Please be patient as the page loads
|
ACCN4_HUMAN
|
||||||
| SwissProt Accessions | Q96FT7, Q53SB7, Q6GMS1, Q6PIN9, Q9NQA4 | Gene names | ACCN4, ASIC4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amiloride-sensitive cation channel 4 (Amiloride-sensitive cation channel 4, pituitary) (Acid-sensing ion channel 4). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ACCN4_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96FT7, Q53SB7, Q6GMS1, Q6PIN9, Q9NQA4 | Gene names | ACCN4, ASIC4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amiloride-sensitive cation channel 4 (Amiloride-sensitive cation channel 4, pituitary) (Acid-sensing ion channel 4). | |||||
|
ACCN4_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.998452 (rank : 2) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7TNS7, Q80XK4 | Gene names | Accn4, Asic4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amiloride-sensitive cation channel 4 (Acid-sensing ion channel 4). | |||||
|
ACCN2_MOUSE
|
||||||
| θ value | 1.74701e-128 (rank : 3) | NC score | 0.982125 (rank : 3) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6NXK8, Q50K97 | Gene names | Accn2, Asic, Asic1, Bnac2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amiloride-sensitive cation channel 2, neuronal (Acid-sensing ion channel) (Acid-sensing ion channel 1) (Brain sodium channel 2). | |||||
|
ACCN2_HUMAN
|
||||||
| θ value | 3.89193e-128 (rank : 4) | NC score | 0.982103 (rank : 4) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P78348, P78349, Q96CV2 | Gene names | ACCN2, ASIC1, BNAC2 | |||
|
Domain Architecture |

|
|||||
| Description | Amiloride-sensitive cation channel 2, neuronal (Acid-sensing ion channel 1) (ASIC1) (Brain sodium channel 2) (BNaC2). | |||||
|
ACCN3_MOUSE
|
||||||
| θ value | 4.7686e-118 (rank : 5) | NC score | 0.981657 (rank : 5) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6X1Y6, Q7TQH4 | Gene names | Accn3, Asic3, Drasic | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amiloride-sensitive cation channel 3 (Acid-sensing ion channel 3) (ASIC3) (Dorsal root ASIC) (DRASIC). | |||||
|
ACCN3_HUMAN
|
||||||
| θ value | 4.03687e-117 (rank : 6) | NC score | 0.981005 (rank : 6) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UHC3, O60263, O75906, Q59FN9, Q9UER8, Q9UHC4 | Gene names | ACCN3, ASIC3, SLNAC1, TNAC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amiloride-sensitive cation channel 3 (Acid-sensing ion channel 3) (ASIC3) (hASIC3) (Testis sodium channel 1) (hTNaC1). | |||||
|
ACCN1_HUMAN
|
||||||
| θ value | 7.12574e-114 (rank : 7) | NC score | 0.979846 (rank : 7) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q16515, Q13553, Q8N3E2 | Gene names | ACCN1, ACCN, BNAC1, MDEG | |||
|
Domain Architecture |
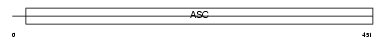
|
|||||
| Description | Amiloride-sensitive cation channel 1, neuronal (Amiloride-sensitive brain sodium channel) (Amiloride-sensitive cation channel neuronal 1) (Acid-sensing ion channel 2) (ASIC2) (Brain sodium channel 1) (BNaC1) (BNC1) (Mammalian degenerin homolog). | |||||
|
ACCN1_MOUSE
|
||||||
| θ value | 7.12574e-114 (rank : 8) | NC score | 0.979825 (rank : 8) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q925H0, Q5SUU2, Q61203 | Gene names | Accn1, Asic2, Bnac1 | |||
|
Domain Architecture |
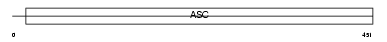
|
|||||
| Description | Amiloride-sensitive cation channel 1, neuronal (Amiloride-sensitive brain sodium channel) (Acid-sensing ion channel 2) (Brain sodium channel 1) (BNaC1). | |||||
|
SCNNB_HUMAN
|
||||||
| θ value | 5.40856e-29 (rank : 9) | NC score | 0.793852 (rank : 9) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P51168, O60891, Q96KG2, Q9UJ32, Q9UMU5 | Gene names | SCNN1B | |||
|
Domain Architecture |

|
|||||
| Description | Amiloride-sensitive sodium channel subunit beta (Epithelial Na+ channel subunit beta) (Beta ENaC) (Nonvoltage-gated sodium channel 1 subunit beta) (SCNEB) (Beta NaCH) (ENaCB). | |||||
|
SCNNA_HUMAN
|
||||||
| θ value | 2.51237e-26 (rank : 10) | NC score | 0.782689 (rank : 11) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P37088, O43271, Q9UM64 | Gene names | SCNN1A, SCNN1 | |||
|
Domain Architecture |

