | GPRC6A, GPCR33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor family C group 6 member A precursor (hGPRC6A) (G-protein coupled receptor 33) (hGPCR33). | |||||
|
TS1R1_MOUSE
|
||||||
| NC score | 0.511171 (rank : 28) | θ value | 0.000158464 (rank : 34) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q99PG6, Q6NS58, Q923J9, Q925I5, Q99PG5 | Gene names | Tas1r1, Gpr70, T1r1, Tr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 1 member 1 precursor (G-protein coupled receptor 70). | |||||
|
TS1R2_HUMAN
|
||||||
| NC score | 0.507661 (rank : 29) | θ value | 2.44474e-05 (rank : 30) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8TE23, Q5TZ19 | Gene names | TAS1R2, GPR71, T1R2, TR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 1 member 2 precursor (G-protein coupled receptor 71) (Sweet taste receptor T1R2). | |||||
|
V2R1B_MOUSE
|
||||||
| NC score | 0.458942 (rank : 30) | θ value | 0.0961366 (rank : 41) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6TAC4, O70409 | Gene names | V2r1b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vomeronasal type-2 receptor 1b precursor (Putative pheromone receptor V2R1b). | |||||
|
GP158_HUMAN
|
||||||
| NC score | 0.248411 (rank : 31) | θ value | 0.00228821 (rank : 35) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q5T848, Q6QR81, Q9ULT3 | Gene names | GPR158, KIAA1136 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 158 precursor. | |||||
|
ANPRC_MOUSE
|
||||||
| NC score | 0.247306 (rank : 32) | θ value | 3.19293e-05 (rank : 31) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P70180, P97804, Q9R025, Q9R027, Q9R028 | Gene names | Npr3 | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic peptide clearance receptor precursor (ANP-C) (ANPRC) (NPR-C) (Atrial natriuretic peptide C-type receptor) (EF-2). | |||||
|
GP158_MOUSE
|
||||||
| NC score | 0.239797 (rank : 33) | θ value | 0.000158464 (rank : 33) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8C419, Q8CHB0 | Gene names | Gpr158, Kiaa1136 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 158 precursor. | |||||
|
ANPRC_HUMAN
|
||||||
| NC score | 0.239653 (rank : 34) | θ value | 9.29e-05 (rank : 32) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P17342 | Gene names | NPR3, ANPRC | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic peptide clearance receptor precursor (ANP-C) (ANPRC) (NPR-C) (Atrial natriuretic peptide C-type receptor). | |||||
|
GP179_HUMAN
|
||||||
| NC score | 0.218523 (rank : 35) | θ value | 4.45536e-07 (rank : 20) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6PRD1 | Gene names | GPR179, GPR158L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 179 precursor (Probable G-protein coupled receptor 158-like 1). | |||||
|
GPC5B_MOUSE
|
||||||
| NC score | 0.182346 (rank : 36) | θ value | 0.279714 (rank : 44) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q923Z0, Q8CCV3 | Gene names | Gprc5b, Raig2 | |||
|
Domain Architecture |

|
|||||
| Description | G-protein coupled receptor family C group 5 member B precursor (Retinoic acid-induced gene 2 protein) (RAIG-2). | |||||
|
GPC5B_HUMAN
|
||||||
| NC score | 0.179566 (rank : 37) | θ value | 0.0330416 (rank : 38) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9NZH0, O75205, Q8NBZ8 | Gene names | GPRC5B, RAIG2 | |||
|
Domain Architecture |

|
|||||
| Description | G-protein coupled receptor family C group 5 member B precursor (Retinoic acid-induced gene 2 protein) (RAIG-2) (A-69G12.1). | |||||
|
NMDZ1_MOUSE
|
||||||
| NC score | 0.146142 (rank : 38) | θ value | 0.00509761 (rank : 37) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P35438, Q8CFS4 | Gene names | Grin1, Glurz1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit zeta 1 precursor (N-methyl-D- aspartate receptor subunit NR1). | |||||
|
NMDZ1_HUMAN
|
||||||
| NC score | 0.145981 (rank : 39) | θ value | 0.00298849 (rank : 36) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q05586, P35437, Q12867, Q12868 | Gene names | GRIN1, NMDAR1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit zeta 1 precursor (N-methyl-D- aspartate receptor subunit NR1). | |||||
|
GPC5C_MOUSE
|
||||||
| NC score | 0.082651 (rank : 40) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8K3J9, Q8K0H0 | Gene names | Gprc5c, Raig3 | |||
|
Domain Architecture |

|
|||||
