Please be patient as the page loads
|
EFHA1_MOUSE
|
||||||
| SwissProt Accessions | Q8CD10, Q3TJU4, Q8K0K2, Q9CUR8 | Gene names | Efha1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand domain-containing family member A1. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
EFHA1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.992191 (rank : 2) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8IYU8, Q8N0T6, Q8NAX8 | Gene names | EFHA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand domain-containing family member A1. | |||||
|
EFHA1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8CD10, Q3TJU4, Q8K0K2, Q9CUR8 | Gene names | Efha1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand domain-containing family member A1. | |||||
|
EFHA2_MOUSE
|
||||||
| θ value | 3.12697e-77 (rank : 3) | NC score | 0.919138 (rank : 3) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9CTY5 | Gene names | Efha2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand domain-containing family member A2. | |||||
|
EFHA2_HUMAN
|
||||||
| θ value | 1.18825e-76 (rank : 4) | NC score | 0.913012 (rank : 4) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q86XE3, Q8IYZ3 | Gene names | EFHA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand domain-containing family member A2. | |||||
|
CMC1_HUMAN
|
||||||
| θ value | 2.36244e-08 (rank : 5) | NC score | 0.261014 (rank : 5) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O75746 | Gene names | SLC25A12, ARALAR1 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding mitochondrial carrier protein Aralar1 (Mitochondrial aspartate glutamate carrier 1) (Solute carrier family 25 member 12). | |||||
|
CMC1_MOUSE
|
||||||
| θ value | 2.36244e-08 (rank : 6) | NC score | 0.254881 (rank : 6) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BH59 | Gene names | Slc25a12, Aralar1 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding mitochondrial carrier protein Aralar1 (Mitochondrial aspartate glutamate carrier 1) (Solute carrier family 25 member 12). | |||||
|
CMC2_MOUSE
|
||||||
| θ value | 3.77169e-06 (rank : 7) | NC score | 0.227043 (rank : 7) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9QXX4, Q9CZF6, Q9DCF5 | Gene names | Slc25a13, Aralar2 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding mitochondrial carrier protein Aralar2 (Mitochondrial aspartate glutamate carrier 2) (Solute carrier family 25 member 13) (Citrin). | |||||
|
CMC2_HUMAN
|
||||||
| θ value | 6.43352e-06 (rank : 8) | NC score | 0.224668 (rank : 8) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UJS0, O14566, O14575, Q9NZW1, Q9UNI7 | Gene names | SLC25A13, ARALAR2 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding mitochondrial carrier protein Aralar2 (Mitochondrial aspartate glutamate carrier 2) (Solute carrier family 25 member 13) (Citrin). | |||||
|
RECO_MOUSE
|
||||||
| θ value | 0.00020696 (rank : 9) | NC score | 0.156991 (rank : 9) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P34057 | Gene names | Rcvrn, Rcv1 | |||
|
Domain Architecture |
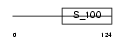
|
|||||
| Description | Recoverin (Cancer-associated retinopathy protein) (Protein CAR) (23 kDa photoreceptor cell-specific protein). | |||||
|
RECO_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 10) | NC score | 0.154694 (rank : 10) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P35243 | Gene names | RCVRN, RCV1 | |||
|
Domain Architecture |
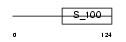
|
|||||
| Description | Recoverin (Cancer-associated retinopathy protein) (Protein CAR). | |||||
|
DUOX1_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 11) | NC score | 0.118476 (rank : 19) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9NRD9, Q6ZMB3, Q6ZR09, Q9NZC1 | Gene names | DUOX1, DUOX, LNOX1, THOX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual oxidase 1 precursor (EC 1.6.3.1) (EC 1.11.1.-) (NADPH thyroid oxidase 1) (Thyroid oxidase 1) (Large NOX 1) (Long NOX 1). | |||||
|
GUC1A_MOUSE
|
||||||
| θ value | 0.125558 (rank : 12) | NC score | 0.128565 (rank : 13) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P43081 | Gene names | Guca1a, Gcap, Gcap1, Guca1 | |||
|
Domain Architecture |
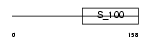
|
|||||
| Description | Guanylyl cyclase-activating protein 1 (GCAP 1) (Guanylate cyclase activator 1A). | |||||
|
CHP1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 13) | NC score | 0.102440 (rank : 25) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q99653 | Gene names | CHP | |||
|
Domain Architecture |
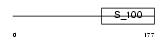
|
|||||
| Description | Calcium-binding protein p22 (Calcium-binding protein CHP) (Calcineurin homologous protein) (Calcineurin B homolog). | |||||
|
CHP1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 14) | NC score | 0.106534 (rank : 22) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P61022, Q62877, Q6ZWQ8 | Gene names | Chp, Sid470 | |||
|
Domain Architecture |
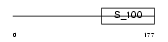
|
|||||
| Description | Calcium-binding protein p22 (Calcium-binding protein CHP) (Calcineurin homologous protein) (Sid 470). | |||||
|
GUC1A_HUMAN
|
||||||
| θ value | 0.62314 (rank : 15) | NC score | 0.122578 (rank : 18) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P43080, Q9NU14 | Gene names | GUCA1A, GCAP, GCAP1, GUCA1 | |||
|
Domain Architecture |
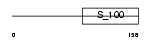
|
|||||
| Description | Guanylyl cyclase-activating protein 1 (GCAP 1) (Guanylate cyclase activator 1A). | |||||
|
HPCL1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 16) | NC score | 0.124989 (rank : 15) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P37235, Q969S5 | Gene names | HPCAL1, BDR1 | |||
|
Domain Architecture |
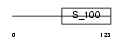
|
|||||
| Description | Hippocalcin-like protein 1 (Visinin-like protein 3) (VILIP-3) (Calcium-binding protein BDR-1) (HLP2). | |||||
|
HPCL1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 17) | NC score | 0.126107 (rank : 14) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P62748, P35333 | Gene names | Hpcal1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hippocalcin-like protein 1 (Visinin-like protein 3) (VILIP-3) (Neural visinin-like protein 3) (NVL-3) (NVP-3). | |||||
|
HPCA_HUMAN
|
||||||
| θ value | 0.813845 (rank : 18) | NC score | 0.128954 (rank : 11) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P84074, P32076, P41211, P70510 | Gene names | HPCA, BDR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuron-specific calcium-binding protein hippocalcin (Calcium-binding protein BDR-2). | |||||
|
HPCA_MOUSE
|
||||||
| θ value | 0.813845 (rank : 19) | NC score | 0.128954 (rank : 12) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P84075, P32076, P41211, P70510 | Gene names | Hpca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuron-specific calcium-binding protein hippocalcin. | |||||
|
TBCD8_HUMAN
|
||||||
| θ value | 0.813845 (rank : 20) | NC score | 0.040079 (rank : 102) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O95759, Q9UQ32 | Gene names | TBC1D8, VRP | |||
|
Domain Architecture |
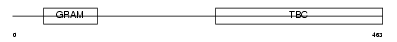
|
|||||
| Description | TBC1 domain family member 8 (Vascular Rab-GAP/TBC-containing protein) (AD 3). | |||||
|
GUC1B_MOUSE
|
||||||
| θ value | 1.06291 (rank : 21) | NC score | 0.115161 (rank : 20) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8VBV8 | Gene names | Guca1b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanylyl cyclase-activating protein 2 (GCAP 2) (Guanylate cyclase activator 1B). | |||||
|
AYT1A_MOUSE
|
||||||
| θ value | 1.81305 (rank : 22) | NC score | 0.098911 (rank : 32) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8BYI6 | Gene names | Aytl1a, Aytl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acyltransferase-like 1-A (EC 2.3.1.-). | |||||
|
AYTL1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 23) | NC score | 0.101191 (rank : 30) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q7L5N7, Q6MZJ6, Q9NX23 | Gene names | AYTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acyltransferase-like 1 (EC 2.3.1.-). | |||||
|
CALL3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 24) | NC score | 0.058077 (rank : 78) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P27482 | Gene names | CALML3 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-like protein 3 (Calmodulin-related protein NB-1) (CaM-like protein) (CLP). | |||||
|
NCALD_HUMAN
|
||||||
| θ value | 2.36792 (rank : 25) | NC score | 0.124040 (rank : 16) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P61601, P29554, Q8IYC3, Q9H0W2 | Gene names | NCALD | |||
|
Domain Architecture |
|
|||||
| Description | Neurocalcin delta. | |||||
|
NCALD_MOUSE
|
||||||
| θ value | 2.36792 (rank : 26) | NC score | 0.124040 (rank : 17) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q91X97, Q3TJS9, Q8BZN9 | Gene names | Ncald, D15Ertd412e | |||
|
Domain Architecture |
|
|||||
| Description | Neurocalcin delta. | |||||
|
PI51A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 27) | NC score | 0.013896 (rank : 103) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99755, Q99754, Q99756 | Gene names | PIP5K1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 5-kinase type-1 alpha (EC 2.7.1.68) (Phosphatidylinositol-4-phosphate 5-kinase type I alpha) (PtdIns(4)P- 5-kinase alpha) (PIP5KIalpha) (68 kDa type I phosphatidylinositol-4- phosphate 5-kinase alpha). | |||||
|
BPA1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 28) | NC score | 0.009345 (rank : 104) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q91ZU6, Q91ZU7 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 1/2/3/4 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
CABP2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 29) | NC score | 0.050090 (rank : 98) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9JLK4, Q9JLK5 | Gene names | Cabp2 | |||
|
Domain Architecture |
|
|||||
| Description | Calcium-binding protein 2 (CaBP2). | |||||
|
CANB1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 30) | NC score | 0.096088 (rank : 35) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P63098, P06705, P15117, Q08044 | Gene names | PPP3R1, CNA2, CNB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcineurin subunit B isoform 1 (Protein phosphatase 2B regulatory subunit 1) (Protein phosphatase 3 regulatory subunit B alpha isoform 1). | |||||
|
CANB1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 31) | NC score | 0.096085 (rank : 36) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q63810 | Gene names | Ppp3r1, Cnb | |||
|
Domain Architecture |
|
|||||
| Description | Calcineurin subunit B isoform 1 (Protein phosphatase 2B regulatory subunit 1) (Protein phosphatase 3 regulatory subunit B alpha isoform 1). | |||||
|
CABP2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 32) | NC score | 0.046342 (rank : 100) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9NPB3 | Gene names | CABP2 | |||
|
Domain Architecture |
|
|||||
| Description | Calcium-binding protein 2 (CaBP2). | |||||
|
CABP4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 33) | NC score | 0.043733 (rank : 101) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P57796, Q8N4Z2, Q8WWY5 | Gene names | CABP4 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 4 (CaBP4). | |||||
|
CABP4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 34) | NC score | 0.046651 (rank : 99) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8VHC5 | Gene names | Cabp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-binding protein 4 (CaBP4). | |||||
|
CANB2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 35) | NC score | 0.106873 (rank : 21) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q96LZ3, Q7Z4V8, Q8WYJ4 | Gene names | PPP3R2, CBLP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcineurin subunit B isoform 2 (Protein phosphatase 2B regulatory subunit 2) (Protein phosphatase 3 regulatory subunit B beta isoform) (Calcineurin B-like protein) (CBLP) (CNBII). | |||||
|
HPCL4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 36) | NC score | 0.102428 (rank : 26) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9UM19, Q5TG97, Q8N611 | Gene names | HPCAL4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hippocalcin-like protein 4 (HLP4). | |||||
|
HPCL4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 37) | NC score | 0.102356 (rank : 27) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8BGZ1 | Gene names | Hpcal4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hippocalcin-like protein 4 (Neural visinin-like protein 2) (NVP-2). | |||||
|
VISL1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 38) | NC score | 0.102116 (rank : 28) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P62760, P28677, P29103, P42323, Q9UM20 | Gene names | VSNL1, VISL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Visinin-like protein 1 (VILIP) (Hippocalcin-like protein 3) (HLP3). | |||||
|
VISL1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 39) | NC score | 0.102116 (rank : 29) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P62761, P28677, P29103, P42323, Q9UM20 | Gene names | Vsnl1, Visl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Visinin-like protein 1 (VILIP) (Neural visinin-like protein 1) (NVL-1) (NVP-1). | |||||
|
AYT1B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 40) | NC score | 0.093257 (rank : 39) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9D5U0 | Gene names | Aytl1b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acyltransferase-like 1-B (EC 2.3.1.-). | |||||
|
EFCB1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 41) | NC score | 0.085192 (rank : 41) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9D3N2, Q8BSW3, Q8C982, Q8CAI0, Q9D527, Q9D5E7 | Gene names | Efcab1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand calcium-binding domain-containing protein 1. | |||||
|
ADT1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 42) | NC score | 0.059250 (rank : 72) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P12235 | Gene names | SLC25A4, ANT1 | |||
|
Domain Architecture |
|
|||||
| Description | ADP/ATP translocase 1 (Adenine nucleotide translocator 1) (ANT 1) (ADP,ATP carrier protein 1) (Solute carrier family 25 member 4) (ADP,ATP carrier protein, heart/skeletal muscle isoform T1). | |||||
|
ADT1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 43) | NC score | 0.058631 (rank : 76) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P48962, Q62164 | Gene names | Slc25a4, Anc1, Ant1 | |||
|
Domain Architecture |
|
|||||
| Description | ADP/ATP translocase 1 (Adenine nucleotide translocator 1) (ANT 1) (ADP,ATP carrier protein 1) (Solute carrier family 25 member 4) (ADP,ATP carrier protein, heart/skeletal muscle isoform T1) (mANC1). | |||||
|
ADT2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 44) | NC score | 0.055103 (rank : 87) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P05141, O43350 | Gene names | SLC25A5, ANT2 | |||
|
Domain Architecture |
|
|||||
| Description | ADP/ATP translocase 2 (Adenine nucleotide translocator 2) (ANT 2) (ADP,ATP carrier protein 2) (Solute carrier family 25 member 5) (ADP,ATP carrier protein, fibroblast isoform). | |||||
|
ADT2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 45) | NC score | 0.055275 (rank : 86) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P51881, Q61311 | Gene names | Slc25a5, Ant2 | |||
|
Domain Architecture |
|
|||||
| Description | ADP/ATP translocase 2 (Adenine nucleotide translocator 2) (ANT 2) (ADP,ATP carrier protein 2) (Solute carrier family 25 member 5). | |||||
|
ADT3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 46) | NC score | 0.056963 (rank : 84) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P12236, Q96C49 | Gene names | SLC25A6, ANT3 | |||
|
Domain Architecture |

