Please be patient as the page loads
|
EXOC4_HUMAN
|
||||||
| SwissProt Accessions | Q96A65, Q9C0G4, Q9H9K0, Q9P102 | Gene names | EXOC4, KIAA1699, SEC8, SEC8L1 | |||
|
Domain Architecture |
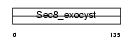
|
|||||
| Description | Exocyst complex component 4 (Exocyst complex component Sec8). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
EXOC4_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q96A65, Q9C0G4, Q9H9K0, Q9P102 | Gene names | EXOC4, KIAA1699, SEC8, SEC8L1 | |||
|
Domain Architecture |
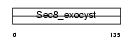
|
|||||
| Description | Exocyst complex component 4 (Exocyst complex component Sec8). | |||||
|
EXOC4_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.990194 (rank : 2) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O35382 | Gene names | Exoc4, Sec8, Sec8l1 | |||
|
Domain Architecture |
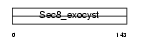
|
|||||
| Description | Exocyst complex component 4 (Exocyst complex component Sec8). | |||||
|
MYO5A_HUMAN
|
||||||
| θ value | 7.1131e-05 (rank : 3) | NC score | 0.089103 (rank : 7) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1092 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9Y4I1, O60653, Q07902, Q16249, Q9UE30, Q9UE31 | Gene names | MYO5A, MYH12 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5A (Myosin Va) (Dilute myosin heavy chain, non-muscle) (Myosin heavy chain 12) (Myoxin). | |||||
|
MYO5A_MOUSE
|
||||||
| θ value | 0.000786445 (rank : 4) | NC score | 0.084689 (rank : 9) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1117 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q99104 | Gene names | Myo5a, Dilute | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5A (Myosin Va) (Dilute myosin heavy chain, non-muscle). | |||||
|
ZW10_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 5) | NC score | 0.153233 (rank : 3) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O54692, Q3TIA5, Q3ULW1, Q921H3 | Gene names | Zw10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere/kinetochore protein zw10 homolog. | |||||
|
TRIPB_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 6) | NC score | 0.090102 (rank : 6) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
GCC2_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 7) | NC score | 0.091847 (rank : 4) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
RBP24_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 8) | NC score | 0.091806 (rank : 5) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
LIPA3_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 9) | NC score | 0.084397 (rank : 10) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O75145, Q9H8B5, Q9UEW4 | Gene names | PPFIA3, KIAA0654 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-3 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-3) (PTPRF-interacting protein alpha-3). | |||||
|
CENPE_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 10) | NC score | 0.068675 (rank : 20) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
LRC40_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 11) | NC score | 0.032753 (rank : 178) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9CRC8, Q78WQ9, Q8BS83 | Gene names | Lrrc40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 40. | |||||
|
CJ118_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 12) | NC score | 0.079179 (rank : 12) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1132 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8C9S4, Q6P3D6, Q8CJC0 | Gene names | Otg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf118 homolog (Oocyte-testis gene 1 protein). | |||||
|
RGNEF_MOUSE
|
||||||
| θ value | 0.21417 (rank : 13) | NC score | 0.056147 (rank : 62) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 641 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P97433 | Gene names | Rgnef, Rhoip2 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-guanine nucleotide exchange factor (Rho-interacting protein 2) (RhoGEF) (RIP2). | |||||
|
GCC2_MOUSE
|
||||||
| θ value | 0.365318 (rank : 14) | NC score | 0.077970 (rank : 14) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
PCNT_MOUSE
|
||||||
| θ value | 0.365318 (rank : 15) | NC score | 0.078528 (rank : 13) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1207 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P48725 | Gene names | Pcnt, Pcnt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pericentrin. | |||||
|
PIBF1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 16) | NC score | 0.074305 (rank : 16) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8WXW3, O95664, Q6U9V2, Q6UG50, Q86V07, Q96SF4 | Gene names | PIBF1, C13orf24, PIBF | |||
|
Domain Architecture |

|
|||||
| Description | Progesterone-induced-blocking factor 1. | |||||
|
GBRT_HUMAN
|
||||||
| θ value | 0.62314 (rank : 17) | NC score | 0.013978 (rank : 194) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UN88, Q9NZK8 | Gene names | GABRQ | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric-acid receptor subunit theta precursor (GABA(A) receptor subunit theta). | |||||
|
KIF5C_HUMAN
|
||||||
| θ value | 0.62314 (rank : 18) | NC score | 0.053426 (rank : 101) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 974 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O60282, O95079 | Gene names | KIF5C, KIAA0531, NKHC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin heavy chain isoform 5C (Kinesin heavy chain neuron-specific 2). | |||||
|
TMCC2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 19) | NC score | 0.068764 (rank : 19) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O75069, Q6ZN09 | Gene names | TMCC2, KIAA0481 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane and coiled-coil domains protein 2 (Cerebral protein 11). | |||||
|
TMCC2_MOUSE
|
||||||
| θ value | 0.62314 (rank : 20) | NC score | 0.068391 (rank : 21) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q80W04, Q6A061 | Gene names | Tmcc2, Kiaa0481 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane and coiled-coil domains protein 2. | |||||
|
CF060_HUMAN
|
||||||
| θ value | 0.813845 (rank : 21) | NC score | 0.074593 (rank : 15) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1270 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8NB25, Q96GY8, Q9H851 | Gene names | C6orf60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf60. | |||||
|
3BP1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 22) | NC score | 0.022025 (rank : 189) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P55194, Q99KK8 | Gene names | Sh3bp1, 3bp1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 1 (3BP-1). | |||||
|
CING_MOUSE
|
||||||
| θ value | 1.38821 (rank : 23) | NC score | 0.065079 (rank : 25) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1432 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P59242 | Gene names | Cgn | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
K22E_HUMAN
|
||||||
| θ value | 1.38821 (rank : 24) | NC score | 0.028375 (rank : 182) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 515 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P35908 | Gene names | KRT2A, KRT2E | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 2 epidermal (Cytokeratin-2e) (K2e) (CK 2e). | |||||
|
SMC3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 25) | NC score | 0.069903 (rank : 18) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1186 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9UQE7, O60464 | Gene names | CSPG6, BAM, BMH, SMC3, SMC3L1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome-associated polypeptide) (hCAP) (Bamacan) (Basement membrane-associated chondroitin proteoglycan). | |||||
|
SMC3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 26) | NC score | 0.070198 (rank : 17) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1187 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9CW03, O35667, Q9QUS3 | Gene names | Cspg6, Bam, Bmh, Mmip1, Smc3, Smc3l1, Smcd | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome segregation protein SmcD) (Bamacan) (Basement membrane-associated chondroitin proteoglycan) (Mad member- interacting protein 1). | |||||
|
SMG1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 27) | NC score | 0.048134 (rank : 169) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BKX6, Q5DU42, Q6ZQC0, Q8BLU4, Q8BWJ5, Q8BXD3 | Gene names | Smg1, Kiaa0421 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SMG1 (EC 2.7.11.1) (SMG-1). | |||||
|
XPOT_HUMAN
|
||||||
| θ value | 1.38821 (rank : 28) | NC score | 0.043872 (rank : 171) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O43592, O43784, Q8WUG2, Q9BVS7 | Gene names | XPOT | |||
|
Domain Architecture |

