Please be patient as the page loads
|
DDX11_MOUSE
|
||||||
| SwissProt Accessions | Q6AXC6 | Gene names | Ddx11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX11 (EC 3.6.1.-) (DEAD/H box protein 11). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
DDX11_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.966030 (rank : 2) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96FC9, Q13333, Q86VQ4, Q86W62, Q92498, Q92770, Q92998, Q92999 | Gene names | DDX11, CHL1, CHLR1, KRG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX11 (EC 3.6.1.-) (DEAD/H box protein 11) (CHL1 homolog) (Keratinocyte growth factor-regulated gene 2 protein) (KRG-2). | |||||
|
DDX11_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q6AXC6 | Gene names | Ddx11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX11 (EC 3.6.1.-) (DEAD/H box protein 11). | |||||
|
FANCJ_MOUSE
|
||||||
| θ value | 2.92676e-75 (rank : 3) | NC score | 0.852179 (rank : 4) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5SXJ3, Q8BJQ8, Q8BKI6 | Gene names | Brip1, Bach1, Fancj | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group J protein homolog (EC 3.6.1.-) (ATP-dependent RNA helicase BRIP1) (Protein FACJ) (BRCA1-interacting protein C-terminal helicase 1) (BRCA1-interacting protein 1) (BRCA1-associated C-terminal helicase 1). | |||||
|
FANCJ_HUMAN
|
||||||
| θ value | 5.51964e-74 (rank : 4) | NC score | 0.855942 (rank : 3) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BX63, Q8NCI5 | Gene names | BRIP1, BACH1, FANCJ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group J protein (EC 3.6.1.-) (ATP-dependent RNA helicase BRIP1) (Protein FACJ) (BRCA1-interacting protein C-terminal helicase 1) (BRCA1-interacting protein 1) (BRCA1-associated C-terminal helicase 1). | |||||
|
RTEL1_HUMAN
|
||||||
| θ value | 2.84138e-62 (rank : 5) | NC score | 0.842785 (rank : 5) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NZ71, Q5JTV4, Q9BW37, Q9H402, Q9H4X6, Q9NX25, Q9NZ73, Q9UPR4, Q9Y4R6 | Gene names | RTEL1, C20orf41, KIAA1088, NHL | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of telomere elongation helicase 1 (EC 3.6.1.-) (Helicase- like protein NHL). | |||||
|
ERCC2_MOUSE
|
||||||
| θ value | 9.83387e-39 (rank : 6) | NC score | 0.781679 (rank : 6) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O08811, Q9DC01 | Gene names | Ercc2, Xpd | |||
|
Domain Architecture |

|
|||||
| Description | TFIIH basal transcription factor complex helicase subunit (EC 3.6.1.-) (DNA-repair protein complementing XP-D cells) (Xeroderma pigmentosum group D-complementing protein) (CXPD) (DNA excision repair protein ERCC-2). | |||||
|
ERCC2_HUMAN
|
||||||
| θ value | 2.19075e-38 (rank : 7) | NC score | 0.779825 (rank : 7) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P18074, Q8N721 | Gene names | ERCC2, XPD, XPDC | |||
|
Domain Architecture |

|
|||||
| Description | TFIIH basal transcription factor complex helicase subunit (EC 3.6.1.-) (DNA-repair protein complementing XP-D cells) (Xeroderma pigmentosum group D-complementing protein) (CXPD) (DNA excision repair protein ERCC-2). | |||||
|
BFSP2_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 8) | NC score | 0.033253 (rank : 10) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6NVD9, Q63832, Q6P5N4, Q8VDD6 | Gene names | Bfsp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phakinin (Beaded filament structural protein 2) (Lens fiber cell beaded filament protein CP 49) (CP49) (49 kDa cytoskeletal protein). | |||||
|
STK10_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 9) | NC score | 0.020118 (rank : 24) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O94804, Q9UIW4 | Gene names | STK10, LOK | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
MY18A_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 10) | NC score | 0.025500 (rank : 14) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
MY18A_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 11) | NC score | 0.027293 (rank : 12) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
TBCD2_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 12) | NC score | 0.045079 (rank : 9) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BYX2 | Gene names | TBC1D2, PARIS1 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 2 (Prostate antigen recognized and identified by SEREX) (PARIS-1). | |||||
|
STK10_MOUSE
|
||||||
| θ value | 0.163984 (rank : 13) | NC score | 0.020572 (rank : 23) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 2132 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O55098 | Gene names | Stk10, Lok | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
AZI1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 14) | NC score | 0.050906 (rank : 8) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 990 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q62036 | Gene names | Azi1, Az1, Azi | |||
|
Domain Architecture |