|
|||||
| Description | ADP/ATP translocase 3 (Adenine nucleotide translocator 2) (ANT 3) (ADP,ATP carrier protein 3) (Solute carrier family 25 member 6) (ADP,ATP carrier protein, isoform T2). | |||||
|
CALL5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 47) | NC score | 0.055492 (rank : 85) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9NZT1, Q8IXU8 | Gene names | CALML5, CLSP | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-like protein 5 (Calmodulin-like skin protein). | |||||
|
CALM4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 48) | NC score | 0.050519 (rank : 95) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9JM83, Q9CR31, Q9D1E9 | Gene names | Calm4, Dd112 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-4 (Calcium-binding protein Dd112). | |||||
|
CANB2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 49) | NC score | 0.104167 (rank : 24) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q63811 | Gene names | Ppp3r2 | |||
|
Domain Architecture |
|
|||||
| Description | Calcineurin subunit B isoform 2 (Protein phosphatase 2B regulatory subunit 2) (Protein phosphatase 3 regulatory subunit B beta isoform). | |||||
|
CETN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 50) | NC score | 0.060375 (rank : 68) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q12798 | Gene names | CETN1, CEN1, CETN | |||
|
Domain Architecture |
|
|||||
| Description | Centrin-1 (Caltractin isoform 2). | |||||
|
CETN1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 51) | NC score | 0.065709 (rank : 57) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P41209, Q3V119, Q9D9G9, Q9DAL6 | Gene names | Cetn1, Calt | |||
|
Domain Architecture |
|
|||||
| Description | Centrin-1 (Caltractin). | |||||
|
CETN2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 52) | NC score | 0.054491 (rank : 88) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9R1K9, Q3UBB4, Q9CWM0 | Gene names | Cetn2, Calt | |||
|
Domain Architecture |

|
|||||
| Description | Centrin-2 (Caltractin isoform 1). | |||||
|
CETN3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 53) | NC score | 0.050868 (rank : 93) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O15182, Q9BS23 | Gene names | CETN3, CEN3 | |||
|
Domain Architecture |
|
|||||
| Description | Centrin-3. | |||||
|
CHP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 54) | NC score | 0.059509 (rank : 70) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O43745 | Gene names | CHP2, HCA520 | |||
|
Domain Architecture |
|
|||||
| Description | Calcineurin B homologous protein 2 (Hepatocellular carcinoma- associated antigen 520). | |||||
|
CHP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 55) | NC score | 0.063634 (rank : 61) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9D869 | Gene names | Chp2, Hca520 | |||
|
Domain Architecture |
|
|||||
| Description | Calcineurin B homologous protein 2 (Hepatocellular carcinoma- associated antigen 520 homolog). | |||||
|
CSEN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 56) | NC score | 0.079794 (rank : 46) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9Y2W7, Q53TJ5, Q96T40, Q9UJ84, Q9UJ85 | Gene names | KCNIP3, CSEN, DREAM, KCHIP3 | |||
|
Domain Architecture |

