Please be patient as the page loads
|
CMC1_MOUSE
|
||||||
| SwissProt Accessions | Q8BH59 | Gene names | Slc25a12, Aralar1 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding mitochondrial carrier protein Aralar1 (Mitochondrial aspartate glutamate carrier 1) (Solute carrier family 25 member 12). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CMC1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999192 (rank : 2) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 102 | |
| SwissProt Accessions | O75746 | Gene names | SLC25A12, ARALAR1 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding mitochondrial carrier protein Aralar1 (Mitochondrial aspartate glutamate carrier 1) (Solute carrier family 25 member 12). | |||||
|
CMC1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 102 | |
| SwissProt Accessions | Q8BH59 | Gene names | Slc25a12, Aralar1 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding mitochondrial carrier protein Aralar1 (Mitochondrial aspartate glutamate carrier 1) (Solute carrier family 25 member 12). | |||||
|
CMC2_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.995516 (rank : 4) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q9UJS0, O14566, O14575, Q9NZW1, Q9UNI7 | Gene names | SLC25A13, ARALAR2 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding mitochondrial carrier protein Aralar2 (Mitochondrial aspartate glutamate carrier 2) (Solute carrier family 25 member 13) (Citrin). | |||||
|
CMC2_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.995522 (rank : 3) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q9QXX4, Q9CZF6, Q9DCF5 | Gene names | Slc25a13, Aralar2 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding mitochondrial carrier protein Aralar2 (Mitochondrial aspartate glutamate carrier 2) (Solute carrier family 25 member 13) (Citrin). | |||||
|
GHC2_HUMAN
|
||||||
| θ value | 3.48141e-52 (rank : 5) | NC score | 0.934136 (rank : 5) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9H1K4 | Gene names | SLC25A18, GC2 | |||
|
Domain Architecture |
|
|||||
| Description | Mitochondrial glutamate carrier 2 (GC-2) (Glutamate/H(+) symporter 2) (Solute carrier family 25 member 18). | |||||
|
GHC1_HUMAN
|
||||||
| θ value | 3.84914e-51 (rank : 6) | NC score | 0.931191 (rank : 6) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9H936, Q8TBU8 | Gene names | SLC25A22, GC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial glutamate carrier 1 (GC-1) (Glutamate/H(+) symporter 1) (Solute carrier family 25 member 22). | |||||
|
GHC1_MOUSE
|
||||||
| θ value | 5.02714e-51 (rank : 7) | NC score | 0.930726 (rank : 7) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9D6M3, Q3TRY1 | Gene names | Slc25a22, Gc1 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial glutamate carrier 1 (GC-1) (Glutamate/H(+) symporter 1) (Solute carrier family 25 member 22). | |||||
|
MCAT_HUMAN
|
||||||
| θ value | 6.61148e-27 (rank : 8) | NC score | 0.865473 (rank : 11) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O43772, Q9UIQ2 | Gene names | SLC25A20, CAC, CACT | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial carnitine/acylcarnitine carrier protein (Carnitine/acylcarnitine translocase) (CAC) (Solute carrier family 25 member 20). | |||||
|
ODC_MOUSE
|
||||||
| θ value | 1.12775e-26 (rank : 9) | NC score | 0.879231 (rank : 8) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8BZ09, Q8BY72 | Gene names | Slc25a21, Odc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial 2-oxodicarboxylate carrier (Solute carrier family 25 member 21). | |||||
|
MCAT_MOUSE
|
||||||
| θ value | 2.12685e-25 (rank : 10) | NC score | 0.859140 (rank : 14) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9Z2Z6, Q3U801 | Gene names | Slc25a20, Cac | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial carnitine/acylcarnitine carrier protein (Carnitine/acylcarnitine translocase) (CAC) (mCAC) (Solute carrier family 25 member 20). | |||||
|
ODC_HUMAN
|
||||||
| θ value | 2.12685e-25 (rank : 11) | NC score | 0.877391 (rank : 9) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9BQT8 | Gene names | SLC25A21, ODC | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial 2-oxodicarboxylate carrier (Solute carrier family 25 member 21). | |||||
|
MCATL_HUMAN
|
||||||
| θ value | 5.23862e-24 (rank : 12) | NC score | 0.863439 (rank : 12) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8N8R3 | Gene names | SLC25A29, C14orf69 | |||
|
Domain Architecture |
|
|||||
| Description | Mitochondrial carnitine/acylcarnitine carrier protein CACL (CACT-like) (Solute carrier family 25 member 29). | |||||
|
MCATL_MOUSE
|
||||||
| θ value | 4.43474e-23 (rank : 13) | NC score | 0.862791 (rank : 13) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8BL03, Q922X4 | Gene names | Slc25a29 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial carnitine/acylcarnitine carrier protein CACL (CACT-like) (Solute carrier family 25 member 29). | |||||
|
S2544_MOUSE
|
||||||
| θ value | 2.20094e-22 (rank : 14) | NC score | 0.852904 (rank : 15) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8BGF9, Q7TSU9 | Gene names | Slc25a44 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Solute carrier family 25 member 44. | |||||
|
UCP5_HUMAN
|
||||||
| θ value | 6.40375e-22 (rank : 15) | NC score | 0.821353 (rank : 21) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O95258, Q9HC60, Q9HC61 | Gene names | SLC25A14, BMCP1, UCP5 | |||
|
Domain Architecture |
|
|||||
| Description | Brain mitochondrial carrier protein 1 (BMCP-1) (Mitochondrial uncoupling protein 5) (UCP 5) (Solute carrier family 25 member 14). | |||||
|
S2544_HUMAN
|
||||||
| θ value | 8.36355e-22 (rank : 16) | NC score | 0.849376 (rank : 16) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q96H78, O75034 | Gene names | SLC25A44, KIAA0446 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Solute carrier family 25 member 44. | |||||
|
UCP5_MOUSE
|
||||||
| θ value | 8.36355e-22 (rank : 17) | NC score | 0.821606 (rank : 20) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9Z2B2, Q9ESI8 | Gene names | Slc25a14, Bmcp1, Ucp5 | |||
|
Domain Architecture |
|
|||||
| Description | Brain mitochondrial carrier protein 1 (BMCP-1) (Mitochondrial uncoupling protein 5) (UCP 5) (Solute carrier family 25 member 14). | |||||
|
M2OM_MOUSE
|
||||||
| θ value | 4.15078e-21 (rank : 18) | NC score | 0.796856 (rank : 29) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9CR62 | Gene names | Slc25a11 | |||
|
Domain Architecture |
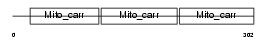
|
|||||
| Description | Mitochondrial 2-oxoglutarate/malate carrier protein (OGCP) (Solute carrier family 25 member 11). | |||||
|
ADT1_HUMAN
|
||||||
| θ value | 5.42112e-21 (rank : 19) | NC score | 0.791189 (rank : 33) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P12235 | Gene names | SLC25A4, ANT1 | |||
|
Domain Architecture |
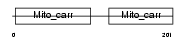
|
|||||
| Description | ADP/ATP translocase 1 (Adenine nucleotide translocator 1) (ANT 1) (ADP,ATP carrier protein 1) (Solute carrier family 25 member 4) (ADP,ATP carrier protein, heart/skeletal muscle isoform T1). | |||||
|
ADT1_MOUSE
|
||||||
| θ value | 5.42112e-21 (rank : 20) | NC score | 0.793965 (rank : 32) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P48962, Q62164 | Gene names | Slc25a4, Anc1, Ant1 | |||
|
Domain Architecture |
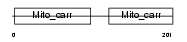
|
|||||
| Description | ADP/ATP translocase 1 (Adenine nucleotide translocator 1) (ANT 1) (ADP,ATP carrier protein 1) (Solute carrier family 25 member 4) (ADP,ATP carrier protein, heart/skeletal muscle isoform T1) (mANC1). | |||||
|
M2OM_HUMAN
|
||||||
| θ value | 4.58923e-20 (rank : 21) | NC score | 0.795477 (rank : 31) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q02978, O75537, Q969P7 | Gene names | SLC25A11, SLC20A4 | |||
|
Domain Architecture |
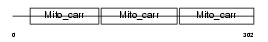
|
|||||
| Description | Mitochondrial 2-oxoglutarate/malate carrier protein (OGCP) (Solute carrier family 25 member 11). | |||||
|
ADT3_HUMAN
|
||||||
| θ value | 1.02238e-19 (rank : 22) | NC score | 0.790961 (rank : 34) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P12236, Q96C49 | Gene names | SLC25A6, ANT3 | |||
|
Domain Architecture |