|
|||||
| Description | Exportin-T (tRNA exportin) (Exportin(tRNA)). | |||||
|
BIR1F_MOUSE
|
||||||
| θ value | 1.81305 (rank : 29) | NC score | 0.035858 (rank : 177) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JIB6, O09121, O09122, P81704 | Gene names | Birc1f, Naip-rs4, Naip6 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1f (Neuronal apoptosis inhibitory protein 6). | |||||
|
BIR1G_MOUSE
|
||||||
| θ value | 1.81305 (rank : 30) | NC score | 0.035916 (rank : 176) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JIB3 | Gene names | Birc1g, Naip7 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1g (Neuronal apoptosis inhibitory protein 7). | |||||
|
MRCKB_HUMAN
|
||||||
| θ value | 1.81305 (rank : 31) | NC score | 0.030028 (rank : 179) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 2239 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9Y5S2, Q2L7A5, Q86TJ1, Q9ULU5 | Gene names | CDC42BPB, KIAA1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
AKAP9_HUMAN
|
||||||
| θ value | 2.36792 (rank : 32) | NC score | 0.067559 (rank : 22) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
CR034_HUMAN
|
||||||
| θ value | 2.36792 (rank : 33) | NC score | 0.086591 (rank : 8) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q5BJE1, Q6ZP67, Q6ZU20 | Gene names | C18orf34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C18orf34. | |||||
|
MYH10_HUMAN
|
||||||
| θ value | 2.36792 (rank : 34) | NC score | 0.065056 (rank : 26) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P35580, Q16087 | Gene names | MYH10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
SMG1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 35) | NC score | 0.051112 (rank : 147) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q96Q15, O43305, Q13284, Q8NFX2, Q96QV0, Q96RW3 | Gene names | SMG1, ATX, KIAA0421, LIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SMG1 (EC 2.7.11.1) (SMG-1) (hSMG-1) (Lambda/iota protein kinase C-interacting protein) (Lambda-interacting protein) (61E3.4). | |||||
|
SYTL4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 36) | NC score | 0.017254 (rank : 191) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96C24, Q5H9J3, Q5JPG8, Q8N9P4, Q9H4R0, Q9H4R1 | Gene names | SYTL4 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 4 (Exophilin-2) (Granuphilin). | |||||
|
OMGP_MOUSE
|
||||||
| θ value | 3.0926 (rank : 37) | NC score | 0.013523 (rank : 195) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q63912 | Gene names | Omg, Omgp | |||
|
Domain Architecture |
|
|||||
| Description | Oligodendrocyte-myelin glycoprotein precursor. | |||||
|
PRP31_MOUSE
|
||||||
| θ value | 3.0926 (rank : 38) | NC score | 0.036833 (rank : 175) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CCF0, Q6P7X2, Q8BQ91, Q8C8U4, Q8C8V5, Q8CCG6, Q8CF52, Q8VBW3 | Gene names | Prpf31, Prp31 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U4/U6 small nuclear ribonucleoprotein Prp31 (Pre-mRNA-processing factor 31) (U4/U6 snRNP 61 kDa protein) (Protein 61K). | |||||
|
SPAG5_MOUSE
|
||||||
| θ value | 3.0926 (rank : 39) | NC score | 0.079839 (rank : 11) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 612 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q7TME2, Q8CH61 | Gene names | Spag5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 5 (Mastrin) (Mitotic spindle-associated protein p126) (MAP126). | |||||
|
SYCP1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 40) | NC score | 0.066058 (rank : 24) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q15431, O14963 | Gene names | SYCP1, SCP1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
TRI34_MOUSE
|
||||||
| θ value | 3.0926 (rank : 41) | NC score | 0.015241 (rank : 193) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99PP6, Q99PP4, Q99PP5 | Gene names | Trim34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 34. | |||||
|
CING_HUMAN
|
||||||
| θ value | 4.03905 (rank : 42) | NC score | 0.066699 (rank : 23) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9P2M7, Q5T386, Q9NR25 | Gene names | CGN, KIAA1319 | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
KIF17_MOUSE
|
||||||
| θ value | 4.03905 (rank : 43) | NC score | 0.028617 (rank : 181) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q99PW8 | Gene names | Kif17 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF17 (MmKIF17). | |||||
|
KIF5C_MOUSE
|
||||||
| θ value | 4.03905 (rank : 44) | NC score | 0.048759 (rank : 167) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P28738, Q9Z2F8 | Gene names | Kif5c, Nkhc2 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain isoform 5C (Kinesin heavy chain neuron-specific 2). | |||||
|
MACF4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 45) | NC score | 0.050382 (rank : 158) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1321 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q96PK2, Q8WXY1 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoform 4. | |||||
|
MRCKB_MOUSE
|
||||||
| θ value | 4.03905 (rank : 46) | NC score | 0.028735 (rank : 180) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 2180 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q7TT50, Q69ZR5, Q80W33 | Gene names | Cdc42bpb, Kiaa1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
MYH10_MOUSE
|
||||||
| θ value | 4.03905 (rank : 47) | NC score | 0.064459 (rank : 27) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1612 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q61879, Q5SV63 | Gene names | Myh10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
SPT16_HUMAN
|
||||||
| θ value | 4.03905 (rank : 48) | NC score | 0.043633 (rank : 172) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y5B9, Q6GMT8, Q6P2F1, Q6PJM1, Q9NRX0 | Gene names | SUPT16H, FACT140, FACTP140 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FACT complex subunit SPT16 (Facilitates chromatin transcription complex subunit SPT16) (hSPT16) (FACT 140 kDa subunit) (FACTp140) (Chromatin-specific transcription elongation factor 140 kDa subunit). | |||||
|
SPT16_MOUSE
|
||||||
| θ value | 4.03905 (rank : 49) | NC score | 0.043306 (rank : 173) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q920B9, Q3TZ48, Q3UFH0, Q921H4 | Gene names | Supt16h, Fact140, Factp140 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FACT complex subunit SPT16 (Facilitates chromatin transcription complex subunit SPT16) (FACT 140 kDa subunit) (FACTp140) (Chromatin- specific transcription elongation factor 140 kDa subunit). | |||||
|
UBE4A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 50) | NC score | 0.038762 (rank : 174) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14139, Q6P5T4, Q7Z639 | Gene names | UBE4A, KIAA0126 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin conjugation factor E4 A. | |||||
|
HMMR_MOUSE
|
||||||
| θ value | 5.27518 (rank : 51) | NC score | 0.063581 (rank : 28) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q00547 | Gene names | Hmmr, Ihabp, Rhamm | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility). | |||||
|
MYH1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 52) | NC score | 0.059156 (rank : 40) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1478 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P12882, Q9Y622 | Gene names | MYH1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1) (Myosin heavy chain IIx/d) (MyHC-IIx/d). | |||||
|
MYH1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 53) | NC score | 0.059077 (rank : 41) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1461 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q5SX40 | Gene names | Myh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1). | |||||
|
MYH2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 54) | NC score | 0.059508 (rank : 39) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1491 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9UKX2, Q14322, Q16229 | Gene names | MYH2, MYHSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, skeletal muscle, adult 2 (Myosin heavy chain IIa) (MyHC-IIa). | |||||
|
OPA1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 55) | NC score | 0.024729 (rank : 186) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O60313 | Gene names | OPA1, KIAA0567 | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-like 120 kDa protein, mitochondrial precursor (Optic atrophy protein 1) [Contains: Dynamin-like 120 kDa protein, form S1]. | |||||
|
OPA1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 56) | NC score | 0.024518 (rank : 187) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P58281, Q3ULA5, Q8BKU7, Q8BLL3, Q8BM08, Q8R3J7 | Gene names | Opa1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-like 120 kDa protein, mitochondrial precursor (Optic atrophy protein 1 homolog) (Large GTP-binding protein) (LargeG) [Contains: Dynamin-like 120 kDa protein, form S1]. | |||||
|
VDP_MOUSE
|
||||||
| θ value | 5.27518 (rank : 57) | NC score | 0.060405 (rank : 38) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 469 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9Z1Z0, Q3T9L9, Q3TH58, Q3U1C7, Q3UMW6, Q91WE7, Q99JZ5 | Gene names | Vdp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | General vesicular transport factor p115 (Transcytosis-associated protein) (TAP) (Vesicle docking protein). | |||||
|
CENPF_HUMAN
|
||||||
| θ value | 6.88961 (rank : 58) | NC score | 0.063005 (rank : 29) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
HOOK2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 59) | NC score | 0.048194 (rank : 168) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 714 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q96ED9, O60562 | Gene names | HOOK2 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 2 (h-hook2) (hHK2). | |||||
|
MY18A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 60) | NC score | 0.058127 (rank : 44) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
REST_MOUSE
|
||||||
| θ value | 6.88961 (rank : 61) | NC score | 0.057438 (rank : 48) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1506 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q922J3, Q571L7 | Gene names | Rsn, Kiaa4046 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin. | |||||
|
DMD_HUMAN
|
||||||
| θ value | 8.99809 (rank : 62) | NC score | 0.047081 (rank : 170) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P11532, Q02295, Q14169, Q14170, Q5JYU0, Q7KZ48 | Gene names | DMD | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
GSTA3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 63) | NC score | 0.007424 (rank : 196) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P30115 | Gene names | Gsta3, Gstyc | |||
|
Domain Architecture |
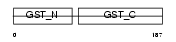
|
|||||
| Description | Glutathione S-transferase A3 (EC 2.5.1.18) (Glutathione S-transferase Yc) (Ya3) (GST class-alpha member 3). | |||||
|
K1914_MOUSE
|
||||||
| θ value | 8.99809 (rank : 64) | NC score | 0.025754 (rank : 184) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5DTU0, Q8BID1, Q8K2G0 | Gene names | Kiaa1914 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1914. | |||||
|
LBC_HUMAN
|
||||||
| θ value | 8.99809 (rank : 65) | NC score | 0.025504 (rank : 185) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q12802 | Gene names | LBC | |||
|
Domain Architecture |
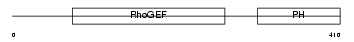
|
|||||
| Description | LBC oncogene (P47) (Lymphoid blast crisis oncogene). | |||||
|
LRC48_MOUSE
|
||||||
| θ value | 8.99809 (rank : 66) | NC score | 0.026901 (rank : 183) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9D5E4 | Gene names | Lrrc48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 48. | |||||
|
MMRN1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 67) | NC score | 0.062220 (rank : 32) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q13201, Q6P3T8 | Gene names | MMRN1, ECM, EMILIN4, MMRN | |||
|
Domain Architecture |

|
|||||
| Description | Multimerin-1 precursor (Endothelial cell multimerin 1) (EMILIN-4) (Elastin microfibril interface located protein 4) (Elastin microfibril interfacer 4). | |||||
|
MYH8_HUMAN
|
||||||
| θ value | 8.99809 (rank : 68) | NC score | 0.057749 (rank : 47) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1610 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P13535, Q14910 | Gene names | MYH8 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
PHAR1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 69) | NC score | 0.019736 (rank : 190) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q2M3X8, Q80VL9, Q8C873 | Gene names | Phactr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatase and actin regulator 1. | |||||
|
PSD12_HUMAN
|
||||||
| θ value | 8.99809 (rank : 70) | NC score | 0.016938 (rank : 192) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O00232, Q53HA2, Q6P053 | Gene names | PSMD12 | |||
|
Domain Architecture |

|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 12 (26S proteasome regulatory subunit p55). | |||||
|
SCEL_MOUSE
|
||||||
| θ value | 8.99809 (rank : 71) | NC score | 0.023871 (rank : 188) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9EQG3, Q9CTT9 | Gene names | Scel | |||
|
Domain Architecture |

|
|||||
| Description | Sciellin. | |||||
|
SM1L2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 72) | NC score | 0.056585 (rank : 57) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 918 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q920F6 | Gene names | Smc1l2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 1-like 2 protein (SMC1beta protein). | |||||
|
APOA4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.050431 (rank : 157) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P06728 | Gene names | Apoa4 | |||
|
Domain Architecture |
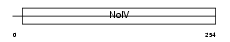
|
|||||
| Description | Apolipoprotein A-IV precursor (Apo-AIV) (ApoA-IV). | |||||
|
AZI1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.055137 (rank : 81) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 990 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q62036 | Gene names | Azi1, Az1, Azi | |||
|
Domain Architecture |

|
|||||
| Description | 5-azacytidine-induced protein 1 (Pre-acrosome localization protein 1). | |||||
|
BICD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.050777 (rank : 152) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 844 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q96G01, O43892, O43893 | Gene names | BICD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 1. | |||||
|
BICD1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.052216 (rank : 119) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 837 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8BR07, O55206, Q8BQ18, Q8BRR8, Q8C4D3, Q8CAK6, Q8R2J4, Q8R2J5, Q8R2J6, Q91YP7 | Gene names | Bicd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 1. | |||||
|
BPAEA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.052326 (rank : 114) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
C102B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.057383 (rank : 49) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q68D86, Q7Z467, Q8NDK7, Q9H5C1 | Gene names | CCDC102B, C18orf14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 102B. | |||||
|
CCD13_HUMAN
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.056089 (rank : 64) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 721 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8IYE1 | Gene names | CCDC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 13. | |||||
|
CCD19_HUMAN
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.053169 (rank : 103) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9UL16 | Gene names | CCDC19, NESG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 19 (Nasopharyngeal epithelium- specific protein 1). | |||||
|
CCD27_MOUSE
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.051343 (rank : 143) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q3V036 | Gene names | Ccdc27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 27. | |||||
|
CCD39_HUMAN
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.056107 (rank : 63) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 868 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UFE4 | Gene names | CCDC39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 39. | |||||
|
CCD39_MOUSE
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.056360 (rank : 61) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1026 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9D5Y1, Q8CDG6 | Gene names | Ccdc39, D3Ertd789e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 39. | |||||
|
CCD41_HUMAN
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.052238 (rank : 118) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1139 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9Y592 | Gene names | CCDC41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41 (NY-REN-58 antigen). | |||||
|
CCD41_MOUSE
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.052089 (rank : 124) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9D5R3, Q3U7X7, Q3UX57, Q80VF0 | Gene names | Ccdc41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41. | |||||
|
CCD46_HUMAN
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.053733 (rank : 97) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1430 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8N8E3, Q6PIB5, Q8NCR4, Q8NFR4 | Gene names | CCDC46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 46. | |||||
|
CCD46_MOUSE
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.054049 (rank : 94) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q5PR68, Q80ZN6, Q99JS4, Q9D9T1 | Gene names | Ccdc46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled coil domain-containing protein 46. | |||||
|
CCD62_HUMAN
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.052837 (rank : 106) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 631 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q6P9F0, Q6ZVF2, Q86VJ0, Q9BYZ5 | Gene names | CCDC62 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 60 (Aaa-protein) (Protein TSP- NY). | |||||
|
CE152_HUMAN
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.060501 (rank : 36) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 976 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O94986, Q6NTA0 | Gene names | CEP152, KIAA0912 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 152 kDa (Cep152 protein). | |||||
|
CE290_HUMAN
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.062077 (rank : 33) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
CEP55_HUMAN
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.052048 (rank : 126) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q53EZ4, Q32WF5, Q3MV20, Q5VY28, Q6N034, Q96H32, Q9NVS7 | Gene names | CEP55, C10orf3, URCC6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome protein 55 (Up-regulated in colon cancer 6). | |||||
|
CEP63_HUMAN
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.055414 (rank : 76) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1005 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q96MT8, Q96CR0, Q9H8F5, Q9H8N0 | Gene names | CEP63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 63 kDa (Cep63 protein). | |||||
|
CI039_HUMAN
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.051512 (rank : 138) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1171 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9NXG0, Q5VYJ0, Q9HAJ5 | Gene names | C9orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf39. | |||||
|
CI093_HUMAN
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.054144 (rank : 92) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q6TFL3, Q5SU58, Q6P1W1, Q6ZR13, Q8N1Z4, Q8N8L3, Q8TBR2 | Gene names | C9orf93 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf93. | |||||
|
CJ118_HUMAN
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.062375 (rank : 31) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q7Z3E2, Q2M2V6, Q6NS91, Q7RTP1, Q8N117, Q8N3G3, Q8N6C2, Q9NWA3 | Gene names | C10orf118 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf118 (CTCL tumor antigen HD-CL-01/L14-2). | |||||
|
CN145_HUMAN
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.050355 (rank : 159) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1196 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q6ZU80, Q96ML4 | Gene names | C14orf145 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf145. | |||||
|
CN145_MOUSE
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.054871 (rank : 84) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8BI22 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf145 homolog. | |||||
|
CP135_HUMAN
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.056404 (rank : 59) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1290 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q66GS9, O75130, Q9H8H7 | Gene names | CEP135, CEP4, KIAA0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
CP135_MOUSE
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.058364 (rank : 43) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1278 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q6P5D4, Q6A030 | Gene names | Cep135, Cep4, Kiaa0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
CP250_HUMAN
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.050773 (rank : 153) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
CP250_MOUSE
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.050950 (rank : 148) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
CR034_MOUSE
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.061924 (rank : 34) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8CDV0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C18orf34 homolog. | |||||
|
CRCM_HUMAN
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.050036 (rank : 166) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 520 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P23508 | Gene names | MCC | |||
|
Domain Architecture |
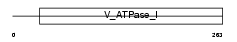
|
|||||
| Description | Colorectal mutant cancer protein (Protein MCC). | |||||
|
CROCC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.052605 (rank : 111) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1409 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q5TZA2, Q2VHY3, Q66GT7, Q7Z2L4, Q7Z5D7 | Gene names | CROCC, KIAA0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
CROCC_MOUSE
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.051920 (rank : 129) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1460 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8CJ40, Q7TQL2, Q80U01, Q8R0B9 | Gene names | Crocc, Kiaa0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
CYLN2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.052366 (rank : 113) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1004 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9UDT6, O14527, O43611 | Gene names | CYLN2, KIAA0291, WBSCR4, WSCR4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic linker protein 2 (Cytoplasmic linker protein 115) (CLIP- 115) (Williams-Beuren syndrome chromosome region 4 protein). | |||||
|
CYLN2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.051511 (rank : 139) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1002 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9Z0H8, Q7TSI9, Q8CHU1, Q9EP81 | Gene names | Cyln2, Kiaa0291 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic linker protein 2 (Cytoplasmic linker protein 115) (CLIP- 115). | |||||
|
DESP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.051380 (rank : 141) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P15924, O75993, Q14189, Q9UHN4 | Gene names | DSP | |||
|
Domain Architecture |