|
|||||
| Description | Amiloride-sensitive sodium channel subunit alpha (Epithelial Na+ channel subunit alpha) (Alpha ENaC) (Nonvoltage-gated sodium channel 1 subunit alpha) (SCNEA) (Alpha NaCH). | |||||
|
SCNNA_MOUSE
|
||||||
| θ value | 7.30988e-26 (rank : 11) | NC score | 0.776214 (rank : 13) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q61180, Q9WU37 | Gene names | Scnn1a | |||
|
Domain Architecture |
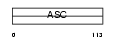
|
|||||
| Description | Amiloride-sensitive sodium channel subunit alpha (Epithelial Na+ channel subunit alpha) (Alpha ENaC) (Nonvoltage-gated sodium channel 1 subunit alpha) (SCNEA) (Alpha NaCH). | |||||
|
SCNND_HUMAN
|
||||||
| θ value | 7.30988e-26 (rank : 12) | NC score | 0.781875 (rank : 12) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P51172, Q5T7L3, Q8NA24 | Gene names | SCNN1D, DNACH | |||
|
Domain Architecture |

|
|||||
| Description | Amiloride-sensitive sodium channel subunit delta (Epithelial Na+ channel subunit delta) (Delta ENaC) (Nonvoltage-gated sodium channel 1 subunit delta) (SCNED) (Delta NaCH). | |||||
|
SCNNB_MOUSE
|
||||||
| θ value | 1.62847e-25 (rank : 13) | NC score | 0.783716 (rank : 10) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9WU38 | Gene names | Scnn1b | |||
|
Domain Architecture |

|
|||||
| Description | Amiloride-sensitive sodium channel subunit beta (Epithelial Na+ channel subunit beta) (Beta ENaC) (Nonvoltage-gated sodium channel 1 subunit beta) (SCNEB) (Beta NaCH). | |||||
|
SCNNG_MOUSE
|
||||||
| θ value | 7.56453e-23 (rank : 14) | NC score | 0.764442 (rank : 14) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9WU39 | Gene names | Scnn1g | |||
|
Domain Architecture |

|
|||||
| Description | Amiloride-sensitive sodium channel gamma-subunit (Epithelial Na+ channel subunit gamma) (Gamma ENaC) (Nonvoltage-gated sodium channel 1 subunit gamma) (SCNEG) (Gamma NaCH). | |||||
|
SCNNG_HUMAN
|
||||||
| θ value | 8.65492e-19 (rank : 15) | NC score | 0.747751 (rank : 15) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P51170, P78437, Q6PCC2, Q93023, Q93024, Q93025, Q93026, Q93027, Q96TD2 | Gene names | SCNN1G | |||
|
Domain Architecture |

|
|||||
| Description | Amiloride-sensitive sodium channel gamma-subunit (Epithelial Na+ channel subunit gamma) (Gamma ENaC) (Nonvoltage-gated sodium channel 1 subunit gamma) (SCNEG) (Gamma NaCH). | |||||
|
K0355_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 16) | NC score | 0.090273 (rank : 16) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15063 | Gene names | KIAA0355 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0355. | |||||
|
EDD1_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 17) | NC score | 0.029346 (rank : 17) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80TP3, Q698K9, Q6PEQ8, Q6PFQ9, Q80VL4, Q810V6, Q9CXE9 | Gene names | Edd1, Edd, Kiaa0896 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase EDD1 (EC 6.3.2.-) (Hyperplastic discs protein homolog). | |||||
|
NPHN_HUMAN
|
||||||
| θ value | 0.813845 (rank : 18) | NC score | 0.009360 (rank : 20) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O60500 | Gene names | NPHS1, NPHN | |||
|
Domain Architecture |