| Description | G-protein coupled receptor family C group 5 member C precursor (Retinoic acid-induced gene 3 protein) (RAIG-3). | |||||
|
GPC5C_HUMAN
|
||||||
| NC score | 0.074536 (rank : 41) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NQ84, Q9NZG5 | Gene names | GPRC5C, RAIG3 | |||
|
Domain Architecture |

|
|||||
| Description | G-protein coupled receptor family C group 5 member C precursor (Retinoic acid-induced gene 3 protein) (RAIG-3). | |||||
|
GUC2F_HUMAN
|
||||||
| NC score | 0.070286 (rank : 42) | θ value | 4.45536e-07 (rank : 21) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 740 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P51841, Q9UJF1 | Gene names | GUCY2F, GUC2F, RETGC2 | |||
|
Domain Architecture |

|
|||||
| Description | Retinal guanylyl cyclase 2 precursor (EC 4.6.1.2) (Guanylate cyclase 2F, retinal) (RETGC-2) (Rod outer segment membrane guanylate cyclase 2) (ROS-GC2) (Guanylate cyclase F) (GC-F). | |||||
|
ANPRB_MOUSE
|
||||||
| NC score | 0.067282 (rank : 43) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6VVW5, Q6VVW3, Q6VVW4, Q8CGA9 | Gene names | Npr2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Atrial natriuretic peptide receptor B precursor (ANP-B) (ANPRB) (GC-B) (Guanylate cyclase B) (EC 4.6.1.2) (NPR-B) (Atrial natriuretic peptide B-type receptor). | |||||
|
ANPRB_HUMAN
|
||||||
| NC score | 0.063429 (rank : 44) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 646 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P20594, O60871, Q9UQ50 | Gene names | NPR2, ANPRB | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic peptide receptor B precursor (ANP-B) (ANPRB) (GC-B) (Guanylate cyclase B) (EC 4.6.1.2) (NPR-B) (Atrial natriuretic peptide B-type receptor). | |||||
|
GPC5D_MOUSE
|
||||||
| NC score | 0.063023 (rank : 45) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9JIL6, Q8CJI0, Q8CJI1 | Gene names | Gprc5d | |||
|
Domain Architecture |

|
|||||
| Description | G-protein coupled receptor family C group 5 member D. | |||||
|
GUC2G_MOUSE
|
||||||
| NC score | 0.062583 (rank : 46) | θ value | 0.0431538 (rank : 39) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6TL19, Q8BWU7 | Gene names | Gucy2g | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanylate cyclase 2G precursor (EC 4.6.1.2) (Guanylyl cyclase receptor G) (mGC-G). | |||||
|
ANPRA_MOUSE
|
||||||
| NC score | 0.058241 (rank : 47) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 600 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P18293 | Gene names | Npr1, Npra | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic peptide receptor A precursor (ANP-A) (ANPRA) (GC-A) (Guanylate cyclase) (EC 4.6.1.2) (NPR-A) (Atrial natriuretic peptide A-type receptor). | |||||
|
DAF_HUMAN
|
||||||
| NC score | 0.058183 (rank : 48) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P08174, P09679, P78361 | Gene names | CD55, CR, DAF | |||
|
Domain Architecture |

|
|||||
| Description | Complement decay-accelerating factor precursor (CD55 antigen). | |||||
|
ANPRA_HUMAN
|
||||||
| NC score | 0.057058 (rank : 49) | θ value | 8.99809 (rank : 91) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 612 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P16066, Q6P4Q3 | Gene names | NPR1, ANPRA | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic peptide receptor A precursor (ANP-A) (ANPRA) (GC-A) (Guanylate cyclase) (EC 4.6.1.2) (NPR-A) (Atrial natriuretic peptide A-type receptor). | |||||
|
RAI3_MOUSE
|
||||||
| NC score | 0.057019 (rank : 50) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BHL4, Q8K067, Q8K1G5 | Gene names | Gprc5a, Rai3, Raig1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoic acid-induced protein 3 (G-protein coupled receptor family C group 5 member A) (Retinoic acid-induced gene 1 protein) (RAIG-1). | |||||
|
K1C10_MOUSE
|
||||||
| NC score | 0.049703 (rank : 51) | θ value | 0.0961366 (rank : 40) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 466 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P02535, P08731, Q8BUX3, Q8BV09, Q8BVU3, Q9CXH6, Q9JKB4 | Gene names | Krt10, Krt1-10 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 10 (Cytokeratin-10) (CK-10) (Keratin-10) (K10) (56 kDa cytokeratin) (Keratin, type I cytoskeletal 59 kDa). | |||||
|
K1C10_HUMAN
|
||||||