|
|||||
| Description | Desmoplakin (DP) (250/210 kDa paraneoplastic pemphigus antigen). | |||||
|
EEA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.052808 (rank : 108) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
EEA1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.053679 (rank : 98) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1637 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8BL66, Q6DIC2 | Gene names | Eea1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early endosome antigen 1. | |||||
|
ERC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.050911 (rank : 149) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1156 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O15083, Q86TK4 | Gene names | ERC2, KIAA0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2. | |||||
|
ERC2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.051283 (rank : 144) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1166 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q6PH08, Q80U20, Q8CCP1 | Gene names | Erc2, Cast1, D14Ertd171e, Kiaa0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2 (CAZ-associated structural protein 1) (CAST1). | |||||
|
FLIP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.051894 (rank : 131) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q7Z7B0, Q5VUL6, Q86TC3, Q8N8B9, Q96SK6, Q9NVI8, Q9ULE5 | Gene names | FILIP1, KIAA1275 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
FLIP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.051745 (rank : 135) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1176 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9CS72 | Gene names | Filip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
FYCO1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.051921 (rank : 128) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8VDC1, Q3V385, Q7TMT0, Q8BJN2 | Gene names | Fyco1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1. | |||||
|
GCC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.053459 (rank : 100) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 887 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q96CN9, Q9H6N7 | Gene names | GCC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP and coiled-coil domain-containing protein 1 (Golgi coiled coil protein 1). | |||||
|
GCC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.051180 (rank : 146) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9D4H2, Q8CCR3 | Gene names | Gcc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP and coiled-coil domain-containing protein 1 (Golgi coiled coil protein 1). | |||||
|
GOGA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.050457 (rank : 156) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1222 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q92805, Q8IYZ9 | Gene names | GOLGA1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
GOGA1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.050180 (rank : 162) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 994 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9CW79, Q80YB0 | Gene names | Golga1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
GOGA2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.052104 (rank : 123) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q921M4 | Gene names | Golga2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgin subfamily A member 2 (Cis-Golgi matrix protein GM130). | |||||
|
GOGA3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.054160 (rank : 90) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q08378, O43241, Q6P9C7, Q86XW3, Q8TDA9, Q8WZA3 | Gene names | GOLGA3 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Golgi complex-associated protein of 170 kDa) (GCP170). | |||||
|
GOGA3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.055133 (rank : 82) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1437 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P55937, Q80VF5, Q8CCK4, Q9QYT2, Q9QYT3 | Gene names | Golga3, Mea2 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Male-enhanced antigen 2) (MEA-2). | |||||
|
GOGA4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.055097 (rank : 83) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
GOGA4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.052940 (rank : 105) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.053819 (rank : 95) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
GRAP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.051895 (rank : 130) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q4V328, Q4V327, Q4V330, Q5HYG1, Q6N046, Q96DH8, Q9NQ43, Q9ULQ3 | Gene names | GRIPAP1, KIAA1167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP1-associated protein 1 (GRASP-1). | |||||
|
GRAP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.056471 (rank : 58) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1234 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8VD04, O35693, Q3T9C3, Q69ZP9 | Gene names | Gripap1, DXImx47e, Kiaa1167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP1-associated protein 1 (GRASP-1) (HCMV-interacting protein). | |||||
|
HMMR_HUMAN
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.058494 (rank : 42) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O75330, Q92767 | Gene names | HMMR, IHABP, RHAMM | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility) (CD168 antigen). | |||||
|
HOOK1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.052073 (rank : 125) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 942 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9UJC3, O60561, Q5TG44 | Gene names | HOOK1 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 1 (h-hook1) (hHK1). | |||||
|
HOOK1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.052953 (rank : 104) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 910 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8BIL5, Q8BIZ2, Q8K0P1, Q8K454, Q9CTN6 | Gene names | Hook1 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 1. | |||||
|
HOOK3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.051405 (rank : 140) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1070 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q86VS8, Q8NBH0, Q9BY13 | Gene names | HOOK3 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 3 (hHK3). | |||||
|
HOOK3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.051723 (rank : 136) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8BUK6, Q8BV48, Q8BW57, Q8BY41 | Gene names | Hook3 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 3. | |||||
|
IFT74_HUMAN
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.057073 (rank : 51) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q96LB3, Q6PGQ8, Q9H643, Q9H8G7 | Gene names | IFT74, CCDC2, CMG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 74 homolog (Coiled-coil domain-containing protein 2) (Capillary morphogenesis protein 1) (CMG-1). | |||||
|
IFT74_MOUSE
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.055283 (rank : 78) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 939 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8BKE9, Q80ZI3, Q9CSY1, Q9CUS0, Q9D9T5 | Gene names | Ift74, Ccdc2, Cmg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 74 homolog (Coiled-coil domain-containing protein 2) (Capillary morphogenesis protein 1) (CMG-1). | |||||
|
IFT81_HUMAN
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.052738 (rank : 109) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8WYA0, Q8NB51, Q9BSV2, Q9UNY8 | Gene names | IFT81, CDV1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 81 (Carnitine deficiency-associated protein expressed in ventricle 1) (CDV-1 protein). | |||||
|
IFT81_MOUSE
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.050813 (rank : 151) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O35594, Q8R4W2, Q9CSA1, Q9EQM1 | Gene names | Ift81, Cdv-1, Cdv1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 81 (Carnitine deficiency-associated protein expressed in ventricle 1) (CDV-1 protein). | |||||
|
K0555_HUMAN
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.054639 (rank : 85) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 880 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q96AA8, O60302 | Gene names | KIAA0555 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0555. | |||||
|
KMHN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.051361 (rank : 142) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 950 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8TBZ0, Q86YI9, Q8N7W0 | Gene names | KMHN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cancer/testis antigen KM-HN-1. | |||||
|
KMHN1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.050203 (rank : 161) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q3V125, Q3TTQ8 | Gene names | Gm172 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cancer/testis antigen KM-HN-1 homolog. | |||||
|
KNTC2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.050547 (rank : 155) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 692 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9D0F1, Q3TQT6, Q3UWM5, Q99P70 | Gene names | Kntc2, Hec1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Hec1 (Kinetochore-associated protein 2). | |||||
|
KTN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.062455 (rank : 30) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q86UP2, Q13999, Q14707, Q15387, Q86W57 | Gene names | KTN1, CG1, KIAA0004 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin (Kinesin receptor) (CG-1 antigen). | |||||
|
KTN1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.054408 (rank : 89) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1324 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q61595, Q8BG49, Q8BHF4, Q8BHM8, Q8C9Y5, Q8CG51, Q8CG52, Q8CG53, Q8CG54, Q8CG55, Q8CG56, Q8CG57, Q8CG58, Q8CG59, Q8CG60, Q8CG61, Q8CG62, Q8CG63 | Gene names | Ktn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin. | |||||
|
LIPA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.051721 (rank : 137) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1294 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q13136, Q13135, Q14567, Q8N4I2 | Gene names | PPFIA1, LIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-1 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-1) (PTPRF-interacting protein alpha-1) (LAR-interacting protein 1) (LIP.1). | |||||
|
LIPA2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.053807 (rank : 96) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1044 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O75334 | Gene names | PPFIA2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-2) (PTPRF-interacting protein alpha-2). | |||||
|
LIPA2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.051770 (rank : 134) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1282 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8BSS9, Q8BN73 | Gene names | Ppfia2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-2) (PTPRF-interacting protein alpha-2). | |||||
|
LIPA3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.053649 (rank : 99) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P60469 | Gene names | Ppfia3 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-3 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-3) (PTPRF-interacting protein alpha-3). | |||||
|
MD1L1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.052253 (rank : 116) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9Y6D9, Q13312, Q75MI0, Q86UM4, Q9UNH0 | Gene names | MAD1L1, MAD1, TXBP181 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1) (Mitotic checkpoint MAD1 protein-homolog) (HsMAD1) (hMAD1) (Tax-binding protein 181). | |||||
|
MD1L1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.051845 (rank : 133) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1001 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9WTX8, Q9WTX9 | Gene names | Mad1l1, Mad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1). | |||||
|
MY18A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.054535 (rank : 88) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
MY18B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.055774 (rank : 70) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1299 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8IUG5, Q8NDI8, Q8TE65, Q8WWS0, Q96KH2, Q96KR8, Q96KR9 | Gene names | MYO18B | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18B (Myosin XVIIIb). | |||||
|
MYH11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.056380 (rank : 60) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
MYH11_MOUSE
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.056693 (rank : 55) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
MYH13_HUMAN
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.057313 (rank : 50) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1504 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9UKX3, O95252 | Gene names | MYH13 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-13 (Myosin heavy chain, skeletal muscle, extraocular) (MyHC- eo). | |||||
|
MYH14_HUMAN
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.055289 (rank : 77) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 995 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q7Z406, Q5CZ75, Q6XYE4, Q76B62, Q8WV23, Q96I22, Q9BT27, Q9BW35, Q9H882 | Gene names | MYH14, KIAA2034 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
MYH14_MOUSE
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.055211 (rank : 80) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1225 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q6URW6, Q80V64, Q80ZE6 | Gene names | Myh14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
MYH3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.055260 (rank : 79) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1660 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P11055, Q15492 | Gene names | MYH3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, fast skeletal muscle, embryonic (Muscle embryonic myosin heavy chain) (SMHCE). | |||||
|
MYH4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.055929 (rank : 65) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1377 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9Y623 | Gene names | MYH4 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal) (Myosin heavy chain IIb) (MyHC-IIb). | |||||
|
MYH4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.055789 (rank : 68) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
MYH6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.055784 (rank : 69) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1426 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P13533, Q13943, Q14906, Q14907 | Gene names | MYH6, MYHCA | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
MYH6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.055732 (rank : 71) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1501 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q02566, Q64258, Q64738 | Gene names | Myh6, Myhca | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
MYH7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.055675 (rank : 73) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P12883, Q14836, Q14837, Q14904, Q16579, Q92679, Q9H1D5, Q9UMM8 | Gene names | MYH7, MYHCB | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle beta isoform (MyHC-beta). | |||||
|
MYH8_MOUSE
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.055638 (rank : 74) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1600 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P13542, Q5SX36 | Gene names | Myh8, Myhsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
MYH9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.055928 (rank : 66) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
MYH9_MOUSE
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.055704 (rank : 72) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
MYO5B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.061200 (rank : 35) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 911 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9ULV0 | Gene names | MYO5B, KIAA1119 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5B (Myosin Vb). | |||||
|
MYO5C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.060434 (rank : 37) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1206 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9NQX4 | Gene names | MYO5C | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5C (Myosin Vc). | |||||
|
NIN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.056662 (rank : 56) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1416 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8N4C6, Q6P0P6, Q9BWU6, Q9C012, Q9C013, Q9C014, Q9H5I6, Q9HAT7, Q9HBY5, Q9HCK7, Q9UH61 | Gene names | NIN, KIAA1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein (hNinein) (Glycogen synthase kinase 3 beta-interacting protein) (GSK3B-interacting protein). | |||||
|
NIN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.054535 (rank : 87) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q61043, Q6ZPM7 | Gene names | Nin, Kiaa1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein. | |||||
|
NUF2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.051941 (rank : 127) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 661 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9BZD4, Q8WU69, Q96HJ4, Q96Q78 | Gene names | CDCA1, NUF2, NUF2R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Nuf2 (hNuf2) (hNuf2R) (hsNuf2) (Cell division cycle-associated protein 1). | |||||
|
NUMA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.052415 (rank : 112) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
OPTN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.050681 (rank : 154) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1120 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8K3K8 | Gene names | Optn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin. | |||||
|
PCNT_HUMAN
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.056906 (rank : 52) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
PEPL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.050168 (rank : 163) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1141 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O60437, O60314, O60454 | Gene names | PPL, KIAA0568 | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin (195 kDa cornified envelope precursor protein) (190 kDa paraneoplastic pemphigus antigen). | |||||
|
RABE1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.054062 (rank : 93) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q15276, O95369, Q8IVX3 | Gene names | RABEP1, RABPT5, RABPT5A | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha) (Rabaptin-4) (NY-REN-17 antigen). | |||||
|
RABE1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.051860 (rank : 132) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1105 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O35551, Q99L08, Q9CRP3, Q9EQF9, Q9JI94 | Gene names | Rabep1, Rab5ep, Rabpt5, Rabpt5a | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha). | |||||
|
RAD50_HUMAN
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.058109 (rank : 45) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q92878, O43254, Q6GMT7, Q6P5X3, Q9UP86 | Gene names | RAD50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (hRAD50). | |||||
|
RAD50_MOUSE
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.056703 (rank : 54) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1366 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P70388, Q6PEB0, Q8C2T7, Q9CU59 | Gene names | Rad50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (mRad50). | |||||
|
RB6I2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.050079 (rank : 165) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q99MI1, Q80TK7, Q8BPL1, Q8C7Y1, Q99MI2 | Gene names | Erc1, Cast2, Kiaa1081, Rab6ip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1) (ERC1) (CAZ-associated structural protein 2) (CAST2). | |||||
|
RBCC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.052250 (rank : 117) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1023 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8TDY2, Q8WVU9, Q92601 | Gene names | RB1CC1, KIAA0203, RBICC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RB1-inducible coiled-coil protein 1. | |||||
|
RBCC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.055865 (rank : 67) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1048 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9ESK9, Q3TXX2, Q61384, Q8BRY9, Q9JK14 | Gene names | Rb1cc1, Cc1, Kiaa0203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RB1-inducible coiled-coil protein 1 (Coiled-coil-forming protein 1) (LaXp180 protein). | |||||
|
REST_HUMAN
|
||||||
| θ value | θ > 10 (rank : 181) | NC score | 0.058058 (rank : 46) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1605 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P30622 | Gene names | RSN, CYLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Restin (Cytoplasmic linker protein 170 alpha-2) (CLIP-170) (Reed- Sternberg intermediate filament-associated protein) (Cytoplasmic linker protein 1). | |||||
|
RFIP3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 182) | NC score | 0.052108 (rank : 122) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1055 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O75154, Q4VXV7, Q9H155, Q9H1G0, Q9NUI0 | Gene names | RAB11FIP3, KIAA0665 | |||
|
Domain Architecture |