|
|||||
| Description | ADP/ATP translocase 3 (Adenine nucleotide translocator 2) (ANT 3) (ADP,ATP carrier protein 3) (Solute carrier family 25 member 6) (ADP,ATP carrier protein, isoform T2). | |||||
|
TXTP_HUMAN
|
||||||
| θ value | 1.02238e-19 (rank : 23) | NC score | 0.873065 (rank : 10) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P53007, Q9BSK6 | Gene names | SLC25A1, SLC20A3 | |||
|
Domain Architecture |
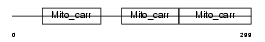
|
|||||
| Description | Tricarboxylate transport protein, mitochondrial precursor (Citrate transport protein) (CTP) (Tricarboxylate carrier protein) (Solute carrier family 25 member 1). | |||||
|
UCP1_HUMAN
|
||||||
| θ value | 2.97466e-19 (rank : 24) | NC score | 0.786744 (rank : 35) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P25874, Q13218, Q4KMZ3, Q68G66 | Gene names | UCP1, SLC25A7, UCP | |||
|
Domain Architecture |
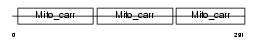
|
|||||
| Description | Mitochondrial brown fat uncoupling protein 1 (UCP 1) (Thermogenin). | |||||
|
ADT2_MOUSE
|
||||||
| θ value | 8.65492e-19 (rank : 25) | NC score | 0.784481 (rank : 38) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P51881, Q61311 | Gene names | Slc25a5, Ant2 | |||
|
Domain Architecture |
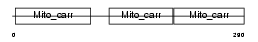
|
|||||
| Description | ADP/ATP translocase 2 (Adenine nucleotide translocator 2) (ANT 2) (ADP,ATP carrier protein 2) (Solute carrier family 25 member 5). | |||||
|
ADT2_HUMAN
|
||||||
| θ value | 1.47631e-18 (rank : 26) | NC score | 0.782586 (rank : 39) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P05141, O43350 | Gene names | SLC25A5, ANT2 | |||
|
Domain Architecture |
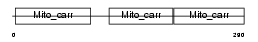
|
|||||
| Description | ADP/ATP translocase 2 (Adenine nucleotide translocator 2) (ANT 2) (ADP,ATP carrier protein 2) (Solute carrier family 25 member 5) (ADP,ATP carrier protein, fibroblast isoform). | |||||
|
DNC_MOUSE
|
||||||
| θ value | 2.5182e-18 (rank : 27) | NC score | 0.829824 (rank : 17) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9DAM5, Q3T9C4 | Gene names | Slc25a19 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial deoxynucleotide carrier (Solute carrier family 25 member 19). | |||||
|
ORNT1_MOUSE
|
||||||
| θ value | 3.28887e-18 (rank : 28) | NC score | 0.803647 (rank : 26) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9WVD5 | Gene names | Slc25a15, Ornt1 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial ornithine transporter 1 (Solute carrier family 25 member 15). | |||||
|
UCP2_HUMAN
|
||||||
| θ value | 4.29542e-18 (rank : 29) | NC score | 0.786256 (rank : 36) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P55851, Q4PJH8 | Gene names | UCP2, SLC25A8 | |||
|
Domain Architecture |
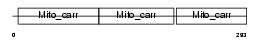
|
|||||
| Description | Mitochondrial uncoupling protein 2 (UCP 2) (UCPH). | |||||
|
UCP2_MOUSE
|
||||||
| θ value | 4.29542e-18 (rank : 30) | NC score | 0.785883 (rank : 37) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P70406, O88285 | Gene names | Ucp2 | |||
|
Domain Architecture |
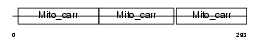
|
|||||
| Description | Mitochondrial uncoupling protein 2 (UCP 2) (UCPH). | |||||
|
ORNT2_HUMAN
|
||||||
| θ value | 1.24977e-17 (rank : 31) | NC score | 0.795974 (rank : 30) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9BXI2, Q8NFZ2 | Gene names | SLC25A2, ORNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial ornithine transporter 2 (Solute carrier family 25 member 2). | |||||
|
ORNT1_HUMAN
|
||||||
| θ value | 2.13179e-17 (rank : 32) | NC score | 0.801299 (rank : 27) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9Y619, Q9HC45 | Gene names | SLC25A15, ORNT1 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial ornithine transporter 1 (Solute carrier family 25 member 15). | |||||
|
GDC_MOUSE
|
||||||
| θ value | 3.63628e-17 (rank : 33) | NC score | 0.826424 (rank : 18) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8C0K5, Q6P6K8 | Gene names | Slc25a16, Gda | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Grave disease carrier protein homolog (GDC) (Mitochondrial solute carrier protein homolog) (Solute carrier family 25 member 16). | |||||
|
GDC_HUMAN
|
||||||
| θ value | 6.20254e-17 (rank : 34) | NC score | 0.821916 (rank : 19) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P16260 | Gene names | SLC25A16, GDA | |||
|
Domain Architecture |
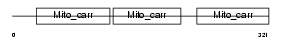
|
|||||
| Description | Grave disease carrier protein (GDC) (Grave disease autoantigen) (GDA) (Mitochondrial solute carrier protein homolog) (Solute carrier family 25 member 16). | |||||
|
MFTC_MOUSE
|
||||||
| θ value | 8.10077e-17 (rank : 35) | NC score | 0.805607 (rank : 25) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8BMG8, Q3TCM5 | Gene names | Slc25a32, Mftc | |||
|
Domain Architecture |
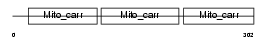
|
|||||
| Description | Mitochondrial folate transporter/carrier (Solute carrier family 25 member 32). | |||||
|
S2539_MOUSE
|
||||||
| θ value | 8.10077e-17 (rank : 36) | NC score | 0.818106 (rank : 22) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9D8K8, Q3TTM8 | Gene names | Slc25a39, D11Ertd333e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Solute carrier family 25 member 39. | |||||
|
MFRN1_MOUSE
|
||||||
| θ value | 1.058e-16 (rank : 37) | NC score | 0.776058 (rank : 43) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q920G8, Q8C367, Q8CEJ7, Q91ZY0, Q9D2G3, Q9D547 | Gene names | Slc25a37, Mfrn, Mscp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitoferrin-1 (Mitochondrial iron transporter 1) (Solute carrier family 25 member 37) (Mitochondrial solute carrier protein). | |||||
|
MFRN2_HUMAN
|
||||||
| θ value | 1.80466e-16 (rank : 38) | NC score | 0.781487 (rank : 40) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q96A46, Q4VBZ0, Q5T777, Q86VX5, Q969G8, Q9H2J3 | Gene names | SLC25A28, MFRN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitoferrin-2 (Mitochondrial iron transporter 2) (Solute carrier family 25 member 28) (Mitochondrial RNA-splicing protein 3/4 homolog) (MRS3/4) (hMRS3/4). | |||||
|
MFRN2_MOUSE
|
||||||
| θ value | 3.07829e-16 (rank : 39) | NC score | 0.779923 (rank : 42) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8R0Z5, Q8K4J8, Q8K4J9, Q8R578 | Gene names | Slc25a28, Mfrn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitoferrin-2 (Mitochondrial iron transporter 2) (Solute carrier family 25 member 28) (Mitochondrial RNA-splicing protein 3/4 homolog) (MRS3/4). | |||||
|
S2539_HUMAN
|
||||||
| θ value | 6.85773e-16 (rank : 40) | NC score | 0.809537 (rank : 24) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9BZJ4, Q9P182, Q9UF66, Q9Y379 | Gene names | SLC25A39 | |||
|
Domain Architecture |
|
|||||
| Description | Solute carrier family 25 member 39. | |||||
|
UCP3_HUMAN
|
||||||
| θ value | 1.16975e-15 (rank : 41) | NC score | 0.774360 (rank : 44) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P55916, O60475, Q96HL3 | Gene names | UCP3, SLC25A9 | |||
|
Domain Architecture |
|
|||||
| Description | Mitochondrial uncoupling protein 3 (UCP 3). | |||||
|
MFTC_HUMAN
|
||||||
| θ value | 1.99529e-15 (rank : 42) | NC score | 0.799025 (rank : 28) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9H2D1, Q96JZ6, Q96SU7 | Gene names | SLC25A32, MFT, MFTC | |||
|
Domain Architecture |
|
|||||
| Description | Mitochondrial folate transporter/carrier (Solute carrier family 25 member 32). | |||||
|
DNC_HUMAN
|
||||||
| θ value | 2.60593e-15 (rank : 43) | NC score | 0.815998 (rank : 23) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9HC21 | Gene names | SLC25A19, DNC, MUP1 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial deoxynucleotide carrier (Solute carrier family 25 member 19) (Mitochondrial uncoupling protein 1). | |||||
|
UCP1_MOUSE
|
||||||
| θ value | 5.8054e-15 (rank : 44) | NC score | 0.758501 (rank : 47) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P12242 | Gene names | Ucp1, Ucp | |||
|
Domain Architecture |
|
|||||
| Description | Mitochondrial brown fat uncoupling protein 1 (UCP 1) (Thermogenin). | |||||
|
UCP3_MOUSE
|
||||||
| θ value | 1.09485e-13 (rank : 45) | NC score | 0.768707 (rank : 45) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P56501, O88293 | Gene names | Ucp3 | |||
|
Domain Architecture |
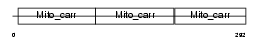
|
|||||
| Description | Mitochondrial uncoupling protein 3 (UCP 3). | |||||
|
DIC_MOUSE
|
||||||
| θ value | 1.42992e-13 (rank : 46) | NC score | 0.747175 (rank : 50) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9QZD8, Q543J6, Q99LJ9, Q9D8G8 | Gene names | Slc25a10, Dic | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial dicarboxylate carrier (Solute carrier family 25 member 10). | |||||
|
UCP4_HUMAN
|
||||||
| θ value | 1.86753e-13 (rank : 47) | NC score | 0.780186 (rank : 41) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O95847 | Gene names | SLC25A27, UCP4 | |||
|
Domain Architecture |
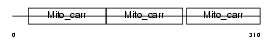
|
|||||
| Description | Mitochondrial uncoupling protein 4 (UCP 4) (Solute carrier family 25 member 27). | |||||
|
MFRN1_HUMAN
|
||||||
| θ value | 4.16044e-13 (rank : 48) | NC score | 0.764496 (rank : 46) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9NYZ2, Q53FT7, Q69YJ8, Q969S1, Q9P0J2 | Gene names | SLC25A37, MFRN, MSCP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitoferrin-1 (Mitochondrial iron transporter 1) (Solute carrier family 25 member 37) (Mitochondrial solute carrier protein). | |||||
|
DIC_HUMAN
|
||||||
| θ value | 2.0648e-12 (rank : 49) | NC score | 0.727085 (rank : 53) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9UBX3, Q96BA1, Q96IP1 | Gene names | SLC25A10, DIC | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial dicarboxylate carrier (Solute carrier family 25 member 10). | |||||
|
MPCP_MOUSE
|
||||||
| θ value | 1.25267e-09 (rank : 50) | NC score | 0.757020 (rank : 48) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8VEM8, Q542V7 | Gene names | Slc25a3 | |||
|
Domain Architecture |
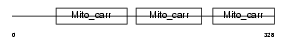
|
|||||
| Description | Phosphate carrier protein, mitochondrial precursor (PTP) (Solute carrier family 25 member 3). | |||||
|
EFHA1_HUMAN
|
||||||
| θ value | 2.79066e-09 (rank : 51) | NC score | 0.264993 (rank : 56) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8IYU8, Q8N0T6, Q8NAX8 | Gene names | EFHA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand domain-containing family member A1. | |||||
|
MPCP_HUMAN
|
||||||
| θ value | 2.79066e-09 (rank : 52) | NC score | 0.754665 (rank : 49) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q00325, Q96A03 | Gene names | SLC25A3, PHC | |||
|
Domain Architecture |
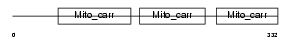
|
|||||
| Description | Phosphate carrier protein, mitochondrial precursor (PTP) (Solute carrier family 25 member 3). | |||||
|
PM34_HUMAN
|
||||||
| θ value | 8.11959e-09 (rank : 53) | NC score | 0.730227 (rank : 52) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O43808, Q5TFL0, Q9UGW8, Q9UGY7 | Gene names | SLC25A17, PMP34 | |||
|
Domain Architecture |
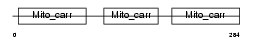
|
|||||
| Description | Peroxisomal membrane protein PMP34 (34 kDa peroxisomal membrane protein) (Solute carrier family 25 member 17). | |||||
|
EFHA1_MOUSE
|
||||||
| θ value | 2.36244e-08 (rank : 54) | NC score | 0.254881 (rank : 57) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8CD10, Q3TJU4, Q8K0K2, Q9CUR8 | Gene names | Efha1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand domain-containing family member A1. | |||||
|
PM34_MOUSE
|
||||||
| θ value | 1.99992e-07 (rank : 55) | NC score | 0.730486 (rank : 51) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O70579 | Gene names | Slc25a17, Pmp34, Pmp35 | |||
|
Domain Architecture |
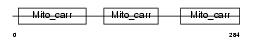
|
|||||
| Description | Peroxisomal membrane protein PMP34 (34 kDa peroxisomal membrane protein) (Solute carrier family 25 member 17). | |||||
|
MCAR1_HUMAN
|
||||||
| θ value | 4.1701e-05 (rank : 56) | NC score | 0.553978 (rank : 55) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9H1U9 | Gene names | MCART1 | |||
|
Domain Architecture |
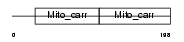
|
|||||
| Description | Mitochondrial carrier triple repeat 1. | |||||
|
EFHA2_MOUSE
|
||||||
| θ value | 9.29e-05 (rank : 57) | NC score | 0.192630 (rank : 59) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9CTY5 | Gene names | Efha2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand domain-containing family member A2. | |||||
|
EFHA2_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 58) | NC score | 0.192086 (rank : 60) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q86XE3, Q8IYZ3 | Gene names | EFHA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand domain-containing family member A2. | |||||
|
MCAR1_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 59) | NC score | 0.570348 (rank : 54) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q5HZI9 | Gene names | Mcart1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial carrier triple repeat 1. | |||||
|
AYT1B_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 60) | NC score | 0.088727 (rank : 64) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9D5U0 | Gene names | Aytl1b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acyltransferase-like 1-B (EC 2.3.1.-). | |||||
|
AYTL1_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 61) | NC score | 0.087383 (rank : 65) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q7L5N7, Q6MZJ6, Q9NX23 | Gene names | AYTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acyltransferase-like 1 (EC 2.3.1.-). | |||||
|
AYT1A_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 62) | NC score | 0.084981 (rank : 66) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8BYI6 | Gene names | Aytl1a, Aytl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acyltransferase-like 1-A (EC 2.3.1.-). | |||||
|
MTCH2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 63) | NC score | 0.220742 (rank : 58) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9Y6C9 | Gene names | MTCH2, MIMP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial carrier homolog 2 (Met-induced mitochondrial protein). | |||||
|
CALL5_HUMAN
|
||||||
| θ value | 0.279714 (rank : 64) | NC score | 0.060661 (rank : 67) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9NZT1, Q8IXU8 | Gene names | CALML5, CLSP | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-like protein 5 (Calmodulin-like skin protein). | |||||
|
TRHY_HUMAN
|
||||||
| θ value | 0.365318 (rank : 65) | NC score | 0.005389 (rank : 101) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
MYL6_HUMAN
|
||||||
| θ value | 0.47712 (rank : 66) | NC score | 0.033958 (rank : 90) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P60660, P16475, P24572, P24573, Q12790, Q6IAZ0, Q6IPY5 | Gene names | MYL6 | |||
|
Domain Architecture |
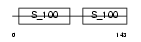
|
|||||
| Description | Myosin light polypeptide 6 (Smooth muscle and nonmuscle myosin light chain alkali 6) (Myosin light chain alkali 3) (Myosin light chain 3) (MLC-3) (LC17). | |||||
|
MYL6_MOUSE
|
||||||
| θ value | 0.47712 (rank : 67) | NC score | 0.033958 (rank : 91) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q60605 | Gene names | Myl6, Myln | |||
|
Domain Architecture |
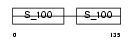
|
|||||
| Description | Myosin light polypeptide 6 (Smooth muscle and nonmuscle myosin light chain alkali 6) (Myosin light chain alkali 3) (Myosin light chain 3) (MLC-3) (LC17). | |||||
|
CALL3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 68) | NC score | 0.039142 (rank : 79) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P27482 | Gene names | CALML3 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-like protein 3 (Calmodulin-related protein NB-1) (CaM-like protein) (CLP). | |||||
|
CALM_HUMAN
|
||||||
| θ value | 1.06291 (rank : 69) | NC score | 0.034444 (rank : 88) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P62158, P02593, P70667, P99014, Q13942, Q53S29, Q61379, Q61380, Q96HK3 | Gene names | CALM1, CALM, CAM, CAM1 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin (CaM). | |||||
|
CALM_MOUSE
|
||||||
| θ value | 1.06291 (rank : 70) | NC score | 0.034444 (rank : 89) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P62204, P02593, P70667, P99014, Q61379, Q61380 | Gene names | Calm1, Calm, Cam, Cam1 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin (CaM). | |||||
|
CETN1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 71) | NC score | 0.045639 (rank : 72) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P41209, Q3V119, Q9D9G9, Q9DAL6 | Gene names | Cetn1, Calt | |||
|
Domain Architecture |
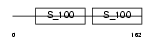
|
|||||
| Description | Centrin-1 (Caltractin). | |||||
|
RECO_MOUSE
|
||||||
| θ value | 1.38821 (rank : 72) | NC score | 0.047772 (rank : 69) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P34057 | Gene names | Rcvrn, Rcv1 | |||
|
Domain Architecture |
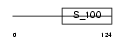
|
|||||
| Description | Recoverin (Cancer-associated retinopathy protein) (Protein CAR) (23 kDa photoreceptor cell-specific protein). | |||||
|
OLF19_MOUSE
|
||||||
| θ value | 1.81305 (rank : 73) | NC score | -0.003775 (rank : 102) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 798 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JHB2, Q9JM32 | Gene names | Olfr19, Mor140-1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Olfactory receptor 19 (Olfactory receptor 140-1) (Odorant receptor M12). | |||||
|
MTCH1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 74) | NC score | 0.185373 (rank : 63) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9NZJ7, Q6PK60, Q6UX45, Q7L465, Q9BW23, Q9NZR6, Q9UJZ5 | Gene names | MTCH1, PSAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial carrier homolog 1 (Presenilin-associated protein). | |||||
|
MYL6B_HUMAN
|
||||||
| θ value | 2.36792 (rank : 75) | NC score | 0.027513 (rank : 92) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P14649 | Gene names | MYL6B, MLC1SA | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light polypeptide 6B (Smooth muscle and nonmuscle myosin light chain alkali 6B) (Myosin light chain 1 slow-twitch muscle A isoform) (MLC1sa). | |||||
|
NCS1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 76) | NC score | 0.042522 (rank : 75) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P62166, P36610, Q9UK26 | Gene names | FREQ, FLUP, NCS1 | |||
|
Domain Architecture |
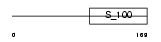
|
|||||
| Description | Neuronal calcium sensor 1 (NCS-1) (Frequenin homolog) (Frequenin-like protein) (Frequenin-like ubiquitous protein). | |||||
|
NCS1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 77) | NC score | 0.042522 (rank : 76) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8BNY6 | Gene names | Freq, Ncs1 | |||
|
Domain Architecture |
|
|||||
| Description | Neuronal calcium sensor 1 (NCS-1) (Frequenin homolog). | |||||
|
DYH11_HUMAN
|
||||||
| θ value | 3.0926 (rank : 78) | NC score | 0.008748 (rank : 98) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96DT5, Q9UJ82 | Gene names | DNAH11 | |||
|
Domain Architecture |

