Please be patient as the page loads
|
RT15_HUMAN
|
||||||
| SwissProt Accessions | P82914, Q9H2K1 | Gene names | MRPS15, RPMS15 | |||
|
Domain Architecture |

|
|||||
| Description | 28S ribosomal protein S15, mitochondrial precursor (S15mt) (MRP-S15). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
RT15_HUMAN
|
||||||
| θ value | 9.26343e-130 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P82914, Q9H2K1 | Gene names | MRPS15, RPMS15 | |||
|
Domain Architecture |

|
|||||
| Description | 28S ribosomal protein S15, mitochondrial precursor (S15mt) (MRP-S15). | |||||
|
RT15_MOUSE
|
||||||
| θ value | 2.02684e-76 (rank : 2) | NC score | 0.930814 (rank : 2) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9DC71, Q7TMG9, Q8C7F1, Q9CVE0, Q9CWX1 | Gene names | Mrps15, Rpms15 | |||
|
Domain Architecture |
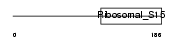
|
|||||
| Description | 28S ribosomal protein S15, mitochondrial precursor (S15mt) (MRP-S15). | |||||
|
SMC2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 3) | NC score | 0.032446 (rank : 10) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O95347, Q9P1P2 | Gene names | SMC2L1, CAPE, SMC2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (hCAP-E) (XCAP-E homolog). | |||||
|
CF170_HUMAN
|
||||||
| θ value | 1.38821 (rank : 4) | NC score | 0.064529 (rank : 3) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96NH3, Q6ZMY4, Q6ZUR7, Q8NB47 | Gene names | C6orf170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf170. | |||||
|
IFT74_HUMAN
|
||||||
| θ value | 1.38821 (rank : 5) | NC score | 0.036573 (rank : 9) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q96LB3, Q6PGQ8, Q9H643, Q9H8G7 | Gene names | IFT74, CCDC2, CMG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 74 homolog (Coiled-coil domain-containing protein 2) (Capillary morphogenesis protein 1) (CMG-1). | |||||
|
PININ_HUMAN
|
||||||
| θ value | 1.81305 (rank : 6) | NC score | 0.049234 (rank : 8) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9H307, O60899, Q53EM7, Q6P5X4, Q7KYL1, Q99738, Q9UHZ9, Q9UQR9 | Gene names | PNN, DRS, MEMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin (140 kDa nuclear and cell adhesion-related phosphoprotein) (Domain-rich serine protein) (DRS-protein) (DRSP) (Melanoma metastasis clone A protein) (Desmosome-associated protein) (SR-like protein) (Nuclear protein SDK3). | |||||
|
PININ_MOUSE
|
||||||
| θ value | 1.81305 (rank : 7) | NC score | 0.050663 (rank : 5) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O35691, Q8CD89, Q8CGU3 | Gene names | Pnn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin. | |||||
|
RAD50_MOUSE
|
||||||
| θ value | 1.81305 (rank : 8) | NC score | 0.028195 (rank : 12) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 1366 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P70388, Q6PEB0, Q8C2T7, Q9CU59 | Gene names | Rad50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (mRad50). | |||||
|
KINH_MOUSE
|
||||||
| θ value | 2.36792 (rank : 9) | NC score | 0.017550 (rank : 23) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q61768, O08711, Q61580 | Gene names | Kif5b, Khcs, Kns1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain (Ubiquitous kinesin heavy chain) (UKHC). | |||||
|
SDCG8_MOUSE
|
||||||
| θ value | 2.36792 (rank : 10) | NC score | 0.049262 (rank : 7) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q80UF4 | Gene names | Sdccag8, Cccap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serologically defined colon cancer antigen 8 (Centrosomal colon cancer autoantigen protein) (mCCCAP). | |||||
|
STMN1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 11) | NC score | 0.050940 (rank : 4) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 554 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P16949 | Gene names | STMN1, LAP18, OP18 | |||
|
Domain Architecture |