|
|||||
| Description | Rab11 family-interacting protein 3 (Rab11-FIP3) (EF hands-containing Rab-interacting protein) (Eferin). | |||||
|
SDCG8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 183) | NC score | 0.051262 (rank : 145) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1002 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q86SQ7, O60527, Q3ZCR6, Q8N5F2, Q9P0F1 | Gene names | SDCCAG8, CCCAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serologically defined colon cancer antigen 8 (Centrosomal colon cancer autoantigen protein) (hCCCAP) (Antigen NY-CO-8). | |||||
|
SM1L2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 184) | NC score | 0.052114 (rank : 121) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1019 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8NDV3, Q5TIC3, Q9Y3G5 | Gene names | SMC1L2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 2 protein (SMC1beta protein). | |||||
|
SMC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 185) | NC score | 0.050204 (rank : 160) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O95347, Q9P1P2 | Gene names | SMC2L1, CAPE, SMC2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (hCAP-E) (XCAP-E homolog). | |||||
|
SMC2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 186) | NC score | 0.050113 (rank : 164) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8CG48, Q61076, Q9CS17, Q9CSD8 | Gene names | Smc2l1, Cape, Fin16, Smc2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (XCAP-E homolog) (FGF-inducible protein 16). | |||||
|
SMC4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 187) | NC score | 0.052178 (rank : 120) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9NTJ3, O95752, Q8NDL4, Q9UNT9 | Gene names | SMC4L1, CAPC, SMC4 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 4-like 1 protein (Chromosome- associated polypeptide C) (hCAP-C) (XCAP-C homolog). | |||||
|
SMC4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 188) | NC score | 0.050834 (rank : 150) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 830 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8CG47, Q8BTS7, Q8BTY9, Q99K21 | Gene names | Smc4l1, Capc, Smc4 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 4-like 1 protein (Chromosome- associated polypeptide C) (XCAP-C homolog). | |||||
|
SPAG5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 189) | NC score | 0.052311 (rank : 115) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 625 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q96R06, O95213, Q9BWE8, Q9NT17, Q9UFE6 | Gene names | SPAG5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 5 (Astrin) (Mitotic spindle-associated protein p126) (MAP126) (Deepest). | |||||
|
SYCP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 190) | NC score | 0.055509 (rank : 75) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1456 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q62209, O09205, P70192, Q62329 | Gene names | Sycp1, Scp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
TAXB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 191) | NC score | 0.052669 (rank : 110) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 829 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q86VP1, O60398, O95770, Q13311, Q9BQG5, Q9UI88 | Gene names | TAX1BP1, T6BP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tax1-binding protein 1 (TRAF6-binding protein). | |||||
|
TAXB1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 192) | NC score | 0.052817 (rank : 107) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q3UKC1, Q91YT6, Q9CVF0, Q9DC45 | Gene names | Tax1bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tax1-binding protein 1 homolog. | |||||
|
TMF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 193) | NC score | 0.054592 (rank : 86) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 987 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P82094 | Gene names | TMF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TATA element modulatory factor (TMF). | |||||
|
TPR_HUMAN
|
||||||
| θ value | θ > 10 (rank : 194) | NC score | 0.053264 (rank : 102) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
TRHY_HUMAN
|
||||||
| θ value | θ > 10 (rank : 195) | NC score | 0.054158 (rank : 91) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
ZW10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 196) | NC score | 0.056889 (rank : 53) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O43264 | Gene names | ZW10 | |||
|
Domain Architecture |

|
|||||
| Description | Centromere/kinetochore protein zw10 homolog. | |||||
|
EXOC4_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q96A65, Q9C0G4, Q9H9K0, Q9P102 | Gene names | EXOC4, KIAA1699, SEC8, SEC8L1 | |||
|
Domain Architecture |
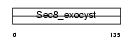
|
|||||
| Description | Exocyst complex component 4 (Exocyst complex component Sec8). | |||||
|
EXOC4_MOUSE
|
||||||
| NC score | 0.990194 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O35382 | Gene names | Exoc4, Sec8, Sec8l1 | |||
|
Domain Architecture |
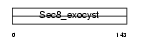
|
|||||
| Description | Exocyst complex component 4 (Exocyst complex component Sec8). | |||||
|
ZW10_MOUSE
|
||||||
| NC score | 0.153233 (rank : 3) | θ value | 0.00134147 (rank : 5) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O54692, Q3TIA5, Q3ULW1, Q921H3 | Gene names | Zw10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere/kinetochore protein zw10 homolog. | |||||
|
GCC2_HUMAN
|
||||||
| NC score | 0.091847 (rank : 4) | θ value | 0.00390308 (rank : 7) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
RBP24_HUMAN
|
||||||
| NC score | 0.091806 (rank : 5) | θ value | 0.00390308 (rank : 8) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
TRIPB_HUMAN
|
||||||
| NC score | 0.090102 (rank : 6) | θ value | 0.00298849 (rank : 6) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
MYO5A_HUMAN
|
||||||
| NC score | 0.089103 (rank : 7) | θ value | 7.1131e-05 (rank : 3) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1092 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9Y4I1, O60653, Q07902, Q16249, Q9UE30, Q9UE31 | Gene names | MYO5A, MYH12 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5A (Myosin Va) (Dilute myosin heavy chain, non-muscle) (Myosin heavy chain 12) (Myoxin). | |||||
|
CR034_HUMAN
|
||||||
| NC score | 0.086591 (rank : 8) | θ value | 2.36792 (rank : 33) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q5BJE1, Q6ZP67, Q6ZU20 | Gene names | C18orf34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C18orf34. | |||||
|
MYO5A_MOUSE
|
||||||
| NC score | 0.084689 (rank : 9) | θ value | 0.000786445 (rank : 4) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1117 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q99104 | Gene names | Myo5a, Dilute | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5A (Myosin Va) (Dilute myosin heavy chain, non-muscle). | |||||
|
LIPA3_HUMAN
|
||||||
| NC score | 0.084397 (rank : 10) | θ value | 0.0330416 (rank : 9) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O75145, Q9H8B5, Q9UEW4 | Gene names | PPFIA3, KIAA0654 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-3 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-3) (PTPRF-interacting protein alpha-3). | |||||
|
SPAG5_MOUSE
|
||||||
| NC score | 0.079839 (rank : 11) | θ value | 3.0926 (rank : 39) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 612 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q7TME2, Q8CH61 | Gene names | Spag5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 5 (Mastrin) (Mitotic spindle-associated protein p126) (MAP126). | |||||
|
CJ118_MOUSE
|
||||||
| NC score | 0.079179 (rank : 12) | θ value | 0.0736092 (rank : 12) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1132 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8C9S4, Q6P3D6, Q8CJC0 | Gene names | Otg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf118 homolog (Oocyte-testis gene 1 protein). | |||||
|
PCNT_MOUSE
|
||||||
| NC score | 0.078528 (rank : 13) | θ value | 0.365318 (rank : 15) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1207 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P48725 | Gene names | Pcnt, Pcnt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pericentrin. | |||||
|
GCC2_MOUSE
|
||||||
| NC score | 0.077970 (rank : 14) | θ value | 0.365318 (rank : 14) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
CF060_HUMAN
|
||||||
| NC score | 0.074593 (rank : 15) | θ value | 0.813845 (rank : 21) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1270 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8NB25, Q96GY8, Q9H851 | Gene names | C6orf60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf60. | |||||
|
PIBF1_HUMAN
|
||||||
| NC score | 0.074305 (rank : 16) | θ value | 0.47712 (rank : 16) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8WXW3, O95664, Q6U9V2, Q6UG50, Q86V07, Q96SF4 | Gene names | PIBF1, C13orf24, PIBF | |||
|
Domain Architecture |