|
|||||
| Description | Ciliary dynein heavy chain 11 (Axonemal beta dynein heavy chain 11). | |||||
|
HPCA_HUMAN
|
||||||
| θ value | 3.0926 (rank : 79) | NC score | 0.047309 (rank : 70) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P84074, P32076, P41211, P70510 | Gene names | HPCA, BDR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuron-specific calcium-binding protein hippocalcin (Calcium-binding protein BDR-2). | |||||
|
HPCA_MOUSE
|
||||||
| θ value | 3.0926 (rank : 80) | NC score | 0.047309 (rank : 71) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P84075, P32076, P41211, P70510 | Gene names | Hpca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuron-specific calcium-binding protein hippocalcin. | |||||
|
HPCL4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 81) | NC score | 0.038125 (rank : 81) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9UM19, Q5TG97, Q8N611 | Gene names | HPCAL4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hippocalcin-like protein 4 (HLP4). | |||||
|
HPCL4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 82) | NC score | 0.038126 (rank : 80) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8BGZ1 | Gene names | Hpcal4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hippocalcin-like protein 4 (Neural visinin-like protein 2) (NVP-2). | |||||
|
KCIP2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 83) | NC score | 0.034935 (rank : 87) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9NS61, Q7Z6F1, Q96K86, Q96T41, Q96T42, Q96T43, Q96T44, Q9H0N4, Q9HD10, Q9HD11, Q9NS60, Q9NY10, Q9NZI1 | Gene names | KCNIP2, KCHIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kv channel-interacting protein 2 (KChIP2) (A-type potassium channel modulatory protein 2) (Potassium channel-interacting protein 2) (Cardiac voltage-gated potassium channel modulatory subunit). | |||||
|
KCIP2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 84) | NC score | 0.035064 (rank : 86) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9JJ69, Q6DTJ2, Q8K1T8, Q8K1T9, Q8K1U0, Q8VHN4, Q8VHN5, Q8VHN6, Q9JJ68 | Gene names | Kcnip2, Kchip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kv channel-interacting protein 2 (KChIP2) (A-type potassium channel modulatory protein 2) (Potassium channel-interacting protein 2). | |||||
|
MTCH2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 85) | NC score | 0.191239 (rank : 61) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q791V5, Q3TPS5, Q99LZ6, Q9D060, Q9D7Y2, Q9QZP3 | Gene names | Mtch2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial carrier homolog 2. | |||||
|
VISL1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 86) | NC score | 0.037929 (rank : 82) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P62760, P28677, P29103, P42323, Q9UM20 | Gene names | VSNL1, VISL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Visinin-like protein 1 (VILIP) (Hippocalcin-like protein 3) (HLP3). | |||||
|
VISL1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 87) | NC score | 0.037929 (rank : 83) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P62761, P28677, P29103, P42323, Q9UM20 | Gene names | Vsnl1, Visl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Visinin-like protein 1 (VILIP) (Neural visinin-like protein 1) (NVL-1) (NVP-1). | |||||
|
CABP7_HUMAN
|
||||||
| θ value | 4.03905 (rank : 88) | NC score | 0.044168 (rank : 73) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q86V35 | Gene names | CABP7 | |||
|
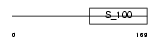
Domain Architecture |
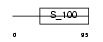
|
|||||
| Description | Calcium-binding protein 7 (CaBP7). | |||||
|
CABP7_MOUSE
|
||||||
| θ value | 4.03905 (rank : 89) | NC score | 0.044168 (rank : 74) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q91ZM8 | Gene names | Cabp7 | |||
|
Domain Architecture |
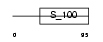
|
|||||
| Description | Calcium-binding protein 7 (CaBP7). | |||||
|
CANB2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 90) | NC score | 0.037920 (rank : 84) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q63811 | Gene names | Ppp3r2 | |||
|
Domain Architecture |
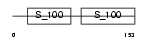
|
|||||
| Description | Calcineurin subunit B isoform 2 (Protein phosphatase 2B regulatory subunit 2) (Protein phosphatase 3 regulatory subunit B beta isoform). | |||||
|
CETN1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 91) | NC score | 0.040623 (rank : 77) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q12798 | Gene names | CETN1, CEN1, CETN | |||
|
Domain Architecture |
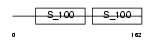
|
|||||
| Description | Centrin-1 (Caltractin isoform 2). | |||||
|
MTCH1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 92) | NC score | 0.185975 (rank : 62) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q791T5, Q3U4W7, Q791V6, Q8R0T0, Q8R1T8, Q8R2T2, Q9QZA5, Q9QZP4 | Gene names | Mtch1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial carrier homolog 1 (Mitochondrial carrier-like protein 1). | |||||
|
PLSI_HUMAN
|
||||||
| θ value | 4.03905 (rank : 93) | NC score | 0.024452 (rank : 96) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q14651 | Gene names | PLS1 | |||
|
Domain Architecture |