|
|||||
| Description | 5-azacytidine-induced protein 1 (Pre-acrosome localization protein 1). | |||||
|
BFSP2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 15) | NC score | 0.025849 (rank : 13) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q13515, Q9HBW5 | Gene names | BFSP2 | |||
|
Domain Architecture |
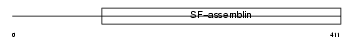
|
|||||
| Description | Phakinin (Beaded filament structural protein 2) (Lens fiber cell beaded filament protein CP 49) (CP49) (49 kDa cytoskeletal protein) (CP 47) (CP47) (Lens intermediate filament-like light) (LIFL-L). | |||||
|
SLK_HUMAN
|
||||||
| θ value | 0.365318 (rank : 16) | NC score | 0.016634 (rank : 28) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1775 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9H2G2, O00211, Q6P1Z4, Q86WU7, Q86WW1, Q92603, Q9NQL0, Q9NQL1 | Gene names | SLK, KIAA0204, STK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (hSLK) (Serine/threonine-protein kinase 2) (CTCL tumor antigen se20-9). | |||||
|
CDYL_MOUSE
|
||||||
| θ value | 0.62314 (rank : 17) | NC score | 0.021077 (rank : 21) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9WTK2 | Gene names | Cdyl | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain Y-like protein (CDY-like). | |||||
|
FBX3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 18) | NC score | 0.031987 (rank : 11) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UK99, Q9NUX2 | Gene names | FBXO3, FBX3 | |||
|
Domain Architecture |

|
|||||
| Description | F-box only protein 3. | |||||
|
NFAT5_MOUSE
|
||||||
| θ value | 1.06291 (rank : 19) | NC score | 0.025319 (rank : 15) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9WV30 | Gene names | Nfat5 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells 5 (T cell transcription factor NFAT5) (NF-AT5) (Rel domain-containing transcription factor NFAT5). | |||||
|
REST_HUMAN
|
||||||
| θ value | 1.06291 (rank : 20) | NC score | 0.022741 (rank : 17) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1605 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P30622 | Gene names | RSN, CYLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Restin (Cytoplasmic linker protein 170 alpha-2) (CLIP-170) (Reed- Sternberg intermediate filament-associated protein) (Cytoplasmic linker protein 1). | |||||
|
SLK_MOUSE
|
||||||
| θ value | 1.06291 (rank : 21) | NC score | 0.014378 (rank : 30) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1798 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O54988, Q80U65, Q8CAU2, Q8CDW2, Q9WU41 | Gene names | Slk, Kiaa0204, Stk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (mSLK) (Serine/threonine-protein kinase 2) (STE20-related kinase SMAK) (Etk4). | |||||
|
DMD_MOUSE
|
||||||
| θ value | 1.38821 (rank : 22) | NC score | 0.022001 (rank : 19) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1111 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P11531, O35653, Q60703 | Gene names | Dmd | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
GOGA4_MOUSE
|
||||||
| θ value | 1.81305 (rank : 23) | NC score | 0.021517 (rank : 20) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
EST1A_MOUSE
|
||||||
| θ value | 2.36792 (rank : 24) | NC score | 0.019595 (rank : 25) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P61406, Q5NC64 | Gene names | Est1a, Kiaa0732 | |||
|
Domain Architecture |