|
|||||
| Description | Nephrin precursor (Renal glomerulus-specific cell adhesion receptor). | |||||
|
MISS_MOUSE
|
||||||
| θ value | 1.38821 (rank : 19) | NC score | 0.019138 (rank : 19) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D7G9, Q7M749 | Gene names | Mapk1ip1, Miss | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAPK-interacting and spindle-stabilizing protein (Mitogen-activated protein kinase 1-interacting protein 1). | |||||
|
FOXM1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 20) | NC score | 0.008989 (rank : 21) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q08050, O43258, O43259, O43260, Q4ZGG7, Q9BRL2 | Gene names | FOXM1, FKHL16, HFH11, MPP2, WIN | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein M1 (Forkhead-related protein FKHL16) (Hepatocyte nuclear factor 3 forkhead homolog 11) (HNF-3/fork-head homolog 11) (HFH-11) (Winged helix factor from INS-1 cells) (M-phase phosphoprotein 2) (MPM-2 reactive phosphoprotein 2) (Transcription factor Trident). | |||||
|
LRRC1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 21) | NC score | 0.003977 (rank : 24) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BTT6, Q9HAC0, Q9NVF1 | Gene names | LRRC1, LANO | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 1 (LAP and no PDZ protein) (LANO adapter protein). | |||||
|
EDD1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 22) | NC score | 0.021912 (rank : 18) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95071, O94970, Q9NPL3 | Gene names | EDD1, EDD, HYD, KIAA0896 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase EDD1 (EC 6.3.2.-) (Hyperplastic discs protein homolog) (hHYD) (Progestin-induced protein). | |||||
|
KV1H_HUMAN
|
||||||
| θ value | 5.27518 (rank : 23) | NC score | 0.002464 (rank : 26) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P01600 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig kappa chain V-I region Hau. | |||||
|
CRTC1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 24) | NC score | 0.008445 (rank : 22) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6UUV9, O75114, Q6Y3A3, Q7LDZ2, Q8IUL3, Q8IZ34, Q8IZL1, Q8N6W3, Q96AI8, Q9H801 | Gene names | CRTC1, KIAA0616, MECT1, TORC1, WAMTP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CREB-regulated transcription coactivator 1 (Transducer of regulated cAMP response element-binding protein 1) (Transducer of CREB protein 1) (Mucoepidermoid carcinoma translocated protein 1). | |||||
|
PDE4A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 25) | NC score | 0.002510 (rank : 25) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O89084, Q8R078, Q9JHQ4, Q9QX48, Q9QX49, Q9QXI8 | Gene names | Pde4a | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent 3',5'-cyclic phosphodiesterase 4A (EC 3.1.4.17). | |||||
|
STAB1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 26) | NC score | 0.004000 (rank : 23) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 546 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NY15, Q8IUH0, Q8IUH1, Q93072 | Gene names | STAB1, FEEL1, KIAA0246 | |||
|
Domain Architecture |

|
|||||
| Description | Stabilin-1 precursor (FEEL-1 protein) (MS-1 antigen). | |||||
|
ACCN4_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96FT7, Q53SB7, Q6GMS1, Q6PIN9, Q9NQA4 | Gene names | ACCN4, ASIC4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amiloride-sensitive cation channel 4 (Amiloride-sensitive cation channel 4, pituitary) (Acid-sensing ion channel 4). | |||||
|
ACCN4_MOUSE
|
||||||
| NC score | 0.998452 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7TNS7, Q80XK4 | Gene names | Accn4, Asic4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amiloride-sensitive cation channel 4 (Acid-sensing ion channel 4). | |||||
|
ACCN2_MOUSE
|
||||||
| NC score | 0.982125 (rank : 3) | θ value | 1.74701e-128 (rank : 3) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6NXK8, Q50K97 | Gene names | Accn2, Asic, Asic1, Bnac2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amiloride-sensitive cation channel 2, neuronal (Acid-sensing ion channel) (Acid-sensing ion channel 1) (Brain sodium channel 2). | |||||
|
ACCN2_HUMAN
|
||||||
| NC score | 0.982103 (rank : 4) | θ value | 3.89193e-128 (rank : 4) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P78348, P78349, Q96CV2 | Gene names | ACCN2, ASIC1, BNAC2 | |||
|
Domain Architecture |

|
|||||
| Description | Amiloride-sensitive cation channel 2, neuronal (Acid-sensing ion channel 1) (ASIC1) (Brain sodium channel 2) (BNaC2). | |||||
|
ACCN3_MOUSE
|
||||||
| NC score | 0.981657 (rank : 5) | θ value | 4.7686e-118 (rank : 5) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6X1Y6, Q7TQH4 | Gene names | Accn3, Asic3, Drasic | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amiloride-sensitive cation channel 3 (Acid-sensing ion channel 3) (ASIC3) (Dorsal root ASIC) (DRASIC). | |||||
|
ACCN3_HUMAN
|
||||||
| NC score | 0.981005 (rank : 6) | θ value | 4.03687e-117 (rank : 6) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UHC3, O60263, O75906, Q59FN9, Q9UER8, Q9UHC4 | Gene names | ACCN3, ASIC3, SLNAC1, TNAC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amiloride-sensitive cation channel 3 (Acid-sensing ion channel 3) (ASIC3) (hASIC3) (Testis sodium channel 1) (hTNaC1). | |||||
|
ACCN1_HUMAN
|
||||||
| NC score | 0.979846 (rank : 7) | θ value | 7.12574e-114 (rank : 7) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q16515, Q13553, Q8N3E2 | Gene names | ACCN1, ACCN, BNAC1, MDEG | |||
|
Domain Architecture |