|
|||||
| Description | Plastin-1 (I-plastin) (Intestine-specific plastin). | |||||
|
ZMY15_HUMAN
|
||||||
| θ value | 4.03905 (rank : 94) | NC score | 0.020194 (rank : 97) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H091 | Gene names | ZMYND15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYND domain-containing protein 15. | |||||
|
GUC1C_HUMAN
|
||||||
| θ value | 5.27518 (rank : 95) | NC score | 0.051657 (rank : 68) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O95843, O95844, Q9UNM0 | Gene names | GUCA1C, GCAP3 | |||
|
Domain Architecture |
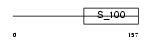
|
|||||
| Description | Guanylyl cyclase-activating protein 3 (GCAP 3) (Guanylate cyclase activator 1C). | |||||
|
CABP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 96) | NC score | 0.025685 (rank : 95) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9NZU7, O95663, Q9NZU8 | Gene names | CABP1 | |||
|
Domain Architecture |
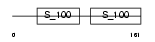
|
|||||
| Description | Calcium-binding protein 1 (CaBP1) (Calbrain). | |||||
|
CABP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 97) | NC score | 0.025690 (rank : 94) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9JLK7, Q9JLK6 | Gene names | Cabp1 | |||
|
Domain Architecture |
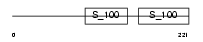
|
|||||
| Description | Calcium-binding protein 1 (CaBP1). | |||||
|
HPCL1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 98) | NC score | 0.037725 (rank : 85) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P62748, P35333 | Gene names | Hpcal1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hippocalcin-like protein 1 (Visinin-like protein 3) (VILIP-3) (Neural visinin-like protein 3) (NVL-3) (NVP-3). | |||||
|
PIWL1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 99) | NC score | 0.007966 (rank : 99) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JMB7 | Gene names | Piwil1, Miwi | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Piwi-like protein 1. | |||||
|
S100G_HUMAN
|
||||||
| θ value | 6.88961 (rank : 100) | NC score | 0.039659 (rank : 78) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P29377 | Gene names | S100G, CABP9K, CALB3, S100D | |||
|
Domain Architecture |
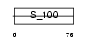
|
|||||
| Description | Protein S100-G (S100 calcium-binding protein G) (Vitamin D-dependent calcium-binding protein, intestinal) (CABP) (Calbindin D9K). | |||||
|
DUOX1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 101) | NC score | 0.025963 (rank : 93) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9NRD9, Q6ZMB3, Q6ZR09, Q9NZC1 | Gene names | DUOX1, DUOX, LNOX1, THOX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual oxidase 1 precursor (EC 1.6.3.1) (EC 1.11.1.-) (NADPH thyroid oxidase 1) (Thyroid oxidase 1) (Large NOX 1) (Long NOX 1). | |||||
|
PIWL1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 102) | NC score | 0.007111 (rank : 100) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96J94, O95404, Q8NA60, Q8TBY5, Q96JD5 | Gene names | PIWIL1, HIWI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Piwi-like protein 1. | |||||
|
CMC1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 102 | |
| SwissProt Accessions | Q8BH59 | Gene names | Slc25a12, Aralar1 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding mitochondrial carrier protein Aralar1 (Mitochondrial aspartate glutamate carrier 1) (Solute carrier family 25 member 12). | |||||
|
CMC1_HUMAN
|
||||||
| NC score | 0.999192 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 102 | |
| SwissProt Accessions | O75746 | Gene names | SLC25A12, ARALAR1 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding mitochondrial carrier protein Aralar1 (Mitochondrial aspartate glutamate carrier 1) (Solute carrier family 25 member 12). | |||||
|
CMC2_MOUSE
|
||||||
| NC score | 0.995522 (rank : 3) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q9QXX4, Q9CZF6, Q9DCF5 | Gene names | Slc25a13, Aralar2 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding mitochondrial carrier protein Aralar2 (Mitochondrial aspartate glutamate carrier 2) (Solute carrier family 25 member 13) (Citrin). | |||||
|
CMC2_HUMAN
|
||||||
| NC score | 0.995516 (rank : 4) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q9UJS0, O14566, O14575, Q9NZW1, Q9UNI7 | Gene names | SLC25A13, ARALAR2 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding mitochondrial carrier protein Aralar2 (Mitochondrial aspartate glutamate carrier 2) (Solute carrier family 25 member 13) (Citrin). | |||||
|
GHC2_HUMAN
|
||||||
| NC score | 0.934136 (rank : 5) | θ value | 3.48141e-52 (rank : 5) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9H1K4 | Gene names | SLC25A18, GC2 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial glutamate carrier 2 (GC-2) (Glutamate/H(+) symporter 2) (Solute carrier family 25 member 18). | |||||
|
GHC1_HUMAN
|
||||||
| NC score | 0.931191 (rank : 6) | θ value | 3.84914e-51 (rank : 6) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9H936, Q8TBU8 | Gene names | SLC25A22, GC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial glutamate carrier 1 (GC-1) (Glutamate/H(+) symporter 1) (Solute carrier family 25 member 22). | |||||
|
GHC1_MOUSE
|
||||||
| NC score | 0.930726 (rank : 7) | θ value | 5.02714e-51 (rank : 7) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9D6M3, Q3TRY1 | Gene names | Slc25a22, Gc1 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial glutamate carrier 1 (GC-1) (Glutamate/H(+) symporter 1) (Solute carrier family 25 member 22). | |||||
|
ODC_MOUSE
|
||||||
| NC score | 0.879231 (rank : 8) | θ value | 1.12775e-26 (rank : 9) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8BZ09, Q8BY72 | Gene names | Slc25a21, Odc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial 2-oxodicarboxylate carrier (Solute carrier family 25 member 21). | |||||
|
ODC_HUMAN
|
||||||
| NC score | 0.877391 (rank : 9) | θ value | 2.12685e-25 (rank : 11) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9BQT8 | Gene names | SLC25A21, ODC | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial 2-oxodicarboxylate carrier (Solute carrier family 25 member 21). | |||||
|
TXTP_HUMAN
|
||||||
| NC score | 0.873065 (rank : 10) | θ value | 1.02238e-19 (rank : 23) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P53007, Q9BSK6 | Gene names | SLC25A1, SLC20A3 | |||
|
Domain Architecture |