|
|||||
| Description | Telomerase-binding protein EST1A (Ever shorter telomeres 1A) (Telomerase subunit EST1A) (EST1-like protein A). | |||||
|
DMD_HUMAN
|
||||||
| θ value | 3.0926 (rank : 25) | NC score | 0.022352 (rank : 18) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P11532, Q02295, Q14169, Q14170, Q5JYU0, Q7KZ48 | Gene names | DMD | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
IL6RA_MOUSE
|
||||||
| θ value | 3.0926 (rank : 26) | NC score | 0.016914 (rank : 26) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P22272 | Gene names | Il6ra, Il6r | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-6 receptor alpha chain precursor (IL-6R-alpha) (IL-6R 1) (CD126 antigen). | |||||
|
NALP5_MOUSE
|
||||||
| θ value | 3.0926 (rank : 27) | NC score | 0.009196 (rank : 33) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9R1M5, Q9JLR2 | Gene names | Nalp5, Mater | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Maternal antigen that embryos require) (Mater protein) (Ooplasm-specific protein 1) (OP1). | |||||
|
TRIM8_HUMAN
|
||||||
| θ value | 3.0926 (rank : 28) | NC score | 0.013138 (rank : 31) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9BZR9, Q9C028 | Gene names | TRIM8, GERP, RNF27 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 8 (RING finger protein 27) (Glioblastoma-expressed RING finger protein). | |||||
|
REST_MOUSE
|
||||||
| θ value | 4.03905 (rank : 29) | NC score | 0.020579 (rank : 22) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1506 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q922J3, Q571L7 | Gene names | Rsn, Kiaa4046 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin. | |||||
|
DYH5_MOUSE
|
||||||
| θ value | 5.27518 (rank : 30) | NC score | 0.011453 (rank : 32) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8VHE6, Q8BWG1 | Gene names | Dnah5, Dnahc5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ciliary dynein heavy chain 5 (Axonemal beta dynein heavy chain 5) (Mdnah5). | |||||
|
EP15_HUMAN
|
||||||
| θ value | 5.27518 (rank : 31) | NC score | 0.015202 (rank : 29) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1021 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P42566 | Gene names | EPS15, AF1P | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15 (Protein Eps15) (AF-1p protein). | |||||
|
CC062_HUMAN
|
||||||
| θ value | 6.88961 (rank : 32) | NC score | 0.016881 (rank : 27) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6ZUJ4, Q6P7E9, Q7Z3X6 | Gene names | C3orf62 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C3orf62. | |||||
|
DEN2D_HUMAN
|
||||||
| θ value | 6.88961 (rank : 33) | NC score | 0.007805 (rank : 36) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H6A0, Q5T5V6, Q9BSU0 | Gene names | DENND2D | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 2D. | |||||
|
NFAT5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 34) | NC score | 0.023027 (rank : 16) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O94916, O95693, Q9UN18 | Gene names | NFAT5, KIAA0827, TONEBP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells 5 (T-cell transcription factor NFAT5) (NF-AT5) (Tonicity-responsive enhancer-binding protein) (TonE- binding protein) (TonEBP). | |||||
|
ROCK1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 35) | NC score | 0.009107 (rank : 34) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 2126 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P70335, Q8C3G4, Q8C7H0 | Gene names | Rock1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK). | |||||
|
TRI62_HUMAN
|
||||||
| θ value | 6.88961 (rank : 36) | NC score | 0.008222 (rank : 35) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9BVG3, Q9NVG0 | Gene names | TRIM62 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 62. | |||||
|
ATBF1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 37) | NC score | 0.004369 (rank : 40) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
BIG2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 38) | NC score | 0.006770 (rank : 38) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y6D5, Q5TFT9, Q9NTS1 | Gene names | ARFGEF2, ARFGEP2, BIG2 | |||
|
Domain Architecture |

|
|||||
| Description | Brefeldin A-inhibited guanine nucleotide-exchange protein 2 (Brefeldin A-inhibited GEP 2). | |||||
|
LRBA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 39) | NC score | 0.006321 (rank : 39) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P50851, Q9H2U3, Q9H2U4 | Gene names | LRBA, BGL, CDC4L, LBA | |||
|
Domain Architecture |