| NC score | 0.049334 (rank : 52) | θ value | 0.125558 (rank : 42) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 526 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P13645, Q14664, Q8N175 | Gene names | KRT10 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 10 (Cytokeratin-10) (CK-10) (Keratin-10) (K10). | |||||
|
NMDE2_MOUSE
|
||||||
| NC score | 0.048531 (rank : 53) | θ value | 0.47712 (rank : 47) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q01097, Q9DCB2 | Gene names | Grin2b | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 2 precursor (N-methyl D- aspartate receptor subtype 2B) (NR2B) (NMDAR2B). | |||||
|
K1C14_MOUSE
|
||||||
| NC score | 0.047961 (rank : 54) | θ value | 8.99809 (rank : 96) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 501 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q61781, Q91VQ4 | Gene names | Krt14, Krt1-14 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 14 (Cytokeratin-14) (CK-14) (Keratin-14) (K14). | |||||
|
K1C14_HUMAN
|
||||||
| NC score | 0.047372 (rank : 55) | θ value | 1.81305 (rank : 60) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P02533, Q14715, Q53XY3, Q9BUE3, Q9UBN2, Q9UBN3, Q9UCY4 | Gene names | KRT14 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 14 (Cytokeratin-14) (CK-14) (Keratin-14) (K14). | |||||
|
K1C15_MOUSE
|
||||||
| NC score | 0.047350 (rank : 56) | θ value | 0.813845 (rank : 48) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 429 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q61414, Q6PEQ0 | Gene names | Krt15, Krt1-15 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 15 (Cytokeratin-15) (CK-15) (Keratin-15) (K15). | |||||
|
K1C17_HUMAN
|
||||||
| NC score | 0.046640 (rank : 57) | θ value | 1.38821 (rank : 56) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 463 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q04695 | Gene names | KRT17 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 17 (Cytokeratin-17) (CK-17) (Keratin-17) (K17) (39.1). | |||||
|
K1C17_MOUSE
|
||||||
| NC score | 0.046350 (rank : 58) | θ value | 1.38821 (rank : 57) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9QWL7, Q61783 | Gene names | Krt17, Krt1-17 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 17 (Cytokeratin-17) (CK-17) (Keratin-17) (K17). | |||||
|
K1C15_HUMAN
|
||||||
| NC score | 0.045997 (rank : 59) | θ value | 1.06291 (rank : 50) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 367 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P19012, Q53XV8, Q9BUG4 | Gene names | KRT15, KRTB | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 15 (Cytokeratin-15) (CK-15) (Keratin-15) (K15). | |||||
|
K1C13_HUMAN
|
||||||
| NC score | 0.045943 (rank : 60) | θ value | 1.06291 (rank : 49) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 386 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P13646, Q53G54, Q6AZK5, Q8N240 | Gene names | KRT13 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 13 (Cytokeratin-13) (CK-13) (Keratin-13) (K13). | |||||
|
NMDE2_HUMAN
|
||||||
| NC score | 0.045638 (rank : 61) | θ value | 0.47712 (rank : 46) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13224, Q12919, Q13220, Q13225, Q9UM56 | Gene names | GRIN2B | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 2 precursor (N-methyl D- aspartate receptor subtype 2B) (NR2B) (NMDAR2B) (N-methyl-D-aspartate receptor subunit 3) (NR3) (hNR3). | |||||
|
K1C12_HUMAN
|
||||||
| NC score | 0.045418 (rank : 62) | θ value | 1.38821 (rank : 54) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q99456 | Gene names | KRT12 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 12 (Cytokeratin-12) (CK-12) (Keratin-12) (K12). | |||||
|
K1C13_MOUSE
|
||||||
| NC score | 0.045287 (rank : 63) | θ value | 1.38821 (rank : 55) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 410 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P08730, Q3V176, Q6PAI1 | Gene names | Krt13, Krt1-13 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 13 (Cytokeratin-13) (CK-13) (Keratin-13) (K13) (47 kDa cytokeratin). | |||||
|
VDP_HUMAN
|
||||||
| NC score | 0.045134 (rank : 64) | θ value | 0.279714 (rank : 45) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O60763 | Gene names | VDP | |||
|
Domain Architecture |

|
|||||
| Description | General vesicular transport factor p115 (Transcytosis-associated protein) (TAP) (Vesicle docking protein). | |||||
|
K1C19_HUMAN
|
||||||