|
|||||
| Description | Tricarboxylate transport protein, mitochondrial precursor (Citrate transport protein) (CTP) (Tricarboxylate carrier protein) (Solute carrier family 25 member 1). | |||||
|
MCAT_HUMAN
|
||||||
| NC score | 0.865473 (rank : 11) | θ value | 6.61148e-27 (rank : 8) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O43772, Q9UIQ2 | Gene names | SLC25A20, CAC, CACT | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial carnitine/acylcarnitine carrier protein (Carnitine/acylcarnitine translocase) (CAC) (Solute carrier family 25 member 20). | |||||
|
MCATL_HUMAN
|
||||||
| NC score | 0.863439 (rank : 12) | θ value | 5.23862e-24 (rank : 12) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8N8R3 | Gene names | SLC25A29, C14orf69 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial carnitine/acylcarnitine carrier protein CACL (CACT-like) (Solute carrier family 25 member 29). | |||||
|
MCATL_MOUSE
|
||||||
| NC score | 0.862791 (rank : 13) | θ value | 4.43474e-23 (rank : 13) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8BL03, Q922X4 | Gene names | Slc25a29 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial carnitine/acylcarnitine carrier protein CACL (CACT-like) (Solute carrier family 25 member 29). | |||||
|
MCAT_MOUSE
|
||||||
| NC score | 0.859140 (rank : 14) | θ value | 2.12685e-25 (rank : 10) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9Z2Z6, Q3U801 | Gene names | Slc25a20, Cac | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial carnitine/acylcarnitine carrier protein (Carnitine/acylcarnitine translocase) (CAC) (mCAC) (Solute carrier family 25 member 20). | |||||
|
S2544_MOUSE
|
||||||
| NC score | 0.852904 (rank : 15) | θ value | 2.20094e-22 (rank : 14) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8BGF9, Q7TSU9 | Gene names | Slc25a44 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Solute carrier family 25 member 44. | |||||
|
S2544_HUMAN
|
||||||
| NC score | 0.849376 (rank : 16) | θ value | 8.36355e-22 (rank : 16) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q96H78, O75034 | Gene names | SLC25A44, KIAA0446 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Solute carrier family 25 member 44. | |||||
|
DNC_MOUSE
|
||||||
| NC score | 0.829824 (rank : 17) | θ value | 2.5182e-18 (rank : 27) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9DAM5, Q3T9C4 | Gene names | Slc25a19 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial deoxynucleotide carrier (Solute carrier family 25 member 19). | |||||
|
GDC_MOUSE
|
||||||
| NC score | 0.826424 (rank : 18) | θ value | 3.63628e-17 (rank : 33) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8C0K5, Q6P6K8 | Gene names | Slc25a16, Gda | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Grave disease carrier protein homolog (GDC) (Mitochondrial solute carrier protein homolog) (Solute carrier family 25 member 16). | |||||
|
GDC_HUMAN
|
||||||
| NC score | 0.821916 (rank : 19) | θ value | 6.20254e-17 (rank : 34) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P16260 | Gene names | SLC25A16, GDA | |||
|
Domain Architecture |

|
|||||
| Description | Grave disease carrier protein (GDC) (Grave disease autoantigen) (GDA) (Mitochondrial solute carrier protein homolog) (Solute carrier family 25 member 16). | |||||
|
UCP5_MOUSE
|
||||||
| NC score | 0.821606 (rank : 20) | θ value | 8.36355e-22 (rank : 17) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9Z2B2, Q9ESI8 | Gene names | Slc25a14, Bmcp1, Ucp5 | |||
|
Domain Architecture |

|
|||||
| Description | Brain mitochondrial carrier protein 1 (BMCP-1) (Mitochondrial uncoupling protein 5) (UCP 5) (Solute carrier family 25 member 14). | |||||
|
UCP5_HUMAN
|
||||||
| NC score | 0.821353 (rank : 21) | θ value | 6.40375e-22 (rank : 15) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O95258, Q9HC60, Q9HC61 | Gene names | SLC25A14, BMCP1, UCP5 | |||
|
Domain Architecture |