|
|||||
| Description | Lipopolysaccharide-responsive and beige-like anchor protein (CDC4-like protein) (Beige-like protein). | |||||
|
MRCKG_MOUSE
|
||||||
| θ value | 8.99809 (rank : 40) | NC score | 0.007419 (rank : 37) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1466 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q80UW5 | Gene names | Cdc42bpg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK gamma (EC 2.7.11.1) (CDC42- binding protein kinase gamma) (Myotonic dystrophy kinase-related CDC42-binding kinase gamma) (Myotonic dystrophy protein kinase-like alpha) (MRCK gamma) (DMPK-like gamma). | |||||
|
DDX11_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q6AXC6 | Gene names | Ddx11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX11 (EC 3.6.1.-) (DEAD/H box protein 11). | |||||
|
DDX11_HUMAN
|
||||||
| NC score | 0.966030 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96FC9, Q13333, Q86VQ4, Q86W62, Q92498, Q92770, Q92998, Q92999 | Gene names | DDX11, CHL1, CHLR1, KRG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX11 (EC 3.6.1.-) (DEAD/H box protein 11) (CHL1 homolog) (Keratinocyte growth factor-regulated gene 2 protein) (KRG-2). | |||||
|
FANCJ_HUMAN
|
||||||
| NC score | 0.855942 (rank : 3) | θ value | 5.51964e-74 (rank : 4) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BX63, Q8NCI5 | Gene names | BRIP1, BACH1, FANCJ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group J protein (EC 3.6.1.-) (ATP-dependent RNA helicase BRIP1) (Protein FACJ) (BRCA1-interacting protein C-terminal helicase 1) (BRCA1-interacting protein 1) (BRCA1-associated C-terminal helicase 1). | |||||
|
FANCJ_MOUSE
|
||||||
| NC score | 0.852179 (rank : 4) | θ value | 2.92676e-75 (rank : 3) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5SXJ3, Q8BJQ8, Q8BKI6 | Gene names | Brip1, Bach1, Fancj | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group J protein homolog (EC 3.6.1.-) (ATP-dependent RNA helicase BRIP1) (Protein FACJ) (BRCA1-interacting protein C-terminal helicase 1) (BRCA1-interacting protein 1) (BRCA1-associated C-terminal helicase 1). | |||||
|
RTEL1_HUMAN
|
||||||
| NC score | 0.842785 (rank : 5) | θ value | 2.84138e-62 (rank : 5) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NZ71, Q5JTV4, Q9BW37, Q9H402, Q9H4X6, Q9NX25, Q9NZ73, Q9UPR4, Q9Y4R6 | Gene names | RTEL1, C20orf41, KIAA1088, NHL | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of telomere elongation helicase 1 (EC 3.6.1.-) (Helicase- like protein NHL). | |||||
|
ERCC2_MOUSE
|
||||||
| NC score | 0.781679 (rank : 6) | θ value | 9.83387e-39 (rank : 6) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O08811, Q9DC01 | Gene names | Ercc2, Xpd | |||
|
Domain Architecture |

|
|||||
| Description | TFIIH basal transcription factor complex helicase subunit (EC 3.6.1.-) (DNA-repair protein complementing XP-D cells) (Xeroderma pigmentosum group D-complementing protein) (CXPD) (DNA excision repair protein ERCC-2). | |||||
|
ERCC2_HUMAN
|
||||||
| NC score | 0.779825 (rank : 7) | θ value | 2.19075e-38 (rank : 7) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P18074, Q8N721 | Gene names | ERCC2, XPD, XPDC | |||
|
Domain Architecture |

|
|||||
| Description | TFIIH basal transcription factor complex helicase subunit (EC 3.6.1.-) (DNA-repair protein complementing XP-D cells) (Xeroderma pigmentosum group D-complementing protein) (CXPD) (DNA excision repair protein ERCC-2). | |||||
|
AZI1_MOUSE
|
||||||
| NC score | 0.050906 (rank : 8) | θ value | 0.279714 (rank : 14) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 990 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q62036 | Gene names | Azi1, Az1, Azi | |||
|
Domain Architecture |

|
|||||
| Description | 5-azacytidine-induced protein 1 (Pre-acrosome localization protein 1). | |||||
|
TBCD2_HUMAN
|
||||||
| NC score | 0.045079 (rank : 9) | θ value | 0.0961366 (rank : 12) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BYX2 | Gene names | TBC1D2, PARIS1 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 2 (Prostate antigen recognized and identified by SEREX) (PARIS-1). | |||||
|
BFSP2_MOUSE
|
||||||
| NC score | 0.033253 (rank : 10) | θ value | 0.0431538 (rank : 8) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6NVD9, Q63832, Q6P5N4, Q8VDD6 | Gene names | Bfsp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phakinin (Beaded filament structural protein 2) (Lens fiber cell beaded filament protein CP 49) (CP49) (49 kDa cytoskeletal protein). | |||||
|
FBX3_HUMAN
|
||||||
| NC score | 0.031987 (rank : 11) | θ value | 1.06291 (rank : 18) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UK99, Q9NUX2 | Gene names | FBXO3, FBX3 | |||
|
Domain Architecture |