|
|||||
| Description | Progesterone-induced-blocking factor 1. | |||||
|
SMC3_MOUSE
|
||||||
| NC score | 0.070198 (rank : 17) | θ value | 1.38821 (rank : 26) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1187 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9CW03, O35667, Q9QUS3 | Gene names | Cspg6, Bam, Bmh, Mmip1, Smc3, Smc3l1, Smcd | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome segregation protein SmcD) (Bamacan) (Basement membrane-associated chondroitin proteoglycan) (Mad member- interacting protein 1). | |||||
|
SMC3_HUMAN
|
||||||
| NC score | 0.069903 (rank : 18) | θ value | 1.38821 (rank : 25) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1186 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9UQE7, O60464 | Gene names | CSPG6, BAM, BMH, SMC3, SMC3L1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome-associated polypeptide) (hCAP) (Bamacan) (Basement membrane-associated chondroitin proteoglycan). | |||||
|
TMCC2_HUMAN
|
||||||
| NC score | 0.068764 (rank : 19) | θ value | 0.62314 (rank : 19) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O75069, Q6ZN09 | Gene names | TMCC2, KIAA0481 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane and coiled-coil domains protein 2 (Cerebral protein 11). | |||||
|
CENPE_HUMAN
|
||||||
| NC score | 0.068675 (rank : 20) | θ value | 0.0563607 (rank : 10) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
TMCC2_MOUSE
|
||||||
| NC score | 0.068391 (rank : 21) | θ value | 0.62314 (rank : 20) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q80W04, Q6A061 | Gene names | Tmcc2, Kiaa0481 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane and coiled-coil domains protein 2. | |||||
|
AKAP9_HUMAN
|
||||||
| NC score | 0.067559 (rank : 22) | θ value | 2.36792 (rank : 32) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
CING_HUMAN
|
||||||
| NC score | 0.066699 (rank : 23) | θ value | 4.03905 (rank : 42) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9P2M7, Q5T386, Q9NR25 | Gene names | CGN, KIAA1319 | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
SYCP1_HUMAN
|
||||||
| NC score | 0.066058 (rank : 24) | θ value | 3.0926 (rank : 40) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q15431, O14963 | Gene names | SYCP1, SCP1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
CING_MOUSE
|
||||||
| NC score | 0.065079 (rank : 25) | θ value | 1.38821 (rank : 23) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1432 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P59242 | Gene names | Cgn | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
MYH10_HUMAN
|
||||||
| NC score | 0.065056 (rank : 26) | θ value | 2.36792 (rank : 34) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P35580, Q16087 | Gene names | MYH10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
MYH10_MOUSE
|
||||||
| NC score | 0.064459 (rank : 27) | θ value | 4.03905 (rank : 47) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1612 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q61879, Q5SV63 | Gene names | Myh10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
HMMR_MOUSE
|
||||||
| NC score | 0.063581 (rank : 28) | θ value | 5.27518 (rank : 51) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q00547 | Gene names | Hmmr, Ihabp, Rhamm | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility). | |||||
|
CENPF_HUMAN
|
||||||
| NC score | 0.063005 (rank : 29) | θ value | 6.88961 (rank : 58) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
KTN1_HUMAN
|
||||||
| NC score | 0.062455 (rank : 30) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q86UP2, Q13999, Q14707, Q15387, Q86W57 | Gene names | KTN1, CG1, KIAA0004 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin (Kinesin receptor) (CG-1 antigen). | |||||
|
CJ118_HUMAN
|
||||||
| NC score | 0.062375 (rank : 31) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q7Z3E2, Q2M2V6, Q6NS91, Q7RTP1, Q8N117, Q8N3G3, Q8N6C2, Q9NWA3 | Gene names | C10orf118 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf118 (CTCL tumor antigen HD-CL-01/L14-2). | |||||
|
MMRN1_HUMAN
|
||||||
| NC score | 0.062220 (rank : 32) | θ value | 8.99809 (rank : 67) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q13201, Q6P3T8 | Gene names | MMRN1, ECM, EMILIN4, MMRN | |||
|
Domain Architecture |

|
|||||
| Description | Multimerin-1 precursor (Endothelial cell multimerin 1) (EMILIN-4) (Elastin microfibril interface located protein 4) (Elastin microfibril interfacer 4). | |||||
|
CE290_HUMAN
|
||||||
| NC score | 0.062077 (rank : 33) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
CR034_MOUSE
|
||||||
| NC score | 0.061924 (rank : 34) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8CDV0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C18orf34 homolog. | |||||
|
MYO5B_HUMAN
|
||||||
| NC score | 0.061200 (rank : 35) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 911 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9ULV0 | Gene names | MYO5B, KIAA1119 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5B (Myosin Vb). | |||||
|
CE152_HUMAN
|
||||||
| NC score | 0.060501 (rank : 36) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 976 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O94986, Q6NTA0 | Gene names | CEP152, KIAA0912 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 152 kDa (Cep152 protein). | |||||
|
MYO5C_HUMAN
|
||||||
| NC score | 0.060434 (rank : 37) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1206 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9NQX4 | Gene names | MYO5C | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5C (Myosin Vc). | |||||
|
VDP_MOUSE
|
||||||
| NC score | 0.060405 (rank : 38) | θ value | 5.27518 (rank : 57) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 469 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9Z1Z0, Q3T9L9, Q3TH58, Q3U1C7, Q3UMW6, Q91WE7, Q99JZ5 | Gene names | Vdp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | General vesicular transport factor p115 (Transcytosis-associated protein) (TAP) (Vesicle docking protein). | |||||
|
MYH2_HUMAN
|
||||||
| NC score | 0.059508 (rank : 39) | θ value | 5.27518 (rank : 54) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1491 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9UKX2, Q14322, Q16229 | Gene names | MYH2, MYHSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, skeletal muscle, adult 2 (Myosin heavy chain IIa) (MyHC-IIa). | |||||
|
MYH1_HUMAN
|
||||||
| NC score | 0.059156 (rank : 40) | θ value | 5.27518 (rank : 52) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1478 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P12882, Q9Y622 | Gene names | MYH1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1) (Myosin heavy chain IIx/d) (MyHC-IIx/d). | |||||
|
MYH1_MOUSE
|
||||||
| NC score | 0.059077 (rank : 41) | θ value | 5.27518 (rank : 53) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1461 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q5SX40 | Gene names | Myh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1). | |||||
|
HMMR_HUMAN
|
||||||
| NC score | 0.058494 (rank : 42) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O75330, Q92767 | Gene names | HMMR, IHABP, RHAMM | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility) (CD168 antigen). | |||||
|
CP135_MOUSE
|
||||||
| NC score | 0.058364 (rank : 43) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1278 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q6P5D4, Q6A030 | Gene names | Cep135, Cep4, Kiaa0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
MY18A_MOUSE
|
||||||
| NC score | 0.058127 (rank : 44) | θ value | 6.88961 (rank : 60) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
RAD50_HUMAN
|
||||||
| NC score | 0.058109 (rank : 45) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q92878, O43254, Q6GMT7, Q6P5X3, Q9UP86 | Gene names | RAD50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (hRAD50). | |||||
|
REST_HUMAN
|
||||||
| NC score | 0.058058 (rank : 46) | θ value | θ > 10 (rank : 181) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1605 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P30622 | Gene names | RSN, CYLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Restin (Cytoplasmic linker protein 170 alpha-2) (CLIP-170) (Reed- Sternberg intermediate filament-associated protein) (Cytoplasmic linker protein 1). | |||||
|
MYH8_HUMAN
|
||||||
| NC score | 0.057749 (rank : 47) | θ value | 8.99809 (rank : 68) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1610 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P13535, Q14910 | Gene names | MYH8 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
REST_MOUSE
|
||||||
| NC score | 0.057438 (rank : 48) | θ value | 6.88961 (rank : 61) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1506 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q922J3, Q571L7 | Gene names | Rsn, Kiaa4046 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin. | |||||
|
C102B_HUMAN
|
||||||
| NC score | 0.057383 (rank : 49) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q68D86, Q7Z467, Q8NDK7, Q9H5C1 | Gene names | CCDC102B, C18orf14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 102B. | |||||
|
MYH13_HUMAN
|
||||||
| NC score | 0.057313 (rank : 50) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1504 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9UKX3, O95252 | Gene names | MYH13 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-13 (Myosin heavy chain, skeletal muscle, extraocular) (MyHC- eo). | |||||
|
IFT74_HUMAN
|
||||||
| NC score | 0.057073 (rank : 51) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q96LB3, Q6PGQ8, Q9H643, Q9H8G7 | Gene names | IFT74, CCDC2, CMG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 74 homolog (Coiled-coil domain-containing protein 2) (Capillary morphogenesis protein 1) (CMG-1). | |||||
|
PCNT_HUMAN
|
||||||
| NC score | 0.056906 (rank : 52) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
ZW10_HUMAN
|
||||||
| NC score | 0.056889 (rank : 53) | θ value | θ > 10 (rank : 196) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O43264 | Gene names | ZW10 | |||
|
Domain Architecture |