|
|||||
| Description | Calsenilin (DRE-antagonist modulator) (DREAM) (Kv channel-interacting protein 3) (KChIP3) (A-type potassium channel modulatory protein 3). | |||||
|
CSEN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 57) | NC score | 0.079493 (rank : 47) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9QXT8, Q924L0, Q99PH9, Q99PI0, Q99PI2, Q99PI3, Q9JHZ5 | Gene names | Kcnip3, Csen, Dream, Kchip3 | |||
|
Domain Architecture |

|
|||||
| Description | Calsenilin (DRE-antagonist modulator) (DREAM) (Kv channel-interacting protein 3) (A-type potassium channel modulatory protein 3) (KChIP3). | |||||
|
DIC_MOUSE
|
||||||
| θ value | θ > 10 (rank : 58) | NC score | 0.050277 (rank : 96) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9QZD8, Q543J6, Q99LJ9, Q9D8G8 | Gene names | Slc25a10, Dic | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial dicarboxylate carrier (Solute carrier family 25 member 10). | |||||
|
DNC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 59) | NC score | 0.052810 (rank : 91) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HC21 | Gene names | SLC25A19, DNC, MUP1 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial deoxynucleotide carrier (Solute carrier family 25 member 19) (Mitochondrial uncoupling protein 1). | |||||
|
DNC_MOUSE
|
||||||
| θ value | θ > 10 (rank : 60) | NC score | 0.057281 (rank : 81) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DAM5, Q3T9C4 | Gene names | Slc25a19 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial deoxynucleotide carrier (Solute carrier family 25 member 19). | |||||
|
DUOX2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 61) | NC score | 0.081627 (rank : 45) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9NRD8, Q9NR02, Q9UHF9 | Gene names | DUOX2, LNOX2, THOX2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual oxidase 2 precursor (EC 1.6.3.1) (EC 1.11.1.-) (NADPH oxidase/peroxidase DUOX2) (NADPH thyroid oxidase 2) (Thyroid oxidase 2) (NADH/NADPH thyroid oxidase p138-tox) (p138 thyroid oxidase) (Large NOX 2) (Long NOX 2). | |||||
|
EFCB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 62) | NC score | 0.081965 (rank : 44) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9HAE3 | Gene names | EFCAB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand calcium-binding domain-containing protein 1. | |||||
|
GDC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 63) | NC score | 0.058968 (rank : 74) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P16260 | Gene names | SLC25A16, GDA | |||
|
Domain Architecture |
|
|||||
| Description | Grave disease carrier protein (GDC) (Grave disease autoantigen) (GDA) (Mitochondrial solute carrier protein homolog) (Solute carrier family 25 member 16). | |||||
|
GDC_MOUSE
|
||||||
| θ value | θ > 10 (rank : 64) | NC score | 0.057693 (rank : 79) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C0K5, Q6P6K8 | Gene names | Slc25a16, Gda | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Grave disease carrier protein homolog (GDC) (Mitochondrial solute carrier protein homolog) (Solute carrier family 25 member 16). | |||||
|
GHC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.094440 (rank : 38) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H936, Q8TBU8 | Gene names | SLC25A22, GC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial glutamate carrier 1 (GC-1) (Glutamate/H(+) symporter 1) (Solute carrier family 25 member 22). | |||||
|
GHC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.095039 (rank : 37) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D6M3, Q3TRY1 | Gene names | Slc25a22, Gc1 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial glutamate carrier 1 (GC-1) (Glutamate/H(+) symporter 1) (Solute carrier family 25 member 22). | |||||
|
GHC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.091641 (rank : 40) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H1K4 | Gene names | SLC25A18, GC2 | |||
|
Domain Architecture |
|
|||||
| Description | Mitochondrial glutamate carrier 2 (GC-2) (Glutamate/H(+) symporter 2) (Solute carrier family 25 member 18). | |||||
|
GUC1B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.104303 (rank : 23) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9UMX6, Q9NU15 | Gene names | GUCA1B, GCAP2 | |||
|
Domain Architecture |
|
|||||
| Description | Guanylyl cyclase-activating protein 2 (GCAP 2) (Guanylate cyclase activator 1B). | |||||
|
GUC1C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.099135 (rank : 31) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O95843, O95844, Q9UNM0 | Gene names | GUCA1C, GCAP3 | |||
|
Domain Architecture |
|
|||||
| Description | Guanylyl cyclase-activating protein 3 (GCAP 3) (Guanylate cyclase activator 1C). | |||||
|
KCIP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.079070 (rank : 51) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9NZI2, Q5U822 | Gene names | KCNIP1, KCHIP1, VABP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kv channel-interacting protein 1 (KChIP1) (A-type potassium channel modulatory protein 1) (Potassium channel-interacting protein 1) (Vesicle APC-binding protein). | |||||
|
KCIP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.079270 (rank : 50) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9JJ57, Q5SSA3, Q6DTJ1, Q8BGJ4, Q8C4K4, Q8CGL1, Q8K1U1, Q8K3M2 | Gene names | Kcnip1, Kchip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kv channel-interacting protein 1 (KChIP1) (A-type potassium channel modulatory protein 1) (Potassium channel-interacting protein 1). | |||||
|
KCIP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.083177 (rank : 42) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9NS61, Q7Z6F1, Q96K86, Q96T41, Q96T42, Q96T43, Q96T44, Q9H0N4, Q9HD10, Q9HD11, Q9NS60, Q9NY10, Q9NZI1 | Gene names | KCNIP2, KCHIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kv channel-interacting protein 2 (KChIP2) (A-type potassium channel modulatory protein 2) (Potassium channel-interacting protein 2) (Cardiac voltage-gated potassium channel modulatory subunit). | |||||
|
KCIP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.082985 (rank : 43) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9JJ69, Q6DTJ2, Q8K1T8, Q8K1T9, Q8K1U0, Q8VHN4, Q8VHN5, Q8VHN6, Q9JJ68 | Gene names | Kcnip2, Kchip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kv channel-interacting protein 2 (KChIP2) (A-type potassium channel modulatory protein 2) (Potassium channel-interacting protein 2). | |||||
|
KCIP4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.079319 (rank : 48) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6PIL6, Q4W5G8, Q8NEU0, Q9BWT2, Q9H294, Q9H2A4 | Gene names | KCNIP4, CALP, KCHIP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kv channel-interacting protein 4 (KChIP4) (A-type potassium channel modulatory protein 4) (Potassium channel-interacting protein 4) (Calsenilin-like protein). | |||||
|
KCIP4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.079300 (rank : 49) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6PHZ8, Q6DTJ3, Q8CAD0, Q8R4I2, Q9EQ01 | Gene names | Kcnip4, Calp, Kchip4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kv channel-interacting protein 4 (KChIP4) (A-type potassium channel modulatory protein 4) (Potassium channel-interacting protein 4) (Calsenilin-like protein). | |||||
|
KIP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.059408 (rank : 71) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q99828, O00693, O00735, Q6IB49, Q96J54, Q99971 | Gene names | CIB1, CIB, KIP, PRKDCIP | |||
|
Domain Architecture |
|
|||||
| Description | Calcium and integrin-binding protein 1 (Calmyrin) (DNA-PKcs- interacting protein) (Kinase-interacting protein) (KIP) (CIB) (SNK- interacting protein 2-28) (SIP2-28). | |||||
|
KIP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.059061 (rank : 73) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9Z0F4, Q3TN80 | Gene names | Cib1, Cib, Kip, Prkdcip | |||
|
Domain Architecture |
|
|||||
| Description | Calcium and integrin-binding protein 1 (Calmyrin) (DNA-PKcs- interacting protein) (Kinase-interacting protein) (KIP) (CIB). | |||||
|
KIP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.050775 (rank : 94) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9Z309 | Gene names | Cib2, Kip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium and integrin-binding protein 2 (Kinase-interacting protein 2) (KIP 2). | |||||
|
M2OM_HUMAN
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.058333 (rank : 77) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q02978, O75537, Q969P7 | Gene names | SLC25A11, SLC20A4 | |||
|
Domain Architecture |
|
|||||
| Description | Mitochondrial 2-oxoglutarate/malate carrier protein (OGCP) (Solute carrier family 25 member 11). | |||||
|
M2OM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.058734 (rank : 75) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9CR62 | Gene names | Slc25a11 | |||
|
Domain Architecture |
|
|||||
| Description | Mitochondrial 2-oxoglutarate/malate carrier protein (OGCP) (Solute carrier family 25 member 11). | |||||
|
MCATL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.064976 (rank : 58) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N8R3 | Gene names | SLC25A29, C14orf69 | |||
|
Domain Architecture |
|
|||||
| Description | Mitochondrial carnitine/acylcarnitine carrier protein CACL (CACT-like) (Solute carrier family 25 member 29). | |||||
|
MCATL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.063955 (rank : 60) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BL03, Q922X4 | Gene names | Slc25a29 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial carnitine/acylcarnitine carrier protein CACL (CACT-like) (Solute carrier family 25 member 29). | |||||
|
MCAT_HUMAN
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.068491 (rank : 56) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O43772, Q9UIQ2 | Gene names | SLC25A20, CAC, CACT | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial carnitine/acylcarnitine carrier protein (Carnitine/acylcarnitine translocase) (CAC) (Solute carrier family 25 member 20). | |||||