|
|||||
| Description | Amiloride-sensitive cation channel 1, neuronal (Amiloride-sensitive brain sodium channel) (Amiloride-sensitive cation channel neuronal 1) (Acid-sensing ion channel 2) (ASIC2) (Brain sodium channel 1) (BNaC1) (BNC1) (Mammalian degenerin homolog). | |||||
|
ACCN1_MOUSE
|
||||||
| NC score | 0.979825 (rank : 8) | θ value | 7.12574e-114 (rank : 8) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q925H0, Q5SUU2, Q61203 | Gene names | Accn1, Asic2, Bnac1 | |||
|
Domain Architecture |

|
|||||
| Description | Amiloride-sensitive cation channel 1, neuronal (Amiloride-sensitive brain sodium channel) (Acid-sensing ion channel 2) (Brain sodium channel 1) (BNaC1). | |||||
|
SCNNB_HUMAN
|
||||||
| NC score | 0.793852 (rank : 9) | θ value | 5.40856e-29 (rank : 9) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P51168, O60891, Q96KG2, Q9UJ32, Q9UMU5 | Gene names | SCNN1B | |||
|
Domain Architecture |

|
|||||
| Description | Amiloride-sensitive sodium channel subunit beta (Epithelial Na+ channel subunit beta) (Beta ENaC) (Nonvoltage-gated sodium channel 1 subunit beta) (SCNEB) (Beta NaCH) (ENaCB). | |||||
|
SCNNB_MOUSE
|
||||||
| NC score | 0.783716 (rank : 10) | θ value | 1.62847e-25 (rank : 13) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9WU38 | Gene names | Scnn1b | |||
|
Domain Architecture |

|
|||||
| Description | Amiloride-sensitive sodium channel subunit beta (Epithelial Na+ channel subunit beta) (Beta ENaC) (Nonvoltage-gated sodium channel 1 subunit beta) (SCNEB) (Beta NaCH). | |||||
|
SCNNA_HUMAN
|
||||||
| NC score | 0.782689 (rank : 11) | θ value | 2.51237e-26 (rank : 10) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P37088, O43271, Q9UM64 | Gene names | SCNN1A, SCNN1 | |||
|
Domain Architecture |

|
|||||
| Description | Amiloride-sensitive sodium channel subunit alpha (Epithelial Na+ channel subunit alpha) (Alpha ENaC) (Nonvoltage-gated sodium channel 1 subunit alpha) (SCNEA) (Alpha NaCH). | |||||
|
SCNND_HUMAN
|
||||||
| NC score | 0.781875 (rank : 12) | θ value | 7.30988e-26 (rank : 12) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P51172, Q5T7L3, Q8NA24 | Gene names | SCNN1D, DNACH | |||
|
Domain Architecture |

|
|||||
| Description | Amiloride-sensitive sodium channel subunit delta (Epithelial Na+ channel subunit delta) (Delta ENaC) (Nonvoltage-gated sodium channel 1 subunit delta) (SCNED) (Delta NaCH). | |||||
|
SCNNA_MOUSE
|
||||||
| NC score | 0.776214 (rank : 13) | θ value | 7.30988e-26 (rank : 11) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q61180, Q9WU37 | Gene names | Scnn1a | |||
|
Domain Architecture |

|
|||||
| Description | Amiloride-sensitive sodium channel subunit alpha (Epithelial Na+ channel subunit alpha) (Alpha ENaC) (Nonvoltage-gated sodium channel 1 subunit alpha) (SCNEA) (Alpha NaCH). | |||||
|
SCNNG_MOUSE
|
||||||
| NC score | 0.764442 (rank : 14) | θ value | 7.56453e-23 (rank : 14) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9WU39 | Gene names | Scnn1g | |||
|
Domain Architecture |

|
|||||
| Description | Amiloride-sensitive sodium channel gamma-subunit (Epithelial Na+ channel subunit gamma) (Gamma ENaC) (Nonvoltage-gated sodium channel 1 subunit gamma) (SCNEG) (Gamma NaCH). | |||||
|
SCNNG_HUMAN
|
||||||
| NC score | 0.747751 (rank : 15) | θ value | 8.65492e-19 (rank : 15) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P51170, P78437, Q6PCC2, Q93023, Q93024, Q93025, Q93026, Q93027, Q96TD2 | Gene names | SCNN1G | |||
|
Domain Architecture |