|
|||||
| Description | Brain mitochondrial carrier protein 1 (BMCP-1) (Mitochondrial uncoupling protein 5) (UCP 5) (Solute carrier family 25 member 14). | |||||
|
S2539_MOUSE
|
||||||
| NC score | 0.818106 (rank : 22) | θ value | 8.10077e-17 (rank : 36) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9D8K8, Q3TTM8 | Gene names | Slc25a39, D11Ertd333e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Solute carrier family 25 member 39. | |||||
|
DNC_HUMAN
|
||||||
| NC score | 0.815998 (rank : 23) | θ value | 2.60593e-15 (rank : 43) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9HC21 | Gene names | SLC25A19, DNC, MUP1 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial deoxynucleotide carrier (Solute carrier family 25 member 19) (Mitochondrial uncoupling protein 1). | |||||
|
S2539_HUMAN
|
||||||
| NC score | 0.809537 (rank : 24) | θ value | 6.85773e-16 (rank : 40) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9BZJ4, Q9P182, Q9UF66, Q9Y379 | Gene names | SLC25A39 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 25 member 39. | |||||
|
MFTC_MOUSE
|
||||||
| NC score | 0.805607 (rank : 25) | θ value | 8.10077e-17 (rank : 35) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8BMG8, Q3TCM5 | Gene names | Slc25a32, Mftc | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial folate transporter/carrier (Solute carrier family 25 member 32). | |||||
|
ORNT1_MOUSE
|
||||||
| NC score | 0.803647 (rank : 26) | θ value | 3.28887e-18 (rank : 28) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9WVD5 | Gene names | Slc25a15, Ornt1 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial ornithine transporter 1 (Solute carrier family 25 member 15). | |||||
|
ORNT1_HUMAN
|
||||||
| NC score | 0.801299 (rank : 27) | θ value | 2.13179e-17 (rank : 32) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9Y619, Q9HC45 | Gene names | SLC25A15, ORNT1 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial ornithine transporter 1 (Solute carrier family 25 member 15). | |||||
|
MFTC_HUMAN
|
||||||
| NC score | 0.799025 (rank : 28) | θ value | 1.99529e-15 (rank : 42) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9H2D1, Q96JZ6, Q96SU7 | Gene names | SLC25A32, MFT, MFTC | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial folate transporter/carrier (Solute carrier family 25 member 32). | |||||
|
M2OM_MOUSE
|
||||||
| NC score | 0.796856 (rank : 29) | θ value | 4.15078e-21 (rank : 18) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9CR62 | Gene names | Slc25a11 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial 2-oxoglutarate/malate carrier protein (OGCP) (Solute carrier family 25 member 11). | |||||
|
ORNT2_HUMAN
|
||||||
| NC score | 0.795974 (rank : 30) | θ value | 1.24977e-17 (rank : 31) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9BXI2, Q8NFZ2 | Gene names | SLC25A2, ORNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial ornithine transporter 2 (Solute carrier family 25 member 2). | |||||
|
M2OM_HUMAN
|
||||||
| NC score | 0.795477 (rank : 31) | θ value | 4.58923e-20 (rank : 21) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q02978, O75537, Q969P7 | Gene names | SLC25A11, SLC20A4 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial 2-oxoglutarate/malate carrier protein (OGCP) (Solute carrier family 25 member 11). | |||||
|
ADT1_MOUSE
|
||||||
| NC score | 0.793965 (rank : 32) | θ value | 5.42112e-21 (rank : 20) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P48962, Q62164 | Gene names | Slc25a4, Anc1, Ant1 | |||
|
Domain Architecture |

|
|||||
| Description | ADP/ATP translocase 1 (Adenine nucleotide translocator 1) (ANT 1) (ADP,ATP carrier protein 1) (Solute carrier family 25 member 4) (ADP,ATP carrier protein, heart/skeletal muscle isoform T1) (mANC1). | |||||
|
ADT1_HUMAN
|
||||||
| NC score | 0.791189 (rank : 33) | θ value | 5.42112e-21 (rank : 19) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P12235 | Gene names | SLC25A4, ANT1 | |||
|
Domain Architecture |

|
|||||
| Description | ADP/ATP translocase 1 (Adenine nucleotide translocator 1) (ANT 1) (ADP,ATP carrier protein 1) (Solute carrier family 25 member 4) (ADP,ATP carrier protein, heart/skeletal muscle isoform T1). | |||||
|
ADT3_HUMAN
|
||||||
| NC score | 0.790961 (rank : 34) | θ value | 1.02238e-19 (rank : 22) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P12236, Q96C49 | Gene names | SLC25A6, ANT3 | |||
|
Domain Architecture |

|
|||||
| Description | ADP/ATP translocase 3 (Adenine nucleotide translocator 2) (ANT 3) (ADP,ATP carrier protein 3) (Solute carrier family 25 member 6) (ADP,ATP carrier protein, isoform T2). | |||||
|
UCP1_HUMAN
|
||||||
| NC score | 0.786744 (rank : 35) | θ value | 2.97466e-19 (rank : 24) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P25874, Q13218, Q4KMZ3, Q68G66 | Gene names | UCP1, SLC25A7, UCP | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial brown fat uncoupling protein 1 (UCP 1) (Thermogenin). | |||||
|
UCP2_HUMAN
|
||||||
| NC score | 0.786256 (rank : 36) | θ value | 4.29542e-18 (rank : 29) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P55851, Q4PJH8 | Gene names | UCP2, SLC25A8 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial uncoupling protein 2 (UCP 2) (UCPH). | |||||
|
UCP2_MOUSE
|
||||||
| NC score | 0.785883 (rank : 37) | θ value | 4.29542e-18 (rank : 30) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P70406, O88285 | Gene names | Ucp2 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial uncoupling protein 2 (UCP 2) (UCPH). | |||||
|
ADT2_MOUSE
|
||||||
| NC score | 0.784481 (rank : 38) | θ value | 8.65492e-19 (rank : 25) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P51881, Q61311 | Gene names | Slc25a5, Ant2 | |||
|
Domain Architecture |

|
|||||
| Description | ADP/ATP translocase 2 (Adenine nucleotide translocator 2) (ANT 2) (ADP,ATP carrier protein 2) (Solute carrier family 25 member 5). | |||||
|
ADT2_HUMAN
|
||||||
| NC score | 0.782586 (rank : 39) | θ value | 1.47631e-18 (rank : 26) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P05141, O43350 | Gene names | SLC25A5, ANT2 | |||
|
Domain Architecture |

|
|||||
| Description | ADP/ATP translocase 2 (Adenine nucleotide translocator 2) (ANT 2) (ADP,ATP carrier protein 2) (Solute carrier family 25 member 5) (ADP,ATP carrier protein, fibroblast isoform). | |||||
|
MFRN2_HUMAN
|
||||||
| NC score | 0.781487 (rank : 40) | θ value | 1.80466e-16 (rank : 38) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q96A46, Q4VBZ0, Q5T777, Q86VX5, Q969G8, Q9H2J3 | Gene names | SLC25A28, MFRN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitoferrin-2 (Mitochondrial iron transporter 2) (Solute carrier family 25 member 28) (Mitochondrial RNA-splicing protein 3/4 homolog) (MRS3/4) (hMRS3/4). | |||||
|
UCP4_HUMAN
|
||||||
| NC score | 0.780186 (rank : 41) | θ value | 1.86753e-13 (rank : 47) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O95847 | Gene names | SLC25A27, UCP4 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial uncoupling protein 4 (UCP 4) (Solute carrier family 25 member 27). | |||||
|
MFRN2_MOUSE
|
||||||
| NC score | 0.779923 (rank : 42) | θ value | 3.07829e-16 (rank : 39) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8R0Z5, Q8K4J8, Q8K4J9, Q8R578 | Gene names | Slc25a28, Mfrn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitoferrin-2 (Mitochondrial iron transporter 2) (Solute carrier family 25 member 28) (Mitochondrial RNA-splicing protein 3/4 homolog) (MRS3/4). | |||||
|
MFRN1_MOUSE
|
||||||
| NC score | 0.776058 (rank : 43) | θ value | 1.058e-16 (rank : 37) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q920G8, Q8C367, Q8CEJ7, Q91ZY0, Q9D2G3, Q9D547 | Gene names | Slc25a37, Mfrn, Mscp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitoferrin-1 (Mitochondrial iron transporter 1) (Solute carrier family 25 member 37) (Mitochondrial solute carrier protein). | |||||
|
UCP3_HUMAN
|
||||||
| NC score | 0.774360 (rank : 44) | θ value | 1.16975e-15 (rank : 41) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P55916, O60475, Q96HL3 | Gene names | UCP3, SLC25A9 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial uncoupling protein 3 (UCP 3). | |||||
|
UCP3_MOUSE
|
||||||
| NC score | 0.768707 (rank : 45) | θ value | 1.09485e-13 (rank : 45) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P56501, O88293 | Gene names | Ucp3 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial uncoupling protein 3 (UCP 3). | |||||
|
MFRN1_HUMAN
|
||||||
| NC score | 0.764496 (rank : 46) | θ value | 4.16044e-13 (rank : 48) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9NYZ2, Q53FT7, Q69YJ8, Q969S1, Q9P0J2 | Gene names | SLC25A37, MFRN, MSCP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitoferrin-1 (Mitochondrial iron transporter 1) (Solute carrier family 25 member 37) (Mitochondrial solute carrier protein). | |||||
|
UCP1_MOUSE
|
||||||
| NC score | 0.758501 (rank : 47) | θ value | 5.8054e-15 (rank : 44) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P12242 | Gene names | Ucp1, Ucp | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial brown fat uncoupling protein 1 (UCP 1) (Thermogenin). | |||||
|
MPCP_MOUSE
|
||||||
| NC score | 0.757020 (rank : 48) | θ value | 1.25267e-09 (rank : 50) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8VEM8, Q542V7 | Gene names | Slc25a3 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphate carrier protein, mitochondrial precursor (PTP) (Solute carrier family 25 member 3). | |||||
|
MPCP_HUMAN
|
||||||
| NC score | 0.754665 (rank : 49) | θ value | 2.79066e-09 (rank : 52) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q00325, Q96A03 | Gene names | SLC25A3, PHC | |||
|
Domain Architecture |