|
MCAT_MOUSE
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.064427 (rank : 59) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Z2Z6, Q3U801 | Gene names | Slc25a20, Cac | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial carnitine/acylcarnitine carrier protein (Carnitine/acylcarnitine translocase) (CAC) (mCAC) (Solute carrier family 25 member 20). | |||||
|
MFRN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.057037 (rank : 83) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NYZ2, Q53FT7, Q69YJ8, Q969S1, Q9P0J2 | Gene names | SLC25A37, MFRN, MSCP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitoferrin-1 (Mitochondrial iron transporter 1) (Solute carrier family 25 member 37) (Mitochondrial solute carrier protein). | |||||
|
MFRN1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.059712 (rank : 69) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q920G8, Q8C367, Q8CEJ7, Q91ZY0, Q9D2G3, Q9D547 | Gene names | Slc25a37, Mfrn, Mscp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitoferrin-1 (Mitochondrial iron transporter 1) (Solute carrier family 25 member 37) (Mitochondrial solute carrier protein). | |||||
|
MFRN2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.057687 (rank : 80) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96A46, Q4VBZ0, Q5T777, Q86VX5, Q969G8, Q9H2J3 | Gene names | SLC25A28, MFRN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitoferrin-2 (Mitochondrial iron transporter 2) (Solute carrier family 25 member 28) (Mitochondrial RNA-splicing protein 3/4 homolog) (MRS3/4) (hMRS3/4). | |||||
|
MFRN2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.057259 (rank : 82) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R0Z5, Q8K4J8, Q8K4J9, Q8R578 | Gene names | Slc25a28, Mfrn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitoferrin-2 (Mitochondrial iron transporter 2) (Solute carrier family 25 member 28) (Mitochondrial RNA-splicing protein 3/4 homolog) (MRS3/4). | |||||
|
MFTC_MOUSE
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.051837 (rank : 92) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BMG8, Q3TCM5 | Gene names | Slc25a32, Mftc | |||
|
Domain Architecture |
|
|||||
| Description | Mitochondrial folate transporter/carrier (Solute carrier family 25 member 32). | |||||
|
MPCP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.050217 (rank : 97) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q00325, Q96A03 | Gene names | SLC25A3, PHC | |||
|
Domain Architecture |
|
|||||
| Description | Phosphate carrier protein, mitochondrial precursor (PTP) (Solute carrier family 25 member 3). | |||||
|
NCS1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.096978 (rank : 33) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P62166, P36610, Q9UK26 | Gene names | FREQ, FLUP, NCS1 | |||
|
Domain Architecture |
|
|||||
| Description | Neuronal calcium sensor 1 (NCS-1) (Frequenin homolog) (Frequenin-like protein) (Frequenin-like ubiquitous protein). | |||||
|
NCS1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.096978 (rank : 34) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8BNY6 | Gene names | Freq, Ncs1 | |||
|
Domain Architecture |
|
|||||
| Description | Neuronal calcium sensor 1 (NCS-1) (Frequenin homolog). | |||||
|
ODC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.068568 (rank : 55) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BQT8 | Gene names | SLC25A21, ODC | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial 2-oxodicarboxylate carrier (Solute carrier family 25 member 21). | |||||
|
ODC_MOUSE
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.069522 (rank : 54) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BZ09, Q8BY72 | Gene names | Slc25a21, Odc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial 2-oxodicarboxylate carrier (Solute carrier family 25 member 21). | |||||
|
ORNT1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.061941 (rank : 66) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y619, Q9HC45 | Gene names | SLC25A15, ORNT1 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial ornithine transporter 1 (Solute carrier family 25 member 15). | |||||
|
ORNT1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.063158 (rank : 63) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9WVD5 | Gene names | Slc25a15, Ornt1 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial ornithine transporter 1 (Solute carrier family 25 member 15). | |||||
|
ORNT2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.061056 (rank : 67) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BXI2, Q8NFZ2 | Gene names | SLC25A2, ORNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial ornithine transporter 2 (Solute carrier family 25 member 2). | |||||
|
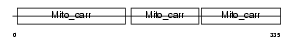
S2539_HUMAN
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.062729 (rank : 65) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BZJ4, Q9P182, Q9UF66, Q9Y379 | Gene names | SLC25A39 | |||
|
Domain Architecture |
|
|||||
| Description | Solute carrier family 25 member 39. | |||||
|
S2539_MOUSE
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.063564 (rank : 62) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D8K8, Q3TTM8 | Gene names | Slc25a39, D11Ertd333e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Solute carrier family 25 member 39. | |||||
|
S2544_HUMAN
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.072934 (rank : 53) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96H78, O75034 | Gene names | SLC25A44, KIAA0446 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Solute carrier family 25 member 44. | |||||
|
S2544_MOUSE
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.073474 (rank : 52) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BGF9, Q7TSU9 | Gene names | Slc25a44 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Solute carrier family 25 member 44. | |||||
|
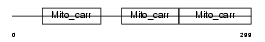
TXTP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.062797 (rank : 64) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P53007, Q9BSK6 | Gene names | SLC25A1, SLC20A3 | |||
|
Domain Architecture |
|
|||||
| Description | Tricarboxylate transport protein, mitochondrial precursor (Citrate transport protein) (CTP) (Tricarboxylate carrier protein) (Solute carrier family 25 member 1). | |||||
|
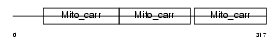
UCP5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.054104 (rank : 89) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95258, Q9HC60, Q9HC61 | Gene names | SLC25A14, BMCP1, UCP5 | |||
|
Domain Architecture |
|
|||||
| Description | Brain mitochondrial carrier protein 1 (BMCP-1) (Mitochondrial uncoupling protein 5) (UCP 5) (Solute carrier family 25 member 14). | |||||
|
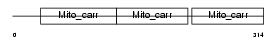
UCP5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.053866 (rank : 90) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Z2B2, Q9ESI8 | Gene names | Slc25a14, Bmcp1, Ucp5 | |||
|
Domain Architecture |
|
|||||
| Description | Brain mitochondrial carrier protein 1 (BMCP-1) (Mitochondrial uncoupling protein 5) (UCP 5) (Solute carrier family 25 member 14). | |||||
|
EFHA1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8CD10, Q3TJU4, Q8K0K2, Q9CUR8 | Gene names | Efha1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand domain-containing family member A1. | |||||
|
EFHA1_HUMAN
|
||||||
| NC score | 0.992191 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8IYU8, Q8N0T6, Q8NAX8 | Gene names | EFHA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand domain-containing family member A1. | |||||
|
EFHA2_MOUSE
|
||||||
| NC score | 0.919138 (rank : 3) | θ value | 3.12697e-77 (rank : 3) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9CTY5 | Gene names | Efha2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand domain-containing family member A2. | |||||
|
EFHA2_HUMAN
|
||||||
| NC score | 0.913012 (rank : 4) | θ value | 1.18825e-76 (rank : 4) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q86XE3, Q8IYZ3 | Gene names | EFHA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand domain-containing family member A2. | |||||
|
CMC1_HUMAN
|
||||||
| NC score | 0.261014 (rank : 5) | θ value | 2.36244e-08 (rank : 5) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O75746 | Gene names | SLC25A12, ARALAR1 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding mitochondrial carrier protein Aralar1 (Mitochondrial aspartate glutamate carrier 1) (Solute carrier family 25 member 12). | |||||
|
CMC1_MOUSE
|
||||||
| NC score | 0.254881 (rank : 6) | θ value | 2.36244e-08 (rank : 6) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BH59 | Gene names | Slc25a12, Aralar1 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding mitochondrial carrier protein Aralar1 (Mitochondrial aspartate glutamate carrier 1) (Solute carrier family 25 member 12). | |||||
|
CMC2_MOUSE
|
||||||
| NC score | 0.227043 (rank : 7) | θ value | 3.77169e-06 (rank : 7) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9QXX4, Q9CZF6, Q9DCF5 | Gene names | Slc25a13, Aralar2 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding mitochondrial carrier protein Aralar2 (Mitochondrial aspartate glutamate carrier 2) (Solute carrier family 25 member 13) (Citrin). | |||||
|
CMC2_HUMAN
|
||||||
| NC score | 0.224668 (rank : 8) | θ value | 6.43352e-06 (rank : 8) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UJS0, O14566, O14575, Q9NZW1, Q9UNI7 | Gene names | SLC25A13, ARALAR2 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding mitochondrial carrier protein Aralar2 (Mitochondrial aspartate glutamate carrier 2) (Solute carrier family 25 member 13) (Citrin). | |||||
|
RECO_MOUSE
|
||||||
| NC score | 0.156991 (rank : 9) | θ value | 0.00020696 (rank : 9) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P34057 | Gene names | Rcvrn, Rcv1 | |||
|
Domain Architecture |