|
|||||
| Description | F-box only protein 3. | |||||
|
MY18A_MOUSE
|
||||||
| NC score | 0.027293 (rank : 12) | θ value | 0.0961366 (rank : 11) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
BFSP2_HUMAN
|
||||||
| NC score | 0.025849 (rank : 13) | θ value | 0.365318 (rank : 15) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q13515, Q9HBW5 | Gene names | BFSP2 | |||
|
Domain Architecture |

|
|||||
| Description | Phakinin (Beaded filament structural protein 2) (Lens fiber cell beaded filament protein CP 49) (CP49) (49 kDa cytoskeletal protein) (CP 47) (CP47) (Lens intermediate filament-like light) (LIFL-L). | |||||
|
MY18A_HUMAN
|
||||||
| NC score | 0.025500 (rank : 14) | θ value | 0.0961366 (rank : 10) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
NFAT5_MOUSE
|
||||||
| NC score | 0.025319 (rank : 15) | θ value | 1.06291 (rank : 19) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9WV30 | Gene names | Nfat5 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells 5 (T cell transcription factor NFAT5) (NF-AT5) (Rel domain-containing transcription factor NFAT5). | |||||
|
NFAT5_HUMAN
|
||||||
| NC score | 0.023027 (rank : 16) | θ value | 6.88961 (rank : 34) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O94916, O95693, Q9UN18 | Gene names | NFAT5, KIAA0827, TONEBP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells 5 (T-cell transcription factor NFAT5) (NF-AT5) (Tonicity-responsive enhancer-binding protein) (TonE- binding protein) (TonEBP). | |||||
|
REST_HUMAN
|
||||||
| NC score | 0.022741 (rank : 17) | θ value | 1.06291 (rank : 20) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1605 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P30622 | Gene names | RSN, CYLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Restin (Cytoplasmic linker protein 170 alpha-2) (CLIP-170) (Reed- Sternberg intermediate filament-associated protein) (Cytoplasmic linker protein 1). | |||||
|
DMD_HUMAN
|
||||||
| NC score | 0.022352 (rank : 18) | θ value | 3.0926 (rank : 25) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P11532, Q02295, Q14169, Q14170, Q5JYU0, Q7KZ48 | Gene names | DMD | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
DMD_MOUSE
|
||||||
| NC score | 0.022001 (rank : 19) | θ value | 1.38821 (rank : 22) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1111 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P11531, O35653, Q60703 | Gene names | Dmd | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
GOGA4_MOUSE
|
||||||
| NC score | 0.021517 (rank : 20) | θ value | 1.81305 (rank : 23) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
CDYL_MOUSE
|
||||||
| NC score | 0.021077 (rank : 21) | θ value | 0.62314 (rank : 17) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9WTK2 | Gene names | Cdyl | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain Y-like protein (CDY-like). | |||||
|
REST_MOUSE
|
||||||
| NC score | 0.020579 (rank : 22) | θ value | 4.03905 (rank : 29) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1506 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q922J3, Q571L7 | Gene names | Rsn, Kiaa4046 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin. | |||||
|
STK10_MOUSE
|
||||||
| NC score | 0.020572 (rank : 23) | θ value | 0.163984 (rank : 13) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 2132 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O55098 | Gene names | Stk10, Lok | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
STK10_HUMAN
|
||||||
| NC score | 0.020118 (rank : 24) | θ value | 0.0736092 (rank : 9) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O94804, Q9UIW4 | Gene names | STK10, LOK | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
EST1A_MOUSE
|
||||||
| NC score | 0.019595 (rank : 25) | θ value | 2.36792 (rank : 24) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P61406, Q5NC64 | Gene names | Est1a, Kiaa0732 | |||
|
Domain Architecture |