|
|||||
| Description | Phosphate carrier protein, mitochondrial precursor (PTP) (Solute carrier family 25 member 3). | |||||
|
DIC_MOUSE
|
||||||
| NC score | 0.747175 (rank : 50) | θ value | 1.42992e-13 (rank : 46) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9QZD8, Q543J6, Q99LJ9, Q9D8G8 | Gene names | Slc25a10, Dic | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial dicarboxylate carrier (Solute carrier family 25 member 10). | |||||
|
PM34_MOUSE
|
||||||
| NC score | 0.730486 (rank : 51) | θ value | 1.99992e-07 (rank : 55) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O70579 | Gene names | Slc25a17, Pmp34, Pmp35 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal membrane protein PMP34 (34 kDa peroxisomal membrane protein) (Solute carrier family 25 member 17). | |||||
|
PM34_HUMAN
|
||||||
| NC score | 0.730227 (rank : 52) | θ value | 8.11959e-09 (rank : 53) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O43808, Q5TFL0, Q9UGW8, Q9UGY7 | Gene names | SLC25A17, PMP34 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal membrane protein PMP34 (34 kDa peroxisomal membrane protein) (Solute carrier family 25 member 17). | |||||
|
DIC_HUMAN
|
||||||
| NC score | 0.727085 (rank : 53) | θ value | 2.0648e-12 (rank : 49) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9UBX3, Q96BA1, Q96IP1 | Gene names | SLC25A10, DIC | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial dicarboxylate carrier (Solute carrier family 25 member 10). | |||||
|
MCAR1_MOUSE
|
||||||
| NC score | 0.570348 (rank : 54) | θ value | 0.00298849 (rank : 59) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q5HZI9 | Gene names | Mcart1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial carrier triple repeat 1. | |||||
|
MCAR1_HUMAN
|
||||||
| NC score | 0.553978 (rank : 55) | θ value | 4.1701e-05 (rank : 56) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9H1U9 | Gene names | MCART1 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial carrier triple repeat 1. | |||||
|
EFHA1_HUMAN
|
||||||
| NC score | 0.264993 (rank : 56) | θ value | 2.79066e-09 (rank : 51) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8IYU8, Q8N0T6, Q8NAX8 | Gene names | EFHA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand domain-containing family member A1. | |||||
|
EFHA1_MOUSE
|
||||||
| NC score | 0.254881 (rank : 57) | θ value | 2.36244e-08 (rank : 54) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8CD10, Q3TJU4, Q8K0K2, Q9CUR8 | Gene names | Efha1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand domain-containing family member A1. | |||||
|
MTCH2_HUMAN
|
||||||
| NC score | 0.220742 (rank : 58) | θ value | 0.21417 (rank : 63) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9Y6C9 | Gene names | MTCH2, MIMP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial carrier homolog 2 (Met-induced mitochondrial protein). | |||||
|
EFHA2_MOUSE
|
||||||
| NC score | 0.192630 (rank : 59) | θ value | 9.29e-05 (rank : 57) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9CTY5 | Gene names | Efha2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand domain-containing family member A2. | |||||
|
EFHA2_HUMAN
|
||||||
| NC score | 0.192086 (rank : 60) | θ value | 0.000121331 (rank : 58) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q86XE3, Q8IYZ3 | Gene names | EFHA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand domain-containing family member A2. | |||||
|
MTCH2_MOUSE
|
||||||
| NC score | 0.191239 (rank : 61) | θ value | 3.0926 (rank : 85) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q791V5, Q3TPS5, Q99LZ6, Q9D060, Q9D7Y2, Q9QZP3 | Gene names | Mtch2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial carrier homolog 2. | |||||
|
MTCH1_MOUSE
|
||||||
| NC score | 0.185975 (rank : 62) | θ value | 4.03905 (rank : 92) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q791T5, Q3U4W7, Q791V6, Q8R0T0, Q8R1T8, Q8R2T2, Q9QZA5, Q9QZP4 | Gene names | Mtch1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial carrier homolog 1 (Mitochondrial carrier-like protein 1). | |||||
|
MTCH1_HUMAN
|
||||||
| NC score | 0.185373 (rank : 63) | θ value | 2.36792 (rank : 74) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9NZJ7, Q6PK60, Q6UX45, Q7L465, Q9BW23, Q9NZR6, Q9UJZ5 | Gene names | MTCH1, PSAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial carrier homolog 1 (Presenilin-associated protein). | |||||
|
AYT1B_MOUSE
|
||||||
| NC score | 0.088727 (rank : 64) | θ value | 0.0148317 (rank : 60) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9D5U0 | Gene names | Aytl1b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acyltransferase-like 1-B (EC 2.3.1.-). | |||||
|
AYTL1_HUMAN
|
||||||
| NC score | 0.087383 (rank : 65) | θ value | 0.0736092 (rank : 61) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q7L5N7, Q6MZJ6, Q9NX23 | Gene names | AYTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acyltransferase-like 1 (EC 2.3.1.-). | |||||
|
AYT1A_MOUSE
|
||||||
| NC score | 0.084981 (rank : 66) | θ value | 0.0961366 (rank : 62) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8BYI6 | Gene names | Aytl1a, Aytl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acyltransferase-like 1-A (EC 2.3.1.-). | |||||
|
CALL5_HUMAN
|
||||||
| NC score | 0.060661 (rank : 67) | θ value | 0.279714 (rank : 64) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9NZT1, Q8IXU8 | Gene names | CALML5, CLSP | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-like protein 5 (Calmodulin-like skin protein). | |||||
|
GUC1C_HUMAN
|
||||||
| NC score | 0.051657 (rank : 68) | θ value | 5.27518 (rank : 95) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O95843, O95844, Q9UNM0 | Gene names | GUCA1C, GCAP3 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylyl cyclase-activating protein 3 (GCAP 3) (Guanylate cyclase activator 1C). | |||||
|
RECO_MOUSE
|
||||||
| NC score | 0.047772 (rank : 69) | θ value | 1.38821 (rank : 72) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P34057 | Gene names | Rcvrn, Rcv1 | |||
|
Domain Architecture |

|
|||||
| Description | Recoverin (Cancer-associated retinopathy protein) (Protein CAR) (23 kDa photoreceptor cell-specific protein). | |||||
|
HPCA_HUMAN
|
||||||
| NC score | 0.047309 (rank : 70) | θ value | 3.0926 (rank : 79) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P84074, P32076, P41211, P70510 | Gene names | HPCA, BDR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuron-specific calcium-binding protein hippocalcin (Calcium-binding protein BDR-2). | |||||
|
HPCA_MOUSE
|
||||||
| NC score | 0.047309 (rank : 71) | θ value | 3.0926 (rank : 80) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P84075, P32076, P41211, P70510 | Gene names | Hpca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuron-specific calcium-binding protein hippocalcin. | |||||
|
CETN1_MOUSE
|
||||||
| NC score | 0.045639 (rank : 72) | θ value | 1.38821 (rank : 71) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P41209, Q3V119, Q9D9G9, Q9DAL6 | Gene names | Cetn1, Calt | |||
|
Domain Architecture |

|
|||||
| Description | Centrin-1 (Caltractin). | |||||
|
CABP7_HUMAN
|
||||||
| NC score | 0.044168 (rank : 73) | θ value | 4.03905 (rank : 88) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q86V35 | Gene names | CABP7 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 7 (CaBP7). | |||||
|
CABP7_MOUSE
|
||||||
| NC score | 0.044168 (rank : 74) | θ value | 4.03905 (rank : 89) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q91ZM8 | Gene names | Cabp7 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 7 (CaBP7). | |||||
|
NCS1_HUMAN
|
||||||
| NC score | 0.042522 (rank : 75) | θ value | 2.36792 (rank : 76) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P62166, P36610, Q9UK26 | Gene names | FREQ, FLUP, NCS1 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal calcium sensor 1 (NCS-1) (Frequenin homolog) (Frequenin-like protein) (Frequenin-like ubiquitous protein). | |||||
|
NCS1_MOUSE
|
||||||
| NC score | 0.042522 (rank : 76) | θ value | 2.36792 (rank : 77) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8BNY6 | Gene names | Freq, Ncs1 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal calcium sensor 1 (NCS-1) (Frequenin homolog). | |||||
|
CETN1_HUMAN
|
||||||
| NC score | 0.040623 (rank : 77) | θ value | 4.03905 (rank : 91) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q12798 | Gene names | CETN1, CEN1, CETN | |||
|
Domain Architecture |