|
|||||
| Description | Centromere/kinetochore protein zw10 homolog. | |||||
|
RAD50_MOUSE
|
||||||
| NC score | 0.056703 (rank : 54) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1366 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P70388, Q6PEB0, Q8C2T7, Q9CU59 | Gene names | Rad50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (mRad50). | |||||
|
MYH11_MOUSE
|
||||||
| NC score | 0.056693 (rank : 55) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
NIN_HUMAN
|
||||||
| NC score | 0.056662 (rank : 56) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1416 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8N4C6, Q6P0P6, Q9BWU6, Q9C012, Q9C013, Q9C014, Q9H5I6, Q9HAT7, Q9HBY5, Q9HCK7, Q9UH61 | Gene names | NIN, KIAA1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein (hNinein) (Glycogen synthase kinase 3 beta-interacting protein) (GSK3B-interacting protein). | |||||
|
SM1L2_MOUSE
|
||||||
| NC score | 0.056585 (rank : 57) | θ value | 8.99809 (rank : 72) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 918 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q920F6 | Gene names | Smc1l2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 1-like 2 protein (SMC1beta protein). | |||||
|
GRAP1_MOUSE
|
||||||
| NC score | 0.056471 (rank : 58) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1234 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8VD04, O35693, Q3T9C3, Q69ZP9 | Gene names | Gripap1, DXImx47e, Kiaa1167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP1-associated protein 1 (GRASP-1) (HCMV-interacting protein). | |||||
|
CP135_HUMAN
|
||||||
| NC score | 0.056404 (rank : 59) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1290 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q66GS9, O75130, Q9H8H7 | Gene names | CEP135, CEP4, KIAA0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
MYH11_HUMAN
|
||||||
| NC score | 0.056380 (rank : 60) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
CCD39_MOUSE
|
||||||
| NC score | 0.056360 (rank : 61) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1026 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9D5Y1, Q8CDG6 | Gene names | Ccdc39, D3Ertd789e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 39. | |||||
|
RGNEF_MOUSE
|
||||||
| NC score | 0.056147 (rank : 62) | θ value | 0.21417 (rank : 13) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 641 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P97433 | Gene names | Rgnef, Rhoip2 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-guanine nucleotide exchange factor (Rho-interacting protein 2) (RhoGEF) (RIP2). | |||||
|
CCD39_HUMAN
|
||||||
| NC score | 0.056107 (rank : 63) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 868 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UFE4 | Gene names | CCDC39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 39. | |||||
|
CCD13_HUMAN
|
||||||
| NC score | 0.056089 (rank : 64) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 721 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8IYE1 | Gene names | CCDC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 13. | |||||
|
MYH4_HUMAN
|
||||||
| NC score | 0.055929 (rank : 65) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1377 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9Y623 | Gene names | MYH4 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal) (Myosin heavy chain IIb) (MyHC-IIb). | |||||
|
MYH9_HUMAN
|
||||||
| NC score | 0.055928 (rank : 66) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
RBCC1_MOUSE
|
||||||
| NC score | 0.055865 (rank : 67) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1048 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9ESK9, Q3TXX2, Q61384, Q8BRY9, Q9JK14 | Gene names | Rb1cc1, Cc1, Kiaa0203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RB1-inducible coiled-coil protein 1 (Coiled-coil-forming protein 1) (LaXp180 protein). | |||||
|
MYH4_MOUSE
|
||||||
| NC score | 0.055789 (rank : 68) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
MYH6_HUMAN
|
||||||
| NC score | 0.055784 (rank : 69) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1426 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P13533, Q13943, Q14906, Q14907 | Gene names | MYH6, MYHCA | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
MY18B_HUMAN
|
||||||
| NC score | 0.055774 (rank : 70) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1299 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8IUG5, Q8NDI8, Q8TE65, Q8WWS0, Q96KH2, Q96KR8, Q96KR9 | Gene names | MYO18B | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18B (Myosin XVIIIb). | |||||
|
MYH6_MOUSE
|
||||||
| NC score | 0.055732 (rank : 71) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1501 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q02566, Q64258, Q64738 | Gene names | Myh6, Myhca | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
MYH9_MOUSE
|
||||||
| NC score | 0.055704 (rank : 72) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
MYH7_HUMAN
|
||||||
| NC score | 0.055675 (rank : 73) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P12883, Q14836, Q14837, Q14904, Q16579, Q92679, Q9H1D5, Q9UMM8 | Gene names | MYH7, MYHCB | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle beta isoform (MyHC-beta). | |||||
|
MYH8_MOUSE
|
||||||
| NC score | 0.055638 (rank : 74) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1600 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P13542, Q5SX36 | Gene names | Myh8, Myhsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
SYCP1_MOUSE
|
||||||
| NC score | 0.055509 (rank : 75) | θ value | θ > 10 (rank : 190) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1456 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q62209, O09205, P70192, Q62329 | Gene names | Sycp1, Scp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
CEP63_HUMAN
|
||||||
| NC score | 0.055414 (rank : 76) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1005 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q96MT8, Q96CR0, Q9H8F5, Q9H8N0 | Gene names | CEP63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 63 kDa (Cep63 protein). | |||||
|
MYH14_HUMAN
|
||||||
| NC score | 0.055289 (rank : 77) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 995 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q7Z406, Q5CZ75, Q6XYE4, Q76B62, Q8WV23, Q96I22, Q9BT27, Q9BW35, Q9H882 | Gene names | MYH14, KIAA2034 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
IFT74_MOUSE
|
||||||
| NC score | 0.055283 (rank : 78) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 939 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8BKE9, Q80ZI3, Q9CSY1, Q9CUS0, Q9D9T5 | Gene names | Ift74, Ccdc2, Cmg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 74 homolog (Coiled-coil domain-containing protein 2) (Capillary morphogenesis protein 1) (CMG-1). | |||||
|
MYH3_HUMAN
|
||||||
| NC score | 0.055260 (rank : 79) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1660 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P11055, Q15492 | Gene names | MYH3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, fast skeletal muscle, embryonic (Muscle embryonic myosin heavy chain) (SMHCE). | |||||
|
MYH14_MOUSE
|
||||||
| NC score | 0.055211 (rank : 80) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1225 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q6URW6, Q80V64, Q80ZE6 | Gene names | Myh14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
AZI1_MOUSE
|
||||||
| NC score | 0.055137 (rank : 81) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 990 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q62036 | Gene names | Azi1, Az1, Azi | |||
|
Domain Architecture |

|
|||||
| Description | 5-azacytidine-induced protein 1 (Pre-acrosome localization protein 1). | |||||
|
GOGA3_MOUSE
|
||||||
| NC score | 0.055133 (rank : 82) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1437 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P55937, Q80VF5, Q8CCK4, Q9QYT2, Q9QYT3 | Gene names | Golga3, Mea2 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Male-enhanced antigen 2) (MEA-2). | |||||
|
GOGA4_HUMAN
|
||||||
| NC score | 0.055097 (rank : 83) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
CN145_MOUSE
|
||||||
| NC score | 0.054871 (rank : 84) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8BI22 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf145 homolog. | |||||
|
K0555_HUMAN
|
||||||
| NC score | 0.054639 (rank : 85) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 880 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q96AA8, O60302 | Gene names | KIAA0555 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0555. | |||||
|
TMF1_HUMAN
|
||||||
| NC score | 0.054592 (rank : 86) | θ value | θ > 10 (rank : 193) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 987 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P82094 | Gene names | TMF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TATA element modulatory factor (TMF). | |||||
|
NIN_MOUSE
|
||||||
| NC score | 0.054535 (rank : 87) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q61043, Q6ZPM7 | Gene names | Nin, Kiaa1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein. | |||||
|
MY18A_HUMAN
|
||||||
| NC score | 0.054535 (rank : 88) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
KTN1_MOUSE
|
||||||
| NC score | 0.054408 (rank : 89) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1324 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q61595, Q8BG49, Q8BHF4, Q8BHM8, Q8C9Y5, Q8CG51, Q8CG52, Q8CG53, Q8CG54, Q8CG55, Q8CG56, Q8CG57, Q8CG58, Q8CG59, Q8CG60, Q8CG61, Q8CG62, Q8CG63 | Gene names | Ktn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin. | |||||
|
GOGA3_HUMAN
|
||||||
| NC score | 0.054160 (rank : 90) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q08378, O43241, Q6P9C7, Q86XW3, Q8TDA9, Q8WZA3 | Gene names | GOLGA3 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Golgi complex-associated protein of 170 kDa) (GCP170). | |||||
|
TRHY_HUMAN
|
||||||
| NC score | 0.054158 (rank : 91) | θ value | θ > 10 (rank : 195) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
CI093_HUMAN
|
||||||
| NC score | 0.054144 (rank : 92) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q6TFL3, Q5SU58, Q6P1W1, Q6ZR13, Q8N1Z4, Q8N8L3, Q8TBR2 | Gene names | C9orf93 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf93. | |||||
|
RABE1_HUMAN
|
||||||
| NC score | 0.054062 (rank : 93) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q15276, O95369, Q8IVX3 | Gene names | RABEP1, RABPT5, RABPT5A | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha) (Rabaptin-4) (NY-REN-17 antigen). | |||||
|
CCD46_MOUSE
|
||||||
| NC score | 0.054049 (rank : 94) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q5PR68, Q80ZN6, Q99JS4, Q9D9T1 | Gene names | Ccdc46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled coil domain-containing protein 46. | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.053819 (rank : 95) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
LIPA2_HUMAN
|
||||||
| NC score | 0.053807 (rank : 96) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1044 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O75334 | Gene names | PPFIA2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-2) (PTPRF-interacting protein alpha-2). | |||||
|
CCD46_HUMAN
|
||||||
| NC score | 0.053733 (rank : 97) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1430 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8N8E3, Q6PIB5, Q8NCR4, Q8NFR4 | Gene names | CCDC46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 46. | |||||
|
EEA1_MOUSE
|
||||||
| NC score | 0.053679 (rank : 98) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1637 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8BL66, Q6DIC2 | Gene names | Eea1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early endosome antigen 1. | |||||
|
LIPA3_MOUSE
|
||||||
| NC score | 0.053649 (rank : 99) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P60469 | Gene names | Ppfia3 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-3 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-3) (PTPRF-interacting protein alpha-3). | |||||
|
GCC1_HUMAN
|
||||||
| NC score | 0.053459 (rank : 100) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 887 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q96CN9, Q9H6N7 | Gene names | GCC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP and coiled-coil domain-containing protein 1 (Golgi coiled coil protein 1). | |||||
|
KIF5C_HUMAN
|
||||||
| NC score | 0.053426 (rank : 101) | θ value | 0.62314 (rank : 18) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 974 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O60282, O95079 | Gene names | KIF5C, KIAA0531, NKHC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin heavy chain isoform 5C (Kinesin heavy chain neuron-specific 2). | |||||
|
TPR_HUMAN
|
||||||
| NC score | 0.053264 (rank : 102) | θ value | θ > 10 (rank : 194) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
CCD19_HUMAN
|
||||||
| NC score | 0.053169 (rank : 103) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9UL16 | Gene names | CCDC19, NESG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 19 (Nasopharyngeal epithelium- specific protein 1). | |||||
|
HOOK1_MOUSE
|
||||||
| NC score | 0.052953 (rank : 104) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 910 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8BIL5, Q8BIZ2, Q8K0P1, Q8K454, Q9CTN6 | Gene names | Hook1 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 1. | |||||
|
GOGA4_MOUSE
|
||||||
| NC score | 0.052940 (rank : 105) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
CCD62_HUMAN
|
||||||
| NC score | 0.052837 (rank : 106) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 631 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q6P9F0, Q6ZVF2, Q86VJ0, Q9BYZ5 | Gene names | CCDC62 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 60 (Aaa-protein) (Protein TSP- NY). | |||||
|
TAXB1_MOUSE
|
||||||
| NC score | 0.052817 (rank : 107) | θ value | θ > 10 (rank : 192) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q3UKC1, Q91YT6, Q9CVF0, Q9DC45 | Gene names | Tax1bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tax1-binding protein 1 homolog. | |||||
|
EEA1_HUMAN
|
||||||
| NC score | 0.052808 (rank : 108) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
IFT81_HUMAN
|
||||||
| NC score | 0.052738 (rank : 109) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8WYA0, Q8NB51, Q9BSV2, Q9UNY8 | Gene names | IFT81, CDV1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 81 (Carnitine deficiency-associated protein expressed in ventricle 1) (CDV-1 protein). | |||||
|
TAXB1_HUMAN
|
||||||
| NC score | 0.052669 (rank : 110) | θ value | θ > 10 (rank : 191) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 829 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q86VP1, O60398, O95770, Q13311, Q9BQG5, Q9UI88 | Gene names | TAX1BP1, T6BP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tax1-binding protein 1 (TRAF6-binding protein). | |||||
|
CROCC_HUMAN
|
||||||
| NC score | 0.052605 (rank : 111) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1409 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q5TZA2, Q2VHY3, Q66GT7, Q7Z2L4, Q7Z5D7 | Gene names | CROCC, KIAA0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
NUMA1_HUMAN
|
||||||
| NC score | 0.052415 (rank : 112) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
CYLN2_HUMAN
|
||||||
| NC score | 0.052366 (rank : 113) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1004 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9UDT6, O14527, O43611 | Gene names | CYLN2, KIAA0291, WBSCR4, WSCR4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic linker protein 2 (Cytoplasmic linker protein 115) (CLIP- 115) (Williams-Beuren syndrome chromosome region 4 protein). | |||||
|
BPAEA_MOUSE
|
||||||
| NC score | 0.052326 (rank : 114) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
SPAG5_HUMAN
|
||||||
| NC score | 0.052311 (rank : 115) | θ value | θ > 10 (rank : 189) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 625 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q96R06, O95213, Q9BWE8, Q9NT17, Q9UFE6 | Gene names | SPAG5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 5 (Astrin) (Mitotic spindle-associated protein p126) (MAP126) (Deepest). | |||||
|
MD1L1_HUMAN
|
||||||
| NC score | 0.052253 (rank : 116) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9Y6D9, Q13312, Q75MI0, Q86UM4, Q9UNH0 | Gene names | MAD1L1, MAD1, TXBP181 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1) (Mitotic checkpoint MAD1 protein-homolog) (HsMAD1) (hMAD1) (Tax-binding protein 181). | |||||
|
RBCC1_HUMAN
|
||||||
| NC score | 0.052250 (rank : 117) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1023 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8TDY2, Q8WVU9, Q92601 | Gene names | RB1CC1, KIAA0203, RBICC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RB1-inducible coiled-coil protein 1. | |||||
|
CCD41_HUMAN
|
||||||
| NC score | 0.052238 (rank : 118) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1139 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9Y592 | Gene names | CCDC41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41 (NY-REN-58 antigen). | |||||
|
BICD1_MOUSE
|
||||||
| NC score | 0.052216 (rank : 119) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 837 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8BR07, O55206, Q8BQ18, Q8BRR8, Q8C4D3, Q8CAK6, Q8R2J4, Q8R2J5, Q8R2J6, Q91YP7 | Gene names | Bicd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 1. | |||||
|
SMC4_HUMAN
|
||||||
| NC score | 0.052178 (rank : 120) | θ value | θ > 10 (rank : 187) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9NTJ3, O95752, Q8NDL4, Q9UNT9 | Gene names | SMC4L1, CAPC, SMC4 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 4-like 1 protein (Chromosome- associated polypeptide C) (hCAP-C) (XCAP-C homolog). | |||||
|
SM1L2_HUMAN
|
||||||
| NC score | 0.052114 (rank : 121) | θ value | θ > 10 (rank : 184) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1019 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8NDV3, Q5TIC3, Q9Y3G5 | Gene names | SMC1L2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 2 protein (SMC1beta protein). | |||||
|
RFIP3_HUMAN
|
||||||
| NC score | 0.052108 (rank : 122) | θ value | θ > 10 (rank : 182) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1055 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O75154, Q4VXV7, Q9H155, Q9H1G0, Q9NUI0 | Gene names | RAB11FIP3, KIAA0665 | |||
|
Domain Architecture |