| NC score | 0.043079 (rank : 65) | θ value | 6.88961 (rank : 88) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 320 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P08727, Q5XG83, Q6NW33, Q7L5M9, Q96A53, Q96FV1, Q9BYF9, Q9P1Y4 | Gene names | KRT19 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 19 (Cytokeratin-19) (CK-19) (Keratin-19) (K19). | |||||
|
K1C19_MOUSE
|
||||||
| NC score | 0.042801 (rank : 66) | θ value | 8.99809 (rank : 97) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P19001 | Gene names | Krt19, Krt1-19 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 19 (Cytokeratin-19) (CK-19) (Keratin-19) (K19). | |||||
|
K1HB_HUMAN
|
||||||
| NC score | 0.041368 (rank : 67) | θ value | 5.27518 (rank : 76) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 470 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q14525, O76010 | Gene names | KRTHA3B, HHA3-II, HKA3B | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cuticular Ha3-II (Hair keratin, type I Ha3-II). | |||||
|
NDE1_MOUSE
|
||||||
| NC score | 0.037073 (rank : 68) | θ value | 1.81305 (rank : 62) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 602 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9CZA6, Q3UBS6, Q3UIC1, Q9ERR0 | Gene names | Nde1, Nude | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear distribution protein nudE homolog 1 (NudE) (mNudE). | |||||
|
NMDE1_HUMAN
|
||||||
| NC score | 0.036046 (rank : 69) | θ value | 5.27518 (rank : 78) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q12879, O00669 | Gene names | GRIN2A | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 1 precursor (N-methyl D- aspartate receptor subtype 2A) (NR2A) (NMDAR2A) (hNR2A). | |||||
|
AINX_HUMAN
|
||||||
| NC score | 0.033804 (rank : 70) | θ value | 6.88961 (rank : 83) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 442 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q16352, Q9BRC5 | Gene names | INA | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-internexin (Alpha-Inx) (66 kDa neurofilament protein) (Neurofilament-66) (NF-66). | |||||
|
AINX_MOUSE
|
||||||
| NC score | 0.033601 (rank : 71) | θ value | 8.99809 (rank : 90) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 448 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P46660, Q61958, Q8VCW5 | Gene names | Ina | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-internexin (Alpha-Inx) (66 kDa neurofilament protein) (Neurofilament-66) (NF-66). | |||||
|
KNTC2_MOUSE
|
||||||
| NC score | 0.032150 (rank : 72) | θ value | 1.38821 (rank : 58) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 692 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9D0F1, Q3TQT6, Q3UWM5, Q99P70 | Gene names | Kntc2, Hec1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Hec1 (Kinetochore-associated protein 2). | |||||
|
CK5P2_MOUSE
|
||||||
| NC score | 0.029622 (rank : 73) | θ value | 5.27518 (rank : 75) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1151 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8K389, Q6PCN1, Q6ZPL0 | Gene names | Cdk5rap2, Kiaa1633 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48). | |||||
|
MYH2_HUMAN
|
||||||
| NC score | 0.029072 (rank : 74) | θ value | 4.03905 (rank : 71) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1491 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9UKX2, Q14322, Q16229 | Gene names | MYH2, MYHSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, skeletal muscle, adult 2 (Myosin heavy chain IIa) (MyHC-IIa). | |||||
|
GCC2_MOUSE
|
||||||
| NC score | 0.028849 (rank : 75) | θ value | 1.38821 (rank : 53) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
MYH9_MOUSE
|
||||||
| NC score | 0.028655 (rank : 76) | θ value | 1.06291 (rank : 52) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
MYH8_MOUSE
|
||||||
| NC score | 0.028576 (rank : 77) | θ value | 1.06291 (rank : 51) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1600 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P13542, Q5SX36 | Gene names | Myh8, Myhsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
MYH1_MOUSE
|
||||||
| NC score | 0.028471 (rank : 78) | θ value | 5.27518 (rank : 77) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1461 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q5SX40 | Gene names | Myh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1). | |||||
|
MYH4_MOUSE
|
||||||