|
|||||
| Description | Centrin-1 (Caltractin isoform 2). | |||||
|
S100G_HUMAN
|
||||||
| NC score | 0.039659 (rank : 78) | θ value | 6.88961 (rank : 100) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P29377 | Gene names | S100G, CABP9K, CALB3, S100D | |||
|
Domain Architecture |
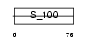
|
|||||
| Description | Protein S100-G (S100 calcium-binding protein G) (Vitamin D-dependent calcium-binding protein, intestinal) (CABP) (Calbindin D9K). | |||||
|
CALL3_HUMAN
|
||||||
| NC score | 0.039142 (rank : 79) | θ value | 0.813845 (rank : 68) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P27482 | Gene names | CALML3 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-like protein 3 (Calmodulin-related protein NB-1) (CaM-like protein) (CLP). | |||||
|
HPCL4_MOUSE
|
||||||
| NC score | 0.038126 (rank : 80) | θ value | 3.0926 (rank : 82) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8BGZ1 | Gene names | Hpcal4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hippocalcin-like protein 4 (Neural visinin-like protein 2) (NVP-2). | |||||
|
HPCL4_HUMAN
|
||||||
| NC score | 0.038125 (rank : 81) | θ value | 3.0926 (rank : 81) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9UM19, Q5TG97, Q8N611 | Gene names | HPCAL4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hippocalcin-like protein 4 (HLP4). | |||||
|
VISL1_HUMAN
|
||||||
| NC score | 0.037929 (rank : 82) | θ value | 3.0926 (rank : 86) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P62760, P28677, P29103, P42323, Q9UM20 | Gene names | VSNL1, VISL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Visinin-like protein 1 (VILIP) (Hippocalcin-like protein 3) (HLP3). | |||||
|
VISL1_MOUSE
|
||||||
| NC score | 0.037929 (rank : 83) | θ value | 3.0926 (rank : 87) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P62761, P28677, P29103, P42323, Q9UM20 | Gene names | Vsnl1, Visl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Visinin-like protein 1 (VILIP) (Neural visinin-like protein 1) (NVL-1) (NVP-1). | |||||
|
CANB2_MOUSE
|
||||||
| NC score | 0.037920 (rank : 84) | θ value | 4.03905 (rank : 90) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q63811 | Gene names | Ppp3r2 | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin subunit B isoform 2 (Protein phosphatase 2B regulatory subunit 2) (Protein phosphatase 3 regulatory subunit B beta isoform). | |||||
|
HPCL1_MOUSE
|
||||||
| NC score | 0.037725 (rank : 85) | θ value | 6.88961 (rank : 98) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P62748, P35333 | Gene names | Hpcal1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hippocalcin-like protein 1 (Visinin-like protein 3) (VILIP-3) (Neural visinin-like protein 3) (NVL-3) (NVP-3). | |||||
|
KCIP2_MOUSE
|
||||||
| NC score | 0.035064 (rank : 86) | θ value | 3.0926 (rank : 84) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9JJ69, Q6DTJ2, Q8K1T8, Q8K1T9, Q8K1U0, Q8VHN4, Q8VHN5, Q8VHN6, Q9JJ68 | Gene names | Kcnip2, Kchip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kv channel-interacting protein 2 (KChIP2) (A-type potassium channel modulatory protein 2) (Potassium channel-interacting protein 2). | |||||
|
KCIP2_HUMAN
|
||||||
| NC score | 0.034935 (rank : 87) | θ value | 3.0926 (rank : 83) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9NS61, Q7Z6F1, Q96K86, Q96T41, Q96T42, Q96T43, Q96T44, Q9H0N4, Q9HD10, Q9HD11, Q9NS60, Q9NY10, Q9NZI1 | Gene names | KCNIP2, KCHIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kv channel-interacting protein 2 (KChIP2) (A-type potassium channel modulatory protein 2) (Potassium channel-interacting protein 2) (Cardiac voltage-gated potassium channel modulatory subunit). | |||||
|
CALM_HUMAN
|
||||||
| NC score | 0.034444 (rank : 88) | θ value | 1.06291 (rank : 69) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P62158, P02593, P70667, P99014, Q13942, Q53S29, Q61379, Q61380, Q96HK3 | Gene names | CALM1, CALM, CAM, CAM1 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin (CaM). | |||||
|
CALM_MOUSE
|
||||||
| NC score | 0.034444 (rank : 89) | θ value | 1.06291 (rank : 70) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P62204, P02593, P70667, P99014, Q61379, Q61380 | Gene names | Calm1, Calm, Cam, Cam1 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin (CaM). | |||||
|
MYL6_HUMAN
|
||||||
| NC score | 0.033958 (rank : 90) | θ value | 0.47712 (rank : 66) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P60660, P16475, P24572, P24573, Q12790, Q6IAZ0, Q6IPY5 | Gene names | MYL6 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light polypeptide 6 (Smooth muscle and nonmuscle myosin light chain alkali 6) (Myosin light chain alkali 3) (Myosin light chain 3) (MLC-3) (LC17). | |||||
|
MYL6_MOUSE
|
||||||
| NC score | 0.033958 (rank : 91) | θ value | 0.47712 (rank : 67) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q60605 | Gene names | Myl6, Myln | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light polypeptide 6 (Smooth muscle and nonmuscle myosin light chain alkali 6) (Myosin light chain alkali 3) (Myosin light chain 3) (MLC-3) (LC17). | |||||
|
MYL6B_HUMAN
|
||||||
| NC score | 0.027513 (rank : 92) | θ value | 2.36792 (rank : 75) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P14649 | Gene names | MYL6B, MLC1SA | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light polypeptide 6B (Smooth muscle and nonmuscle myosin light chain alkali 6B) (Myosin light chain 1 slow-twitch muscle A isoform) (MLC1sa). | |||||
|
DUOX1_HUMAN
|
||||||
| NC score | 0.025963 (rank : 93) | θ value | 8.99809 (rank : 101) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9NRD9, Q6ZMB3, Q6ZR09, Q9NZC1 | Gene names | DUOX1, DUOX, LNOX1, THOX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual oxidase 1 precursor (EC 1.6.3.1) (EC 1.11.1.-) (NADPH thyroid oxidase 1) (Thyroid oxidase 1) (Large NOX 1) (Long NOX 1). | |||||
|
CABP1_MOUSE
|
||||||
| NC score | 0.025690 (rank : 94) | θ value | 6.88961 (rank : 97) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9JLK7, Q9JLK6 | Gene names | Cabp1 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 1 (CaBP1). | |||||
|
CABP1_HUMAN
|
||||||
| NC score | 0.025685 (rank : 95) | θ value | 6.88961 (rank : 96) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9NZU7, O95663, Q9NZU8 | Gene names | CABP1 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 1 (CaBP1) (Calbrain). | |||||
|
PLSI_HUMAN
|
||||||
| NC score | 0.024452 (rank : 96) | θ value | 4.03905 (rank : 93) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q14651 | Gene names | PLS1 | |||
|
Domain Architecture |

|
|||||
| Description | Plastin-1 (I-plastin) (Intestine-specific plastin). | |||||
|
ZMY15_HUMAN
|
||||||
| NC score | 0.020194 (rank : 97) | θ value | 4.03905 (rank : 94) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H091 | Gene names | ZMYND15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYND domain-containing protein 15. | |||||
|
DYH11_HUMAN
|
||||||
| NC score | 0.008748 (rank : 98) | θ value | 3.0926 (rank : 78) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96DT5, Q9UJ82 | Gene names | DNAH11 | |||
|
Domain Architecture |

|
|||||
| Description | Ciliary dynein heavy chain 11 (Axonemal beta dynein heavy chain 11). | |||||
|
PIWL1_MOUSE
|
||||||
| NC score | 0.007966 (rank : 99) | θ value | 6.88961 (rank : 99) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JMB7 | Gene names | Piwil1, Miwi | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Piwi-like protein 1. | |||||
|
PIWL1_HUMAN
|
||||||
| NC score | 0.007111 (rank : 100) | θ value | 8.99809 (rank : 102) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96J94, O95404, Q8NA60, Q8TBY5, Q96JD5 | Gene names | PIWIL1, HIWI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Piwi-like protein 1. | |||||
|
TRHY_HUMAN
|
||||||
| NC score | 0.005389 (rank : 101) | θ value | 0.365318 (rank : 65) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
OLF19_MOUSE
|
||||||
| NC score | -0.003775 (rank : 102) | θ value | 1.81305 (rank : 73) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 798 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JHB2, Q9JM32 | Gene names | Olfr19, Mor140-1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Olfactory receptor 19 (Olfactory receptor 140-1) (Odorant receptor M12). | |||||