|
|||||
| Description | Telomerase-binding protein EST1A (Ever shorter telomeres 1A) (Telomerase subunit EST1A) (EST1-like protein A). | |||||
|
IL6RA_MOUSE
|
||||||
| NC score | 0.016914 (rank : 26) | θ value | 3.0926 (rank : 26) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P22272 | Gene names | Il6ra, Il6r | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-6 receptor alpha chain precursor (IL-6R-alpha) (IL-6R 1) (CD126 antigen). | |||||
|
CC062_HUMAN
|
||||||
| NC score | 0.016881 (rank : 27) | θ value | 6.88961 (rank : 32) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6ZUJ4, Q6P7E9, Q7Z3X6 | Gene names | C3orf62 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C3orf62. | |||||
|
SLK_HUMAN
|
||||||
| NC score | 0.016634 (rank : 28) | θ value | 0.365318 (rank : 16) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1775 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9H2G2, O00211, Q6P1Z4, Q86WU7, Q86WW1, Q92603, Q9NQL0, Q9NQL1 | Gene names | SLK, KIAA0204, STK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (hSLK) (Serine/threonine-protein kinase 2) (CTCL tumor antigen se20-9). | |||||
|
EP15_HUMAN
|
||||||
| NC score | 0.015202 (rank : 29) | θ value | 5.27518 (rank : 31) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1021 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P42566 | Gene names | EPS15, AF1P | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15 (Protein Eps15) (AF-1p protein). | |||||
|
SLK_MOUSE
|
||||||
| NC score | 0.014378 (rank : 30) | θ value | 1.06291 (rank : 21) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1798 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O54988, Q80U65, Q8CAU2, Q8CDW2, Q9WU41 | Gene names | Slk, Kiaa0204, Stk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (mSLK) (Serine/threonine-protein kinase 2) (STE20-related kinase SMAK) (Etk4). | |||||
|
TRIM8_HUMAN
|
||||||
| NC score | 0.013138 (rank : 31) | θ value | 3.0926 (rank : 28) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9BZR9, Q9C028 | Gene names | TRIM8, GERP, RNF27 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 8 (RING finger protein 27) (Glioblastoma-expressed RING finger protein). | |||||
|
DYH5_MOUSE
|
||||||
| NC score | 0.011453 (rank : 32) | θ value | 5.27518 (rank : 30) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8VHE6, Q8BWG1 | Gene names | Dnah5, Dnahc5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ciliary dynein heavy chain 5 (Axonemal beta dynein heavy chain 5) (Mdnah5). | |||||
|
NALP5_MOUSE
|
||||||
| NC score | 0.009196 (rank : 33) | θ value | 3.0926 (rank : 27) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9R1M5, Q9JLR2 | Gene names | Nalp5, Mater | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Maternal antigen that embryos require) (Mater protein) (Ooplasm-specific protein 1) (OP1). | |||||
|
ROCK1_MOUSE
|
||||||
| NC score | 0.009107 (rank : 34) | θ value | 6.88961 (rank : 35) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 2126 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P70335, Q8C3G4, Q8C7H0 | Gene names | Rock1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK). | |||||
|
TRI62_HUMAN
|
||||||
| NC score | 0.008222 (rank : 35) | θ value | 6.88961 (rank : 36) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9BVG3, Q9NVG0 | Gene names | TRIM62 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 62. | |||||
|
DEN2D_HUMAN
|
||||||
| NC score | 0.007805 (rank : 36) | θ value | 6.88961 (rank : 33) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H6A0, Q5T5V6, Q9BSU0 | Gene names | DENND2D | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 2D. | |||||
|
MRCKG_MOUSE
|
||||||
| NC score | 0.007419 (rank : 37) | θ value | 8.99809 (rank : 40) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1466 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q80UW5 | Gene names | Cdc42bpg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK gamma (EC 2.7.11.1) (CDC42- binding protein kinase gamma) (Myotonic dystrophy kinase-related CDC42-binding kinase gamma) (Myotonic dystrophy protein kinase-like alpha) (MRCK gamma) (DMPK-like gamma). | |||||
|
BIG2_HUMAN
|
||||||
| NC score | 0.006770 (rank : 38) | θ value | 8.99809 (rank : 38) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y6D5, Q5TFT9, Q9NTS1 | Gene names | ARFGEF2, ARFGEP2, BIG2 | |||
|
Domain Architecture |

|
|||||
| Description | Brefeldin A-inhibited guanine nucleotide-exchange protein 2 (Brefeldin A-inhibited GEP 2). | |||||
|
LRBA_HUMAN
|
||||||
| NC score | 0.006321 (rank : 39) | θ value | 8.99809 (rank : 39) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P50851, Q9H2U3, Q9H2U4 | Gene names | LRBA, BGL, CDC4L, LBA | |||
|
Domain Architecture |

|
|||||
| Description | Lipopolysaccharide-responsive and beige-like anchor protein (CDC4-like protein) (Beige-like protein). | |||||
|
ATBF1_MOUSE
|
||||||
| NC score | 0.004369 (rank : 40) | θ value | 8.99809 (rank : 37) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||