|
|||||
| Description | Recoverin (Cancer-associated retinopathy protein) (Protein CAR) (23 kDa photoreceptor cell-specific protein). | |||||
|
RECO_HUMAN
|
||||||
| NC score | 0.154694 (rank : 10) | θ value | 0.00509761 (rank : 10) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P35243 | Gene names | RCVRN, RCV1 | |||
|
Domain Architecture |

|
|||||
| Description | Recoverin (Cancer-associated retinopathy protein) (Protein CAR). | |||||
|
HPCA_HUMAN
|
||||||
| NC score | 0.128954 (rank : 11) | θ value | 0.813845 (rank : 18) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P84074, P32076, P41211, P70510 | Gene names | HPCA, BDR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuron-specific calcium-binding protein hippocalcin (Calcium-binding protein BDR-2). | |||||
|
HPCA_MOUSE
|
||||||
| NC score | 0.128954 (rank : 12) | θ value | 0.813845 (rank : 19) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P84075, P32076, P41211, P70510 | Gene names | Hpca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuron-specific calcium-binding protein hippocalcin. | |||||
|
GUC1A_MOUSE
|
||||||
| NC score | 0.128565 (rank : 13) | θ value | 0.125558 (rank : 12) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P43081 | Gene names | Guca1a, Gcap, Gcap1, Guca1 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylyl cyclase-activating protein 1 (GCAP 1) (Guanylate cyclase activator 1A). | |||||
|
HPCL1_MOUSE
|
||||||
| NC score | 0.126107 (rank : 14) | θ value | 0.62314 (rank : 17) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P62748, P35333 | Gene names | Hpcal1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hippocalcin-like protein 1 (Visinin-like protein 3) (VILIP-3) (Neural visinin-like protein 3) (NVL-3) (NVP-3). | |||||
|
HPCL1_HUMAN
|
||||||
| NC score | 0.124989 (rank : 15) | θ value | 0.62314 (rank : 16) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P37235, Q969S5 | Gene names | HPCAL1, BDR1 | |||
|
Domain Architecture |

|
|||||
| Description | Hippocalcin-like protein 1 (Visinin-like protein 3) (VILIP-3) (Calcium-binding protein BDR-1) (HLP2). | |||||
|
NCALD_HUMAN
|
||||||
| NC score | 0.124040 (rank : 16) | θ value | 2.36792 (rank : 25) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P61601, P29554, Q8IYC3, Q9H0W2 | Gene names | NCALD | |||
|
Domain Architecture |

|
|||||
| Description | Neurocalcin delta. | |||||
|
NCALD_MOUSE
|
||||||
| NC score | 0.124040 (rank : 17) | θ value | 2.36792 (rank : 26) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q91X97, Q3TJS9, Q8BZN9 | Gene names | Ncald, D15Ertd412e | |||
|
Domain Architecture |