|
|||||
| Description | Stathmin (Phosphoprotein p19) (pp19) (Oncoprotein 18) (Op18) (Leukemia-associated phosphoprotein p18) (pp17) (Prosolin) (Metablastin) (Protein Pr22). | |||||
|
NEBU_HUMAN
|
||||||
| θ value | 3.0926 (rank : 12) | NC score | 0.027166 (rank : 14) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P20929, Q15346 | Gene names | NEB | |||
|
Domain Architecture |
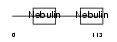
|
|||||
| Description | Nebulin. | |||||
|
SRGP2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 13) | NC score | 0.026192 (rank : 16) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O43295, Q8IX13, Q8IZV8 | Gene names | SRGAP3, ARHGAP14, KIAA0411, MEGAP, SRGAP2 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 3 (srGAP3) (srGAP2) (WAVE- associated Rac GTPase-activating protein) (WRP) (Mental disorder- associated GAP) (Rho-GTPase-activating protein 14). | |||||
|
SRGP2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 14) | NC score | 0.026362 (rank : 15) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q812A2, Q80U09, Q8BKP4, Q8BLD0, Q925I2 | Gene names | Srgap3, Arhgap14, Kiaa0411, Srgap2 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 3 (srGAP3) (srGAP2) (WAVE- associated Rac GTPase-activating protein) (WRP) (Rho-GTPase-activating protein 14). | |||||
|
STMN1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 15) | NC score | 0.049650 (rank : 6) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P54227 | Gene names | Stmn1, Lag, Lap18, Pr22 | |||
|
Domain Architecture |

|
|||||
| Description | Stathmin (Phosphoprotein p19) (pp19) (Oncoprotein 18) (Op18) (Leukemia-associated phosphoprotein p18) (pp17) (Prosolin) (Metablastin) (Pr22 protein) (Leukemia-associated gene protein). | |||||
|
WDR67_HUMAN
|
||||||
| θ value | 3.0926 (rank : 16) | NC score | 0.025558 (rank : 17) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 782 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96DN5, Q3MIR6, Q8TBP9 | Gene names | WDR67 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 67. | |||||
|
RNF8_MOUSE
|
||||||
| θ value | 5.27518 (rank : 17) | NC score | 0.021572 (rank : 19) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VC56, Q9JK13 | Gene names | Rnf8 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin ligase protein RNF8 (EC 6.3.2.-) (RING finger protein 8) (AIP37) (ActA-interacting protein 37) (LaXp180). | |||||
|
TITIN_HUMAN
|
||||||
| θ value | 5.27518 (rank : 18) | NC score | 0.005485 (rank : 26) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
XYLT1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 19) | NC score | 0.028920 (rank : 11) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86Y38, Q9H1B6 | Gene names | XYLT1, XT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Xylosyltransferase 1 (EC 2.4.2.26) (Xylosyltransferase I) (XylT-I) (XT-I) (Peptide O-xylosyltransferase 1). | |||||
|
BORIS_HUMAN
|
||||||
| θ value | 6.88961 (rank : 20) | NC score | 0.001990 (rank : 27) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 743 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NI51, Q5JUG4, Q9BZ30, Q9NQJ3 | Gene names | CTCFL, BORIS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional repressor CTCFL (CCCTC-binding factor) (Brother of the regulator of imprinted sites) (Zinc finger protein CTCF-T) (CTCF paralog). | |||||
|
TXLNB_MOUSE
|
||||||
| θ value | 6.88961 (rank : 21) | NC score | 0.023408 (rank : 18) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 640 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8VBT1, Q3UVB8, Q8BUK2 | Gene names | Txlnb, Mdp77 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-taxilin (Muscle-derived protein 77). | |||||
|
XYLT1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 22) | NC score | 0.027772 (rank : 13) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q811B1, Q9EPL1 | Gene names | Xylt1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Xylosyltransferase 1 (EC 2.4.2.26) (Xylosyltransferase I) (Peptide O- xylosyltransferase 1). | |||||
|
CASP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 23) | NC score | 0.020313 (rank : 21) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 1152 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P70403, Q7TQJ4, Q91WN6 | Gene names | Cutl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
CR034_MOUSE
|
||||||
| θ value | 8.99809 (rank : 24) | NC score | 0.020595 (rank : 20) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8CDV0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C18orf34 homolog. | |||||
|
CTRO_HUMAN
|
||||||
| θ value | 8.99809 (rank : 25) | NC score | 0.011012 (rank : 25) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 2092 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O14578, Q6XUH8, Q86UQ9, Q9UPZ7 | Gene names | CIT, KIAA0949, STK21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
CTRO_MOUSE
|
||||||
| θ value | 8.99809 (rank : 26) | NC score | 0.011614 (rank : 24) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 2344 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P49025, O88528, O88937, O88938, Q3UM99, Q8CIJ1 | Gene names | Cit | |||
|
Domain Architecture |