|
|||||
| Description | Rab11 family-interacting protein 3 (Rab11-FIP3) (EF hands-containing Rab-interacting protein) (Eferin). | |||||
|
GOGA2_MOUSE
|
||||||
| NC score | 0.052104 (rank : 123) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q921M4 | Gene names | Golga2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgin subfamily A member 2 (Cis-Golgi matrix protein GM130). | |||||
|
CCD41_MOUSE
|
||||||
| NC score | 0.052089 (rank : 124) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9D5R3, Q3U7X7, Q3UX57, Q80VF0 | Gene names | Ccdc41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41. | |||||
|
HOOK1_HUMAN
|
||||||
| NC score | 0.052073 (rank : 125) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 942 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9UJC3, O60561, Q5TG44 | Gene names | HOOK1 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 1 (h-hook1) (hHK1). | |||||
|
CEP55_HUMAN
|
||||||
| NC score | 0.052048 (rank : 126) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q53EZ4, Q32WF5, Q3MV20, Q5VY28, Q6N034, Q96H32, Q9NVS7 | Gene names | CEP55, C10orf3, URCC6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome protein 55 (Up-regulated in colon cancer 6). | |||||
|
NUF2_HUMAN
|
||||||
| NC score | 0.051941 (rank : 127) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 661 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9BZD4, Q8WU69, Q96HJ4, Q96Q78 | Gene names | CDCA1, NUF2, NUF2R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Nuf2 (hNuf2) (hNuf2R) (hsNuf2) (Cell division cycle-associated protein 1). | |||||
|
FYCO1_MOUSE
|
||||||
| NC score | 0.051921 (rank : 128) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8VDC1, Q3V385, Q7TMT0, Q8BJN2 | Gene names | Fyco1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1. | |||||
|
CROCC_MOUSE
|
||||||
| NC score | 0.051920 (rank : 129) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1460 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8CJ40, Q7TQL2, Q80U01, Q8R0B9 | Gene names | Crocc, Kiaa0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
GRAP1_HUMAN
|
||||||
| NC score | 0.051895 (rank : 130) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q4V328, Q4V327, Q4V330, Q5HYG1, Q6N046, Q96DH8, Q9NQ43, Q9ULQ3 | Gene names | GRIPAP1, KIAA1167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP1-associated protein 1 (GRASP-1). | |||||
|
FLIP1_HUMAN
|
||||||
| NC score | 0.051894 (rank : 131) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q7Z7B0, Q5VUL6, Q86TC3, Q8N8B9, Q96SK6, Q9NVI8, Q9ULE5 | Gene names | FILIP1, KIAA1275 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
RABE1_MOUSE
|
||||||
| NC score | 0.051860 (rank : 132) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1105 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O35551, Q99L08, Q9CRP3, Q9EQF9, Q9JI94 | Gene names | Rabep1, Rab5ep, Rabpt5, Rabpt5a | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha). | |||||
|
MD1L1_MOUSE
|
||||||
| NC score | 0.051845 (rank : 133) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1001 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9WTX8, Q9WTX9 | Gene names | Mad1l1, Mad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1). | |||||
|
LIPA2_MOUSE
|
||||||
| NC score | 0.051770 (rank : 134) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1282 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8BSS9, Q8BN73 | Gene names | Ppfia2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-2) (PTPRF-interacting protein alpha-2). | |||||
|
FLIP1_MOUSE
|
||||||
| NC score | 0.051745 (rank : 135) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1176 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9CS72 | Gene names | Filip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
HOOK3_MOUSE
|
||||||
| NC score | 0.051723 (rank : 136) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8BUK6, Q8BV48, Q8BW57, Q8BY41 | Gene names | Hook3 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 3. | |||||
|
LIPA1_HUMAN
|
||||||
| NC score | 0.051721 (rank : 137) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1294 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q13136, Q13135, Q14567, Q8N4I2 | Gene names | PPFIA1, LIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-1 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-1) (PTPRF-interacting protein alpha-1) (LAR-interacting protein 1) (LIP.1). | |||||
|
CI039_HUMAN
|
||||||
| NC score | 0.051512 (rank : 138) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1171 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9NXG0, Q5VYJ0, Q9HAJ5 | Gene names | C9orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf39. | |||||
|
CYLN2_MOUSE
|
||||||
| NC score | 0.051511 (rank : 139) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1002 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9Z0H8, Q7TSI9, Q8CHU1, Q9EP81 | Gene names | Cyln2, Kiaa0291 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic linker protein 2 (Cytoplasmic linker protein 115) (CLIP- 115). | |||||
|
HOOK3_HUMAN
|
||||||
| NC score | 0.051405 (rank : 140) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1070 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q86VS8, Q8NBH0, Q9BY13 | Gene names | HOOK3 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 3 (hHK3). | |||||
|
DESP_HUMAN
|
||||||
| NC score | 0.051380 (rank : 141) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P15924, O75993, Q14189, Q9UHN4 | Gene names | DSP | |||
|
Domain Architecture |

|
|||||
| Description | Desmoplakin (DP) (250/210 kDa paraneoplastic pemphigus antigen). | |||||
|
KMHN1_HUMAN
|
||||||
| NC score | 0.051361 (rank : 142) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 950 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8TBZ0, Q86YI9, Q8N7W0 | Gene names | KMHN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cancer/testis antigen KM-HN-1. | |||||
|
CCD27_MOUSE
|
||||||
| NC score | 0.051343 (rank : 143) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q3V036 | Gene names | Ccdc27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 27. | |||||
|
ERC2_MOUSE
|
||||||
| NC score | 0.051283 (rank : 144) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1166 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q6PH08, Q80U20, Q8CCP1 | Gene names | Erc2, Cast1, D14Ertd171e, Kiaa0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2 (CAZ-associated structural protein 1) (CAST1). | |||||
|
SDCG8_HUMAN
|
||||||
| NC score | 0.051262 (rank : 145) | θ value | θ > 10 (rank : 183) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1002 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q86SQ7, O60527, Q3ZCR6, Q8N5F2, Q9P0F1 | Gene names | SDCCAG8, CCCAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serologically defined colon cancer antigen 8 (Centrosomal colon cancer autoantigen protein) (hCCCAP) (Antigen NY-CO-8). | |||||
|
GCC1_MOUSE
|
||||||
| NC score | 0.051180 (rank : 146) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9D4H2, Q8CCR3 | Gene names | Gcc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP and coiled-coil domain-containing protein 1 (Golgi coiled coil protein 1). | |||||
|
SMG1_HUMAN
|
||||||
| NC score | 0.051112 (rank : 147) | θ value | 2.36792 (rank : 35) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q96Q15, O43305, Q13284, Q8NFX2, Q96QV0, Q96RW3 | Gene names | SMG1, ATX, KIAA0421, LIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SMG1 (EC 2.7.11.1) (SMG-1) (hSMG-1) (Lambda/iota protein kinase C-interacting protein) (Lambda-interacting protein) (61E3.4). | |||||
|
CP250_MOUSE
|
||||||
| NC score | 0.050950 (rank : 148) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
ERC2_HUMAN
|
||||||
| NC score | 0.050911 (rank : 149) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1156 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O15083, Q86TK4 | Gene names | ERC2, KIAA0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2. | |||||
|
SMC4_MOUSE
|
||||||
| NC score | 0.050834 (rank : 150) | θ value | θ > 10 (rank : 188) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 830 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8CG47, Q8BTS7, Q8BTY9, Q99K21 | Gene names | Smc4l1, Capc, Smc4 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 4-like 1 protein (Chromosome- associated polypeptide C) (XCAP-C homolog). | |||||
|
IFT81_MOUSE
|
||||||
| NC score | 0.050813 (rank : 151) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O35594, Q8R4W2, Q9CSA1, Q9EQM1 | Gene names | Ift81, Cdv-1, Cdv1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 81 (Carnitine deficiency-associated protein expressed in ventricle 1) (CDV-1 protein). | |||||
|
BICD1_HUMAN
|
||||||
| NC score | 0.050777 (rank : 152) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 844 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q96G01, O43892, O43893 | Gene names | BICD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 1. | |||||
|
CP250_HUMAN
|
||||||
| NC score | 0.050773 (rank : 153) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
OPTN_MOUSE
|
||||||
| NC score | 0.050681 (rank : 154) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1120 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8K3K8 | Gene names | Optn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin. | |||||
|
KNTC2_MOUSE
|
||||||
| NC score | 0.050547 (rank : 155) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 692 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9D0F1, Q3TQT6, Q3UWM5, Q99P70 | Gene names | Kntc2, Hec1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Hec1 (Kinetochore-associated protein 2). | |||||
|
GOGA1_HUMAN
|
||||||
| NC score | 0.050457 (rank : 156) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1222 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q92805, Q8IYZ9 | Gene names | GOLGA1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
APOA4_MOUSE
|
||||||
| NC score | 0.050431 (rank : 157) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P06728 | Gene names | Apoa4 | |||
|
Domain Architecture |