|
|||||
| Description | Neurocalcin delta. | |||||
|
GUC1A_HUMAN
|
||||||
| NC score | 0.122578 (rank : 18) | θ value | 0.62314 (rank : 15) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P43080, Q9NU14 | Gene names | GUCA1A, GCAP, GCAP1, GUCA1 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylyl cyclase-activating protein 1 (GCAP 1) (Guanylate cyclase activator 1A). | |||||
|
DUOX1_HUMAN
|
||||||
| NC score | 0.118476 (rank : 19) | θ value | 0.0431538 (rank : 11) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9NRD9, Q6ZMB3, Q6ZR09, Q9NZC1 | Gene names | DUOX1, DUOX, LNOX1, THOX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual oxidase 1 precursor (EC 1.6.3.1) (EC 1.11.1.-) (NADPH thyroid oxidase 1) (Thyroid oxidase 1) (Large NOX 1) (Long NOX 1). | |||||
|
GUC1B_MOUSE
|
||||||
| NC score | 0.115161 (rank : 20) | θ value | 1.06291 (rank : 21) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8VBV8 | Gene names | Guca1b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanylyl cyclase-activating protein 2 (GCAP 2) (Guanylate cyclase activator 1B). | |||||
|
CANB2_HUMAN
|
||||||
| NC score | 0.106873 (rank : 21) | θ value | 6.88961 (rank : 35) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q96LZ3, Q7Z4V8, Q8WYJ4 | Gene names | PPP3R2, CBLP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcineurin subunit B isoform 2 (Protein phosphatase 2B regulatory subunit 2) (Protein phosphatase 3 regulatory subunit B beta isoform) (Calcineurin B-like protein) (CBLP) (CNBII). | |||||
|
CHP1_MOUSE
|
||||||
| NC score | 0.106534 (rank : 22) | θ value | 0.47712 (rank : 14) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P61022, Q62877, Q6ZWQ8 | Gene names | Chp, Sid470 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein p22 (Calcium-binding protein CHP) (Calcineurin homologous protein) (Sid 470). | |||||
|
GUC1B_HUMAN
|
||||||
| NC score | 0.104303 (rank : 23) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9UMX6, Q9NU15 | Gene names | GUCA1B, GCAP2 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylyl cyclase-activating protein 2 (GCAP 2) (Guanylate cyclase activator 1B). | |||||
|
CANB2_MOUSE
|
||||||
| NC score | 0.104167 (rank : 24) | θ value | θ > 10 (rank : 49) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q63811 | Gene names | Ppp3r2 | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin subunit B isoform 2 (Protein phosphatase 2B regulatory subunit 2) (Protein phosphatase 3 regulatory subunit B beta isoform). | |||||
|
CHP1_HUMAN
|
||||||
| NC score | 0.102440 (rank : 25) | θ value | 0.21417 (rank : 13) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q99653 | Gene names | CHP | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein p22 (Calcium-binding protein CHP) (Calcineurin homologous protein) (Calcineurin B homolog). | |||||
|
HPCL4_HUMAN
|
||||||
| NC score | 0.102428 (rank : 26) | θ value | 6.88961 (rank : 36) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9UM19, Q5TG97, Q8N611 | Gene names | HPCAL4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hippocalcin-like protein 4 (HLP4). | |||||
|
HPCL4_MOUSE
|
||||||
| NC score | 0.102356 (rank : 27) | θ value | 6.88961 (rank : 37) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8BGZ1 | Gene names | Hpcal4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hippocalcin-like protein 4 (Neural visinin-like protein 2) (NVP-2). | |||||
|
VISL1_HUMAN
|
||||||
| NC score | 0.102116 (rank : 28) | θ value | 6.88961 (rank : 38) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P62760, P28677, P29103, P42323, Q9UM20 | Gene names | VSNL1, VISL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Visinin-like protein 1 (VILIP) (Hippocalcin-like protein 3) (HLP3). | |||||
|
VISL1_MOUSE
|
||||||
| NC score | 0.102116 (rank : 29) | θ value | 6.88961 (rank : 39) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P62761, P28677, P29103, P42323, Q9UM20 | Gene names | Vsnl1, Visl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Visinin-like protein 1 (VILIP) (Neural visinin-like protein 1) (NVL-1) (NVP-1). | |||||
|
AYTL1_HUMAN
|
||||||
| NC score | 0.101191 (rank : 30) | θ value | 1.81305 (rank : 23) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q7L5N7, Q6MZJ6, Q9NX23 | Gene names | AYTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acyltransferase-like 1 (EC 2.3.1.-). | |||||
|
GUC1C_HUMAN
|
||||||
| NC score | 0.099135 (rank : 31) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O95843, O95844, Q9UNM0 | Gene names | GUCA1C, GCAP3 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylyl cyclase-activating protein 3 (GCAP 3) (Guanylate cyclase activator 1C). | |||||
|
AYT1A_MOUSE
|
||||||
| NC score | 0.098911 (rank : 32) | θ value | 1.81305 (rank : 22) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8BYI6 | Gene names | Aytl1a, Aytl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acyltransferase-like 1-A (EC 2.3.1.-). | |||||
|
NCS1_HUMAN
|
||||||
| NC score | 0.096978 (rank : 33) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P62166, P36610, Q9UK26 | Gene names | FREQ, FLUP, NCS1 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal calcium sensor 1 (NCS-1) (Frequenin homolog) (Frequenin-like protein) (Frequenin-like ubiquitous protein). | |||||
|
NCS1_MOUSE
|
||||||
| NC score | 0.096978 (rank : 34) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8BNY6 | Gene names | Freq, Ncs1 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal calcium sensor 1 (NCS-1) (Frequenin homolog). | |||||
|
CANB1_HUMAN
|
||||||
| NC score | 0.096088 (rank : 35) | θ value | 4.03905 (rank : 30) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P63098, P06705, P15117, Q08044 | Gene names | PPP3R1, CNA2, CNB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcineurin subunit B isoform 1 (Protein phosphatase 2B regulatory subunit 1) (Protein phosphatase 3 regulatory subunit B alpha isoform 1). | |||||
|
CANB1_MOUSE
|
||||||
| NC score | 0.096085 (rank : 36) | θ value | 4.03905 (rank : 31) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q63810 | Gene names | Ppp3r1, Cnb | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin subunit B isoform 1 (Protein phosphatase 2B regulatory subunit 1) (Protein phosphatase 3 regulatory subunit B alpha isoform 1). | |||||
|
GHC1_MOUSE
|
||||||
| NC score | 0.095039 (rank : 37) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D6M3, Q3TRY1 | Gene names | Slc25a22, Gc1 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial glutamate carrier 1 (GC-1) (Glutamate/H(+) symporter 1) (Solute carrier family 25 member 22). | |||||
|
GHC1_HUMAN
|
||||||
| NC score | 0.094440 (rank : 38) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H936, Q8TBU8 | Gene names | SLC25A22, GC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial glutamate carrier 1 (GC-1) (Glutamate/H(+) symporter 1) (Solute carrier family 25 member 22). | |||||
|
AYT1B_MOUSE
|
||||||
| NC score | 0.093257 (rank : 39) | θ value | 8.99809 (rank : 40) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9D5U0 | Gene names | Aytl1b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acyltransferase-like 1-B (EC 2.3.1.-). | |||||
|
GHC2_HUMAN
|
||||||
| NC score | 0.091641 (rank : 40) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H1K4 | Gene names | SLC25A18, GC2 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial glutamate carrier 2 (GC-2) (Glutamate/H(+) symporter 2) (Solute carrier family 25 member 18). | |||||
|
EFCB1_MOUSE
|
||||||
| NC score | 0.085192 (rank : 41) | θ value | 8.99809 (rank : 41) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9D3N2, Q8BSW3, Q8C982, Q8CAI0, Q9D527, Q9D5E7 | Gene names | Efcab1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand calcium-binding domain-containing protein 1. | |||||
|
KCIP2_HUMAN
|
||||||
| NC score | 0.083177 (rank : 42) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9NS61, Q7Z6F1, Q96K86, Q96T41, Q96T42, Q96T43, Q96T44, Q9H0N4, Q9HD10, Q9HD11, Q9NS60, Q9NY10, Q9NZI1 | Gene names | KCNIP2, KCHIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kv channel-interacting protein 2 (KChIP2) (A-type potassium channel modulatory protein 2) (Potassium channel-interacting protein 2) (Cardiac voltage-gated potassium channel modulatory subunit). | |||||
|
KCIP2_MOUSE
|
||||||
| NC score | 0.082985 (rank : 43) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9JJ69, Q6DTJ2, Q8K1T8, Q8K1T9, Q8K1U0, Q8VHN4, Q8VHN5, Q8VHN6, Q9JJ68 | Gene names | Kcnip2, Kchip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kv channel-interacting protein 2 (KChIP2) (A-type potassium channel modulatory protein 2) (Potassium channel-interacting protein 2). | |||||
|
EFCB1_HUMAN
|
||||||
| NC score | 0.081965 (rank : 44) | θ value | θ > 10 (rank : 62) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9HAE3 | Gene names | EFCAB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand calcium-binding domain-containing protein 1. | |||||
|
DUOX2_HUMAN
|
||||||
| NC score | 0.081627 (rank : 45) | θ value | θ > 10 (rank : 61) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9NRD8, Q9NR02, Q9UHF9 | Gene names | DUOX2, LNOX2, THOX2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual oxidase 2 precursor (EC 1.6.3.1) (EC 1.11.1.-) (NADPH oxidase/peroxidase DUOX2) (NADPH thyroid oxidase 2) (Thyroid oxidase 2) (NADH/NADPH thyroid oxidase p138-tox) (p138 thyroid oxidase) (Large NOX 2) (Long NOX 2). | |||||
|
CSEN_HUMAN
|
||||||
| NC score | 0.079794 (rank : 46) | θ value | θ > 10 (rank : 56) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9Y2W7, Q53TJ5, Q96T40, Q9UJ84, Q9UJ85 | Gene names | KCNIP3, CSEN, DREAM, KCHIP3 | |||
|
Domain Architecture |

|
|||||
| Description | Calsenilin (DRE-antagonist modulator) (DREAM) (Kv channel-interacting protein 3) (KChIP3) (A-type potassium channel modulatory protein 3). | |||||
|
CSEN_MOUSE
|
||||||
| NC score | 0.079493 (rank : 47) | θ value | θ > 10 (rank : 57) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9QXT8, Q924L0, Q99PH9, Q99PI0, Q99PI2, Q99PI3, Q9JHZ5 | Gene names | Kcnip3, Csen, Dream, Kchip3 | |||
|
Domain Architecture |

|
|||||
| Description | Calsenilin (DRE-antagonist modulator) (DREAM) (Kv channel-interacting protein 3) (A-type potassium channel modulatory protein 3) (KChIP3). | |||||
|
KCIP4_HUMAN
|
||||||
| NC score | 0.079319 (rank : 48) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6PIL6, Q4W5G8, Q8NEU0, Q9BWT2, Q9H294, Q9H2A4 | Gene names | KCNIP4, CALP, KCHIP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kv channel-interacting protein 4 (KChIP4) (A-type potassium channel modulatory protein 4) (Potassium channel-interacting protein 4) (Calsenilin-like protein). | |||||
|
KCIP4_MOUSE
|
||||||
| NC score | 0.079300 (rank : 49) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6PHZ8, Q6DTJ3, Q8CAD0, Q8R4I2, Q9EQ01 | Gene names | Kcnip4, Calp, Kchip4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kv channel-interacting protein 4 (KChIP4) (A-type potassium channel modulatory protein 4) (Potassium channel-interacting protein 4) (Calsenilin-like protein). | |||||
|
KCIP1_MOUSE
|
||||||
| NC score | 0.079270 (rank : 50) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9JJ57, Q5SSA3, Q6DTJ1, Q8BGJ4, Q8C4K4, Q8CGL1, Q8K1U1, Q8K3M2 | Gene names | Kcnip1, Kchip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kv channel-interacting protein 1 (KChIP1) (A-type potassium channel modulatory protein 1) (Potassium channel-interacting protein 1). | |||||
|
KCIP1_HUMAN
|
||||||
| NC score | 0.079070 (rank : 51) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9NZI2, Q5U822 | Gene names | KCNIP1, KCHIP1, VABP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kv channel-interacting protein 1 (KChIP1) (A-type potassium channel modulatory protein 1) (Potassium channel-interacting protein 1) (Vesicle APC-binding protein). | |||||
|
S2544_MOUSE
|
||||||
| NC score | 0.073474 (rank : 52) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BGF9, Q7TSU9 | Gene names | Slc25a44 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Solute carrier family 25 member 44. | |||||
|
S2544_HUMAN
|
||||||
| NC score | 0.072934 (rank : 53) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96H78, O75034 | Gene names | SLC25A44, KIAA0446 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Solute carrier family 25 member 44. | |||||
|
ODC_MOUSE
|
||||||
| NC score | 0.069522 (rank : 54) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BZ09, Q8BY72 | Gene names | Slc25a21, Odc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial 2-oxodicarboxylate carrier (Solute carrier family 25 member 21). | |||||
|
ODC_HUMAN
|
||||||
| NC score | 0.068568 (rank : 55) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BQT8 | Gene names | SLC25A21, ODC | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial 2-oxodicarboxylate carrier (Solute carrier family 25 member 21). | |||||
|
MCAT_HUMAN
|
||||||
| NC score | 0.068491 (rank : 56) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O43772, Q9UIQ2 | Gene names | SLC25A20, CAC, CACT | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial carnitine/acylcarnitine carrier protein (Carnitine/acylcarnitine translocase) (CAC) (Solute carrier family 25 member 20). | |||||
|
CETN1_MOUSE
|
||||||
| NC score | 0.065709 (rank : 57) | θ value | θ > 10 (rank : 51) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P41209, Q3V119, Q9D9G9, Q9DAL6 | Gene names | Cetn1, Calt | |||
|
Domain Architecture |