| NC score | 0.028035 (rank : 79) | θ value | 3.0926 (rank : 67) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
MYH9_HUMAN
|
||||||
| NC score | 0.027623 (rank : 80) | θ value | 2.36792 (rank : 64) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
CE290_HUMAN
|
||||||
| NC score | 0.027328 (rank : 81) | θ value | 5.27518 (rank : 74) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
NDE1_HUMAN
|
||||||
| NC score | 0.026888 (rank : 82) | θ value | 6.88961 (rank : 89) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 823 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9NXR1, Q49AQ2 | Gene names | NDE1, NUDE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear distribution protein nudE homolog 1 (NudE). | |||||
|
HOOK3_MOUSE
|
||||||
| NC score | 0.026225 (rank : 83) | θ value | 3.0926 (rank : 65) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8BUK6, Q8BV48, Q8BW57, Q8BY41 | Gene names | Hook3 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 3. | |||||
|
KTN1_HUMAN
|
||||||
| NC score | 0.025867 (rank : 84) | θ value | 4.03905 (rank : 70) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q86UP2, Q13999, Q14707, Q15387, Q86W57 | Gene names | KTN1, CG1, KIAA0004 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin (Kinesin receptor) (CG-1 antigen). | |||||
|
NIN_MOUSE
|
||||||
| NC score | 0.025591 (rank : 85) | θ value | 8.99809 (rank : 99) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q61043, Q6ZPM7 | Gene names | Nin, Kiaa1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein. | |||||
|
MYH14_MOUSE
|
||||||
| NC score | 0.025148 (rank : 86) | θ value | 8.99809 (rank : 98) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1225 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q6URW6, Q80V64, Q80ZE6 | Gene names | Myh14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
CCD41_HUMAN
|
||||||
| NC score | 0.024695 (rank : 87) | θ value | 6.88961 (rank : 85) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1139 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9Y592 | Gene names | CCDC41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41 (NY-REN-58 antigen). | |||||
|
CCD27_MOUSE
|
||||||
| NC score | 0.024338 (rank : 88) | θ value | 6.88961 (rank : 84) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q3V036 | Gene names | Ccdc27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 27. | |||||
|
HOOK3_HUMAN
|
||||||
| NC score | 0.023839 (rank : 89) | θ value | 6.88961 (rank : 87) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1070 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q86VS8, Q8NBH0, Q9BY13 | Gene names | HOOK3 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 3 (hHK3). | |||||
|
UACA_HUMAN
|
||||||
| NC score | 0.023838 (rank : 90) | θ value | 3.0926 (rank : 68) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1622 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9BZF9, Q8N3B8, Q9HCL1, Q9NWC6 | Gene names | UACA, KIAA1561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uveal autoantigen with coiled-coil domains and ankyrin repeats protein. | |||||
|
EEA1_HUMAN
|
||||||
| NC score | 0.023142 (rank : 91) | θ value | 6.88961 (rank : 86) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
GOGA4_MOUSE
|
||||||
| NC score | 0.022889 (rank : 92) | θ value | 8.99809 (rank : 95) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
ERC2_HUMAN
|
||||||
| NC score | 0.022759 (rank : 93) | θ value | 8.99809 (rank : 92) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1156 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O15083, Q86TK4 | Gene names | ERC2, KIAA0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2. | |||||
|
SYTL1_HUMAN
|
||||||
| NC score | 0.022657 (rank : 94) | θ value | 0.125558 (rank : 43) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IYJ3, Q96BB6, Q96GU6, Q96S89, Q96SI0 | Gene names | SYTL1, SLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 1 (Exophilin-7) (Protein JFC1) (SB146). | |||||
|
ERC2_MOUSE
|
||||||
| NC score | 0.022372 (rank : 95) | θ value | 8.99809 (rank : 93) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1166 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q6PH08, Q80U20, Q8CCP1 | Gene names | Erc2, Cast1, D14Ertd171e, Kiaa0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2 (CAZ-associated structural protein 1) (CAST1). | |||||
|
KIF5A_MOUSE
|
||||||