|
|||||
| Description | Apolipoprotein A-IV precursor (Apo-AIV) (ApoA-IV). | |||||
|
MACF4_HUMAN
|
||||||
| NC score | 0.050382 (rank : 158) | θ value | 4.03905 (rank : 45) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1321 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q96PK2, Q8WXY1 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoform 4. | |||||
|
CN145_HUMAN
|
||||||
| NC score | 0.050355 (rank : 159) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1196 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q6ZU80, Q96ML4 | Gene names | C14orf145 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf145. | |||||
|
SMC2_HUMAN
|
||||||
| NC score | 0.050204 (rank : 160) | θ value | θ > 10 (rank : 185) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O95347, Q9P1P2 | Gene names | SMC2L1, CAPE, SMC2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (hCAP-E) (XCAP-E homolog). | |||||
|
KMHN1_MOUSE
|
||||||
| NC score | 0.050203 (rank : 161) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q3V125, Q3TTQ8 | Gene names | Gm172 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cancer/testis antigen KM-HN-1 homolog. | |||||
|
GOGA1_MOUSE
|
||||||
| NC score | 0.050180 (rank : 162) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 994 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9CW79, Q80YB0 | Gene names | Golga1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
PEPL_HUMAN
|
||||||
| NC score | 0.050168 (rank : 163) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1141 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O60437, O60314, O60454 | Gene names | PPL, KIAA0568 | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin (195 kDa cornified envelope precursor protein) (190 kDa paraneoplastic pemphigus antigen). | |||||
|
SMC2_MOUSE
|
||||||
| NC score | 0.050113 (rank : 164) | θ value | θ > 10 (rank : 186) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8CG48, Q61076, Q9CS17, Q9CSD8 | Gene names | Smc2l1, Cape, Fin16, Smc2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (XCAP-E homolog) (FGF-inducible protein 16). | |||||
|
RB6I2_MOUSE
|
||||||
| NC score | 0.050079 (rank : 165) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q99MI1, Q80TK7, Q8BPL1, Q8C7Y1, Q99MI2 | Gene names | Erc1, Cast2, Kiaa1081, Rab6ip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1) (ERC1) (CAZ-associated structural protein 2) (CAST2). | |||||
|
CRCM_HUMAN
|
||||||
| NC score | 0.050036 (rank : 166) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 520 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P23508 | Gene names | MCC | |||
|
Domain Architecture |

|
|||||
| Description | Colorectal mutant cancer protein (Protein MCC). | |||||
|
KIF5C_MOUSE
|
||||||
| NC score | 0.048759 (rank : 167) | θ value | 4.03905 (rank : 44) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P28738, Q9Z2F8 | Gene names | Kif5c, Nkhc2 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain isoform 5C (Kinesin heavy chain neuron-specific 2). | |||||
|
HOOK2_HUMAN
|
||||||
| NC score | 0.048194 (rank : 168) | θ value | 6.88961 (rank : 59) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 714 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q96ED9, O60562 | Gene names | HOOK2 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 2 (h-hook2) (hHK2). | |||||
|
SMG1_MOUSE
|
||||||
| NC score | 0.048134 (rank : 169) | θ value | 1.38821 (rank : 27) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BKX6, Q5DU42, Q6ZQC0, Q8BLU4, Q8BWJ5, Q8BXD3 | Gene names | Smg1, Kiaa0421 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SMG1 (EC 2.7.11.1) (SMG-1). | |||||
|
DMD_HUMAN
|
||||||
| NC score | 0.047081 (rank : 170) | θ value | 8.99809 (rank : 62) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P11532, Q02295, Q14169, Q14170, Q5JYU0, Q7KZ48 | Gene names | DMD | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
XPOT_HUMAN
|
||||||
| NC score | 0.043872 (rank : 171) | θ value | 1.38821 (rank : 28) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O43592, O43784, Q8WUG2, Q9BVS7 | Gene names | XPOT | |||
|
Domain Architecture |

|
|||||
| Description | Exportin-T (tRNA exportin) (Exportin(tRNA)). | |||||
|
SPT16_HUMAN
|
||||||
| NC score | 0.043633 (rank : 172) | θ value | 4.03905 (rank : 48) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y5B9, Q6GMT8, Q6P2F1, Q6PJM1, Q9NRX0 | Gene names | SUPT16H, FACT140, FACTP140 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FACT complex subunit SPT16 (Facilitates chromatin transcription complex subunit SPT16) (hSPT16) (FACT 140 kDa subunit) (FACTp140) (Chromatin-specific transcription elongation factor 140 kDa subunit). | |||||
|
SPT16_MOUSE
|
||||||
| NC score | 0.043306 (rank : 173) | θ value | 4.03905 (rank : 49) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q920B9, Q3TZ48, Q3UFH0, Q921H4 | Gene names | Supt16h, Fact140, Factp140 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FACT complex subunit SPT16 (Facilitates chromatin transcription complex subunit SPT16) (FACT 140 kDa subunit) (FACTp140) (Chromatin- specific transcription elongation factor 140 kDa subunit). | |||||
|
UBE4A_HUMAN
|
||||||
| NC score | 0.038762 (rank : 174) | θ value | 4.03905 (rank : 50) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14139, Q6P5T4, Q7Z639 | Gene names | UBE4A, KIAA0126 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin conjugation factor E4 A. | |||||
|
PRP31_MOUSE
|
||||||
| NC score | 0.036833 (rank : 175) | θ value | 3.0926 (rank : 38) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CCF0, Q6P7X2, Q8BQ91, Q8C8U4, Q8C8V5, Q8CCG6, Q8CF52, Q8VBW3 | Gene names | Prpf31, Prp31 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U4/U6 small nuclear ribonucleoprotein Prp31 (Pre-mRNA-processing factor 31) (U4/U6 snRNP 61 kDa protein) (Protein 61K). | |||||
|
BIR1G_MOUSE
|
||||||
| NC score | 0.035916 (rank : 176) | θ value | 1.81305 (rank : 30) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JIB3 | Gene names | Birc1g, Naip7 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1g (Neuronal apoptosis inhibitory protein 7). | |||||
|
BIR1F_MOUSE
|
||||||
| NC score | 0.035858 (rank : 177) | θ value | 1.81305 (rank : 29) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JIB6, O09121, O09122, P81704 | Gene names | Birc1f, Naip-rs4, Naip6 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1f (Neuronal apoptosis inhibitory protein 6). | |||||
|
LRC40_MOUSE
|
||||||
| NC score | 0.032753 (rank : 178) | θ value | 0.0563607 (rank : 11) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9CRC8, Q78WQ9, Q8BS83 | Gene names | Lrrc40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 40. | |||||
|
MRCKB_HUMAN
|
||||||
| NC score | 0.030028 (rank : 179) | θ value | 1.81305 (rank : 31) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 2239 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9Y5S2, Q2L7A5, Q86TJ1, Q9ULU5 | Gene names | CDC42BPB, KIAA1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
MRCKB_MOUSE
|
||||||
| NC score | 0.028735 (rank : 180) | θ value | 4.03905 (rank : 46) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 2180 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q7TT50, Q69ZR5, Q80W33 | Gene names | Cdc42bpb, Kiaa1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
KIF17_MOUSE
|
||||||
| NC score | 0.028617 (rank : 181) | θ value | 4.03905 (rank : 43) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q99PW8 | Gene names | Kif17 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF17 (MmKIF17). | |||||
|
K22E_HUMAN
|
||||||
| NC score | 0.028375 (rank : 182) | θ value | 1.38821 (rank : 24) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 515 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P35908 | Gene names | KRT2A, KRT2E | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 2 epidermal (Cytokeratin-2e) (K2e) (CK 2e). | |||||
|
LRC48_MOUSE
|
||||||
| NC score | 0.026901 (rank : 183) | θ value | 8.99809 (rank : 66) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9D5E4 | Gene names | Lrrc48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 48. | |||||
|
K1914_MOUSE
|
||||||
| NC score | 0.025754 (rank : 184) | θ value | 8.99809 (rank : 64) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5DTU0, Q8BID1, Q8K2G0 | Gene names | Kiaa1914 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1914. | |||||
|
LBC_HUMAN
|
||||||
| NC score | 0.025504 (rank : 185) | θ value | 8.99809 (rank : 65) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q12802 | Gene names | LBC | |||
|
Domain Architecture |

|
|||||
| Description | LBC oncogene (P47) (Lymphoid blast crisis oncogene). | |||||
|
OPA1_HUMAN
|
||||||
| NC score | 0.024729 (rank : 186) | θ value | 5.27518 (rank : 55) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O60313 | Gene names | OPA1, KIAA0567 | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-like 120 kDa protein, mitochondrial precursor (Optic atrophy protein 1) [Contains: Dynamin-like 120 kDa protein, form S1]. | |||||
|
OPA1_MOUSE
|
||||||
| NC score | 0.024518 (rank : 187) | θ value | 5.27518 (rank : 56) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P58281, Q3ULA5, Q8BKU7, Q8BLL3, Q8BM08, Q8R3J7 | Gene names | Opa1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-like 120 kDa protein, mitochondrial precursor (Optic atrophy protein 1 homolog) (Large GTP-binding protein) (LargeG) [Contains: Dynamin-like 120 kDa protein, form S1]. | |||||
|
SCEL_MOUSE
|
||||||
| NC score | 0.023871 (rank : 188) | θ value | 8.99809 (rank : 71) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9EQG3, Q9CTT9 | Gene names | Scel | |||
|
Domain Architecture |

|
|||||
| Description | Sciellin. | |||||
|
3BP1_MOUSE
|
||||||
| NC score | 0.022025 (rank : 189) | θ value | 1.38821 (rank : 22) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P55194, Q99KK8 | Gene names | Sh3bp1, 3bp1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 1 (3BP-1). | |||||
|
PHAR1_MOUSE
|
||||||
| NC score | 0.019736 (rank : 190) | θ value | 8.99809 (rank : 69) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q2M3X8, Q80VL9, Q8C873 | Gene names | Phactr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatase and actin regulator 1. | |||||
|
SYTL4_HUMAN
|
||||||
| NC score | 0.017254 (rank : 191) | θ value | 2.36792 (rank : 36) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96C24, Q5H9J3, Q5JPG8, Q8N9P4, Q9H4R0, Q9H4R1 | Gene names | SYTL4 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 4 (Exophilin-2) (Granuphilin). | |||||
|
PSD12_HUMAN
|
||||||
| NC score | 0.016938 (rank : 192) | θ value | 8.99809 (rank : 70) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O00232, Q53HA2, Q6P053 | Gene names | PSMD12 | |||
|
Domain Architecture |

|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 12 (26S proteasome regulatory subunit p55). | |||||
|
TRI34_MOUSE
|
||||||
| NC score | 0.015241 (rank : 193) | θ value | 3.0926 (rank : 41) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99PP6, Q99PP4, Q99PP5 | Gene names | Trim34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 34. | |||||
|
GBRT_HUMAN
|
||||||
| NC score | 0.013978 (rank : 194) | θ value | 0.62314 (rank : 17) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UN88, Q9NZK8 | Gene names | GABRQ | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric-acid receptor subunit theta precursor (GABA(A) receptor subunit theta). | |||||
|
OMGP_MOUSE
|
||||||
| NC score | 0.013523 (rank : 195) | θ value | 3.0926 (rank : 37) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q63912 | Gene names | Omg, Omgp | |||
|
Domain Architecture |

|
|||||
| Description | Oligodendrocyte-myelin glycoprotein precursor. | |||||
|
GSTA3_MOUSE
|
||||||
| NC score | 0.007424 (rank : 196) | θ value | 8.99809 (rank : 63) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P30115 | Gene names | Gsta3, Gstyc | |||
|
Domain Architecture |

|
|||||
| Description | Glutathione S-transferase A3 (EC 2.5.1.18) (Glutathione S-transferase Yc) (Ya3) (GST class-alpha member 3). | |||||