|
|||||
| Description | Amiloride-sensitive sodium channel gamma-subunit (Epithelial Na+ channel subunit gamma) (Gamma ENaC) (Nonvoltage-gated sodium channel 1 subunit gamma) (SCNEG) (Gamma NaCH). | |||||
|
K0355_HUMAN
|
||||||
| NC score | 0.090273 (rank : 16) | θ value | 0.0431538 (rank : 16) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15063 | Gene names | KIAA0355 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0355. | |||||
|
EDD1_MOUSE
|
||||||
| NC score | 0.029346 (rank : 17) | θ value | 0.0736092 (rank : 17) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80TP3, Q698K9, Q6PEQ8, Q6PFQ9, Q80VL4, Q810V6, Q9CXE9 | Gene names | Edd1, Edd, Kiaa0896 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase EDD1 (EC 6.3.2.-) (Hyperplastic discs protein homolog). | |||||
|
EDD1_HUMAN
|
||||||
| NC score | 0.021912 (rank : 18) | θ value | 5.27518 (rank : 22) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95071, O94970, Q9NPL3 | Gene names | EDD1, EDD, HYD, KIAA0896 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase EDD1 (EC 6.3.2.-) (Hyperplastic discs protein homolog) (hHYD) (Progestin-induced protein). | |||||
|
MISS_MOUSE
|
||||||
| NC score | 0.019138 (rank : 19) | θ value | 1.38821 (rank : 19) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D7G9, Q7M749 | Gene names | Mapk1ip1, Miss | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAPK-interacting and spindle-stabilizing protein (Mitogen-activated protein kinase 1-interacting protein 1). | |||||
|
NPHN_HUMAN
|
||||||
| NC score | 0.009360 (rank : 20) | θ value | 0.813845 (rank : 18) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O60500 | Gene names | NPHS1, NPHN | |||
|
Domain Architecture |

|
|||||
| Description | Nephrin precursor (Renal glomerulus-specific cell adhesion receptor). | |||||
|
FOXM1_HUMAN
|
||||||
| NC score | 0.008989 (rank : 21) | θ value | 1.81305 (rank : 20) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q08050, O43258, O43259, O43260, Q4ZGG7, Q9BRL2 | Gene names | FOXM1, FKHL16, HFH11, MPP2, WIN | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein M1 (Forkhead-related protein FKHL16) (Hepatocyte nuclear factor 3 forkhead homolog 11) (HNF-3/fork-head homolog 11) (HFH-11) (Winged helix factor from INS-1 cells) (M-phase phosphoprotein 2) (MPM-2 reactive phosphoprotein 2) (Transcription factor Trident). | |||||
|
CRTC1_HUMAN
|
||||||
| NC score | 0.008445 (rank : 22) | θ value | 6.88961 (rank : 24) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6UUV9, O75114, Q6Y3A3, Q7LDZ2, Q8IUL3, Q8IZ34, Q8IZL1, Q8N6W3, Q96AI8, Q9H801 | Gene names | CRTC1, KIAA0616, MECT1, TORC1, WAMTP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CREB-regulated transcription coactivator 1 (Transducer of regulated cAMP response element-binding protein 1) (Transducer of CREB protein 1) (Mucoepidermoid carcinoma translocated protein 1). | |||||
|
STAB1_HUMAN
|
||||||
| NC score | 0.004000 (rank : 23) | θ value | 8.99809 (rank : 26) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 546 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NY15, Q8IUH0, Q8IUH1, Q93072 | Gene names | STAB1, FEEL1, KIAA0246 | |||
|
Domain Architecture |

|
|||||
| Description | Stabilin-1 precursor (FEEL-1 protein) (MS-1 antigen). | |||||
|
LRRC1_HUMAN
|
||||||
| NC score | 0.003977 (rank : 24) | θ value | 2.36792 (rank : 21) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BTT6, Q9HAC0, Q9NVF1 | Gene names | LRRC1, LANO | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 1 (LAP and no PDZ protein) (LANO adapter protein). | |||||
|
PDE4A_MOUSE
|
||||||
| NC score | 0.002510 (rank : 25) | θ value | 8.99809 (rank : 25) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O89084, Q8R078, Q9JHQ4, Q9QX48, Q9QX49, Q9QXI8 | Gene names | Pde4a | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent 3',5'-cyclic phosphodiesterase 4A (EC 3.1.4.17). | |||||
|
KV1H_HUMAN
|
||||||
| NC score | 0.002464 (rank : 26) | θ value | 5.27518 (rank : 23) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P01600 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig kappa chain V-I region Hau. | |||||