| NC score | 0.020711 (rank : 96) | θ value | 1.81305 (rank : 61) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 859 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P33175, Q5DTP1, Q6PDY7, Q9Z2F9 | Gene names | Kif5a, Kiaa4086, Kif5, Nkhc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin heavy chain isoform 5A (Neuronal kinesin heavy chain) (NKHC) (Kinesin heavy chain neuron-specific 1). | |||||
|
FLIP1_MOUSE
|
||||||
| NC score | 0.020408 (rank : 97) | θ value | 8.99809 (rank : 94) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1176 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9CS72 | Gene names | Filip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
KIF5A_HUMAN
|
||||||
| NC score | 0.018904 (rank : 98) | θ value | 4.03905 (rank : 69) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 843 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q12840, Q4LE26 | Gene names | KIF5A, NKHC1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain isoform 5A (Neuronal kinesin heavy chain) (NKHC) (Kinesin heavy chain neuron-specific 1). | |||||
|
SPTB2_MOUSE
|
||||||
| NC score | 0.018010 (rank : 99) | θ value | 5.27518 (rank : 80) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q62261, Q5SQL8 | Gene names | Sptbn1, Spnb-2, Spnb2, Sptb2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
SPTB2_HUMAN
|
||||||
| NC score | 0.017180 (rank : 100) | θ value | 5.27518 (rank : 79) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q01082, O60837, Q16057 | Gene names | SPTBN1, SPTB2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
WWTR1_MOUSE
|
||||||
| NC score | 0.014648 (rank : 101) | θ value | 4.03905 (rank : 73) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9EPK5, Q99KI4 | Gene names | Wwtr1, Taz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing transcription regulator protein 1 (Transcriptional coactivator with PDZ-binding motif). | |||||
|
ITB5_HUMAN
|
||||||
| NC score | 0.013588 (rank : 102) | θ value | 1.81305 (rank : 59) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P18084 | Gene names | ITGB5 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-5 precursor. | |||||
|
WASF3_MOUSE
|
||||||
| NC score | 0.013519 (rank : 103) | θ value | 8.99809 (rank : 101) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8VHI6 | Gene names | Wasf3, Wave3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 3 (WASP-family protein member 3) (Protein WAVE-3). | |||||
|
WASF3_HUMAN
|
||||||
| NC score | 0.013330 (rank : 104) | θ value | 8.99809 (rank : 100) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UPY6, O94974 | Gene names | WASF3, KIAA0900, SCAR3, WAVE3 | |||
|
Domain Architecture |

|
|||||
| Description | Wiskott-Aldrich syndrome protein family member 3 (WASP-family protein member 3) (Protein WAVE-3) (Verprolin homology domain-containing protein 3). | |||||
|
WASF2_HUMAN
|
||||||
| NC score | 0.012991 (rank : 105) | θ value | 5.27518 (rank : 81) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y6W5, O60794, Q9UDY7 | Gene names | WASF2, WAVE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 2 (WASP-family protein member 2) (Protein WAVE-2) (Verprolin homology domain-containing protein 2). | |||||
|
ITB5_MOUSE
|
||||||
| NC score | 0.012785 (rank : 106) | θ value | 2.36792 (rank : 63) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O70309, O70308, O88347 | Gene names | Itgb5 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-5 precursor. | |||||
|
ROCK1_MOUSE
|
||||||
| NC score | 0.010153 (rank : 107) | θ value | 4.03905 (rank : 72) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 2126 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P70335, Q8C3G4, Q8C7H0 | Gene names | Rock1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK). | |||||
|
LAMA1_HUMAN
|
||||||
| NC score | 0.008006 (rank : 108) | θ value | 3.0926 (rank : 66) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 804 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P25391 | Gene names | LAMA1, LAMA | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-1 chain precursor (Laminin A chain). | |||||
|
ZNF8_HUMAN
|
||||||
| NC score | -0.003342 (rank : 109) | θ value | 5.27518 (rank : 82) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P17098, Q6PI99 | Gene names | ZNF8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 8 (Zinc finger protein HF.18). | |||||