|
|||||
| Description | Centrin-1 (Caltractin). | |||||
|
MCATL_HUMAN
|
||||||
| NC score | 0.064976 (rank : 58) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N8R3 | Gene names | SLC25A29, C14orf69 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial carnitine/acylcarnitine carrier protein CACL (CACT-like) (Solute carrier family 25 member 29). | |||||
|
MCAT_MOUSE
|
||||||
| NC score | 0.064427 (rank : 59) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Z2Z6, Q3U801 | Gene names | Slc25a20, Cac | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial carnitine/acylcarnitine carrier protein (Carnitine/acylcarnitine translocase) (CAC) (mCAC) (Solute carrier family 25 member 20). | |||||
|
MCATL_MOUSE
|
||||||
| NC score | 0.063955 (rank : 60) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BL03, Q922X4 | Gene names | Slc25a29 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial carnitine/acylcarnitine carrier protein CACL (CACT-like) (Solute carrier family 25 member 29). | |||||
|
CHP2_MOUSE
|
||||||
| NC score | 0.063634 (rank : 61) | θ value | θ > 10 (rank : 55) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9D869 | Gene names | Chp2, Hca520 | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin B homologous protein 2 (Hepatocellular carcinoma- associated antigen 520 homolog). | |||||
|
S2539_MOUSE
|
||||||
| NC score | 0.063564 (rank : 62) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D8K8, Q3TTM8 | Gene names | Slc25a39, D11Ertd333e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Solute carrier family 25 member 39. | |||||
|
ORNT1_MOUSE
|
||||||
| NC score | 0.063158 (rank : 63) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9WVD5 | Gene names | Slc25a15, Ornt1 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial ornithine transporter 1 (Solute carrier family 25 member 15). | |||||
|
TXTP_HUMAN
|
||||||
| NC score | 0.062797 (rank : 64) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P53007, Q9BSK6 | Gene names | SLC25A1, SLC20A3 | |||
|
Domain Architecture |

|
|||||
| Description | Tricarboxylate transport protein, mitochondrial precursor (Citrate transport protein) (CTP) (Tricarboxylate carrier protein) (Solute carrier family 25 member 1). | |||||
|
S2539_HUMAN
|
||||||
| NC score | 0.062729 (rank : 65) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BZJ4, Q9P182, Q9UF66, Q9Y379 | Gene names | SLC25A39 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 25 member 39. | |||||
|
ORNT1_HUMAN
|
||||||
| NC score | 0.061941 (rank : 66) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y619, Q9HC45 | Gene names | SLC25A15, ORNT1 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial ornithine transporter 1 (Solute carrier family 25 member 15). | |||||
|
ORNT2_HUMAN
|
||||||
| NC score | 0.061056 (rank : 67) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BXI2, Q8NFZ2 | Gene names | SLC25A2, ORNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial ornithine transporter 2 (Solute carrier family 25 member 2). | |||||
|
CETN1_HUMAN
|
||||||
| NC score | 0.060375 (rank : 68) | θ value | θ > 10 (rank : 50) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q12798 | Gene names | CETN1, CEN1, CETN | |||
|
Domain Architecture |

|
|||||
| Description | Centrin-1 (Caltractin isoform 2). | |||||
|
MFRN1_MOUSE
|
||||||
| NC score | 0.059712 (rank : 69) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q920G8, Q8C367, Q8CEJ7, Q91ZY0, Q9D2G3, Q9D547 | Gene names | Slc25a37, Mfrn, Mscp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitoferrin-1 (Mitochondrial iron transporter 1) (Solute carrier family 25 member 37) (Mitochondrial solute carrier protein). | |||||
|
CHP2_HUMAN
|
||||||
| NC score | 0.059509 (rank : 70) | θ value | θ > 10 (rank : 54) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O43745 | Gene names | CHP2, HCA520 | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin B homologous protein 2 (Hepatocellular carcinoma- associated antigen 520). | |||||
|
KIP1_HUMAN
|
||||||
| NC score | 0.059408 (rank : 71) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q99828, O00693, O00735, Q6IB49, Q96J54, Q99971 | Gene names | CIB1, CIB, KIP, PRKDCIP | |||
|
Domain Architecture |

|
|||||
| Description | Calcium and integrin-binding protein 1 (Calmyrin) (DNA-PKcs- interacting protein) (Kinase-interacting protein) (KIP) (CIB) (SNK- interacting protein 2-28) (SIP2-28). | |||||
|
ADT1_HUMAN
|
||||||
| NC score | 0.059250 (rank : 72) | θ value | θ > 10 (rank : 42) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P12235 | Gene names | SLC25A4, ANT1 | |||
|
Domain Architecture |

|
|||||
| Description | ADP/ATP translocase 1 (Adenine nucleotide translocator 1) (ANT 1) (ADP,ATP carrier protein 1) (Solute carrier family 25 member 4) (ADP,ATP carrier protein, heart/skeletal muscle isoform T1). | |||||
|
KIP1_MOUSE
|
||||||
| NC score | 0.059061 (rank : 73) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9Z0F4, Q3TN80 | Gene names | Cib1, Cib, Kip, Prkdcip | |||
|
Domain Architecture |

|
|||||
| Description | Calcium and integrin-binding protein 1 (Calmyrin) (DNA-PKcs- interacting protein) (Kinase-interacting protein) (KIP) (CIB). | |||||
|
GDC_HUMAN
|
||||||
| NC score | 0.058968 (rank : 74) | θ value | θ > 10 (rank : 63) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P16260 | Gene names | SLC25A16, GDA | |||
|
Domain Architecture |

|
|||||
| Description | Grave disease carrier protein (GDC) (Grave disease autoantigen) (GDA) (Mitochondrial solute carrier protein homolog) (Solute carrier family 25 member 16). | |||||
|
M2OM_MOUSE
|
||||||
| NC score | 0.058734 (rank : 75) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9CR62 | Gene names | Slc25a11 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial 2-oxoglutarate/malate carrier protein (OGCP) (Solute carrier family 25 member 11). | |||||
|
ADT1_MOUSE
|
||||||
| NC score | 0.058631 (rank : 76) | θ value | θ > 10 (rank : 43) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P48962, Q62164 | Gene names | Slc25a4, Anc1, Ant1 | |||
|
Domain Architecture |

|
|||||
| Description | ADP/ATP translocase 1 (Adenine nucleotide translocator 1) (ANT 1) (ADP,ATP carrier protein 1) (Solute carrier family 25 member 4) (ADP,ATP carrier protein, heart/skeletal muscle isoform T1) (mANC1). | |||||
|
M2OM_HUMAN
|
||||||
| NC score | 0.058333 (rank : 77) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q02978, O75537, Q969P7 | Gene names | SLC25A11, SLC20A4 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial 2-oxoglutarate/malate carrier protein (OGCP) (Solute carrier family 25 member 11). | |||||
|
CALL3_HUMAN
|
||||||
| NC score | 0.058077 (rank : 78) | θ value | 2.36792 (rank : 24) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P27482 | Gene names | CALML3 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-like protein 3 (Calmodulin-related protein NB-1) (CaM-like protein) (CLP). | |||||
|
GDC_MOUSE
|
||||||
| NC score | 0.057693 (rank : 79) | θ value | θ > 10 (rank : 64) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C0K5, Q6P6K8 | Gene names | Slc25a16, Gda | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Grave disease carrier protein homolog (GDC) (Mitochondrial solute carrier protein homolog) (Solute carrier family 25 member 16). | |||||
|
MFRN2_HUMAN
|
||||||
| NC score | 0.057687 (rank : 80) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96A46, Q4VBZ0, Q5T777, Q86VX5, Q969G8, Q9H2J3 | Gene names | SLC25A28, MFRN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitoferrin-2 (Mitochondrial iron transporter 2) (Solute carrier family 25 member 28) (Mitochondrial RNA-splicing protein 3/4 homolog) (MRS3/4) (hMRS3/4). | |||||
|
DNC_MOUSE
|
||||||
| NC score | 0.057281 (rank : 81) | θ value | θ > 10 (rank : 60) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DAM5, Q3T9C4 | Gene names | Slc25a19 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial deoxynucleotide carrier (Solute carrier family 25 member 19). | |||||
|
MFRN2_MOUSE
|
||||||
| NC score | 0.057259 (rank : 82) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R0Z5, Q8K4J8, Q8K4J9, Q8R578 | Gene names | Slc25a28, Mfrn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitoferrin-2 (Mitochondrial iron transporter 2) (Solute carrier family 25 member 28) (Mitochondrial RNA-splicing protein 3/4 homolog) (MRS3/4). | |||||
|
MFRN1_HUMAN
|
||||||
| NC score | 0.057037 (rank : 83) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NYZ2, Q53FT7, Q69YJ8, Q969S1, Q9P0J2 | Gene names | SLC25A37, MFRN, MSCP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitoferrin-1 (Mitochondrial iron transporter 1) (Solute carrier family 25 member 37) (Mitochondrial solute carrier protein). | |||||
|
ADT3_HUMAN
|
||||||
| NC score | 0.056963 (rank : 84) | θ value | θ > 10 (rank : 46) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P12236, Q96C49 | Gene names | SLC25A6, ANT3 | |||
|
Domain Architecture |

|
|||||
| Description | ADP/ATP translocase 3 (Adenine nucleotide translocator 2) (ANT 3) (ADP,ATP carrier protein 3) (Solute carrier family 25 member 6) (ADP,ATP carrier protein, isoform T2). | |||||
|
CALL5_HUMAN
|
||||||
| NC score | 0.055492 (rank : 85) | θ value | θ > 10 (rank : 47) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9NZT1, Q8IXU8 | Gene names | CALML5, CLSP | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-like protein 5 (Calmodulin-like skin protein). | |||||
|
ADT2_MOUSE
|
||||||
| NC score | 0.055275 (rank : 86) | θ value | θ > 10 (rank : 45) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P51881, Q61311 | Gene names | Slc25a5, Ant2 | |||
|
Domain Architecture |

|
|||||
| Description | ADP/ATP translocase 2 (Adenine nucleotide translocator 2) (ANT 2) (ADP,ATP carrier protein 2) (Solute carrier family 25 member 5). | |||||
|
ADT2_HUMAN
|
||||||
| NC score | 0.055103 (rank : 87) | θ value | θ > 10 (rank : 44) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P05141, O43350 | Gene names | SLC25A5, ANT2 | |||
|
Domain Architecture |

|
|||||
| Description | ADP/ATP translocase 2 (Adenine nucleotide translocator 2) (ANT 2) (ADP,ATP carrier protein 2) (Solute carrier family 25 member 5) (ADP,ATP carrier protein, fibroblast isoform). | |||||
|
CETN2_MOUSE
|
||||||
| NC score | 0.054491 (rank : 88) | θ value | θ > 10 (rank : 52) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9R1K9, Q3UBB4, Q9CWM0 | Gene names | Cetn2, Calt | |||
|
Domain Architecture |

|
|||||
| Description | Centrin-2 (Caltractin isoform 1). | |||||
|
UCP5_HUMAN
|
||||||
| NC score | 0.054104 (rank : 89) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95258, Q9HC60, Q9HC61 | Gene names | SLC25A14, BMCP1, UCP5 | |||
|
Domain Architecture |

|
|||||
| Description | Brain mitochondrial carrier protein 1 (BMCP-1) (Mitochondrial uncoupling protein 5) (UCP 5) (Solute carrier family 25 member 14). | |||||
|
UCP5_MOUSE
|
||||||
| NC score | 0.053866 (rank : 90) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Z2B2, Q9ESI8 | Gene names | Slc25a14, Bmcp1, Ucp5 | |||
|
Domain Architecture |

|
|||||
| Description | Brain mitochondrial carrier protein 1 (BMCP-1) (Mitochondrial uncoupling protein 5) (UCP 5) (Solute carrier family 25 member 14). | |||||
|
DNC_HUMAN
|
||||||
| NC score | 0.052810 (rank : 91) | θ value | θ > 10 (rank : 59) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HC21 | Gene names | SLC25A19, DNC, MUP1 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial deoxynucleotide carrier (Solute carrier family 25 member 19) (Mitochondrial uncoupling protein 1). | |||||
|
MFTC_MOUSE
|
||||||
| NC score | 0.051837 (rank : 92) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BMG8, Q3TCM5 | Gene names | Slc25a32, Mftc | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial folate transporter/carrier (Solute carrier family 25 member 32). | |||||
|
CETN3_HUMAN
|
||||||
| NC score | 0.050868 (rank : 93) | θ value | θ > 10 (rank : 53) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O15182, Q9BS23 | Gene names | CETN3, CEN3 | |||
|
Domain Architecture |

|
|||||
| Description | Centrin-3. | |||||
|
KIP2_MOUSE
|
||||||
| NC score | 0.050775 (rank : 94) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9Z309 | Gene names | Cib2, Kip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium and integrin-binding protein 2 (Kinase-interacting protein 2) (KIP 2). | |||||
|
CALM4_MOUSE
|
||||||
| NC score | 0.050519 (rank : 95) | θ value | θ > 10 (rank : 48) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9JM83, Q9CR31, Q9D1E9 | Gene names | Calm4, Dd112 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-4 (Calcium-binding protein Dd112). | |||||
|
DIC_MOUSE
|
||||||
| NC score | 0.050277 (rank : 96) | θ value | θ > 10 (rank : 58) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9QZD8, Q543J6, Q99LJ9, Q9D8G8 | Gene names | Slc25a10, Dic | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial dicarboxylate carrier (Solute carrier family 25 member 10). | |||||
|
MPCP_HUMAN
|
||||||
| NC score | 0.050217 (rank : 97) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q00325, Q96A03 | Gene names | SLC25A3, PHC | |||
|
Domain Architecture |

|
|||||
| Description | Phosphate carrier protein, mitochondrial precursor (PTP) (Solute carrier family 25 member 3). | |||||
|
CABP2_MOUSE
|
||||||
| NC score | 0.050090 (rank : 98) | θ value | 4.03905 (rank : 29) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9JLK4, Q9JLK5 | Gene names | Cabp2 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 2 (CaBP2). | |||||
|
CABP4_MOUSE
|
||||||
| NC score | 0.046651 (rank : 99) | θ value | 6.88961 (rank : 34) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8VHC5 | Gene names | Cabp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-binding protein 4 (CaBP4). | |||||
|
CABP2_HUMAN
|
||||||
| NC score | 0.046342 (rank : 100) | θ value | 5.27518 (rank : 32) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9NPB3 | Gene names | CABP2 | |||
|
Domain Architecture |
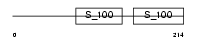
|
|||||
| Description | Calcium-binding protein 2 (CaBP2). | |||||
|
CABP4_HUMAN
|
||||||
| NC score | 0.043733 (rank : 101) | θ value | 6.88961 (rank : 33) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P57796, Q8N4Z2, Q8WWY5 | Gene names | CABP4 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 4 (CaBP4). | |||||
|
TBCD8_HUMAN
|
||||||
| NC score | 0.040079 (rank : 102) | θ value | 0.813845 (rank : 20) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O95759, Q9UQ32 | Gene names | TBC1D8, VRP | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 8 (Vascular Rab-GAP/TBC-containing protein) (AD 3). | |||||
|
PI51A_HUMAN
|
||||||
| NC score | 0.013896 (rank : 103) | θ value | 3.0926 (rank : 27) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99755, Q99754, Q99756 | Gene names | PIP5K1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 5-kinase type-1 alpha (EC 2.7.1.68) (Phosphatidylinositol-4-phosphate 5-kinase type I alpha) (PtdIns(4)P- 5-kinase alpha) (PIP5KIalpha) (68 kDa type I phosphatidylinositol-4- phosphate 5-kinase alpha). | |||||
|
BPA1_MOUSE
|
||||||
| NC score | 0.009345 (rank : 104) | θ value | 4.03905 (rank : 28) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q91ZU6, Q91ZU7 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 1/2/3/4 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||