|
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
CUTL1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 27) | NC score | 0.019380 (rank : 22) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P53564, O08994, P70301, Q571L6, Q91ZD2 | Gene names | Cutl1, Cux, Cux1, Kiaa4047 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP) (Homeobox protein Cux). | |||||
|
RT15_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 9.26343e-130 (rank : 1) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P82914, Q9H2K1 | Gene names | MRPS15, RPMS15 | |||
|
Domain Architecture |

|
|||||
| Description | 28S ribosomal protein S15, mitochondrial precursor (S15mt) (MRP-S15). | |||||
|
RT15_MOUSE
|
||||||
| NC score | 0.930814 (rank : 2) | θ value | 2.02684e-76 (rank : 2) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9DC71, Q7TMG9, Q8C7F1, Q9CVE0, Q9CWX1 | Gene names | Mrps15, Rpms15 | |||
|
Domain Architecture |

|
|||||
| Description | 28S ribosomal protein S15, mitochondrial precursor (S15mt) (MRP-S15). | |||||
|
CF170_HUMAN
|
||||||
| NC score | 0.064529 (rank : 3) | θ value | 1.38821 (rank : 4) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96NH3, Q6ZMY4, Q6ZUR7, Q8NB47 | Gene names | C6orf170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf170. | |||||
|
STMN1_HUMAN
|
||||||
| NC score | 0.050940 (rank : 4) | θ value | 2.36792 (rank : 11) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 554 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P16949 | Gene names | STMN1, LAP18, OP18 | |||
|
Domain Architecture |

|
|||||
| Description | Stathmin (Phosphoprotein p19) (pp19) (Oncoprotein 18) (Op18) (Leukemia-associated phosphoprotein p18) (pp17) (Prosolin) (Metablastin) (Protein Pr22). | |||||
|
PININ_MOUSE
|
||||||
| NC score | 0.050663 (rank : 5) | θ value | 1.81305 (rank : 7) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O35691, Q8CD89, Q8CGU3 | Gene names | Pnn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin. | |||||
|
STMN1_MOUSE
|
||||||
| NC score | 0.049650 (rank : 6) | θ value | 3.0926 (rank : 15) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P54227 | Gene names | Stmn1, Lag, Lap18, Pr22 | |||
|
Domain Architecture |

|
|||||
| Description | Stathmin (Phosphoprotein p19) (pp19) (Oncoprotein 18) (Op18) (Leukemia-associated phosphoprotein p18) (pp17) (Prosolin) (Metablastin) (Pr22 protein) (Leukemia-associated gene protein). | |||||
|
SDCG8_MOUSE
|
||||||
| NC score | 0.049262 (rank : 7) | θ value | 2.36792 (rank : 10) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q80UF4 | Gene names | Sdccag8, Cccap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serologically defined colon cancer antigen 8 (Centrosomal colon cancer autoantigen protein) (mCCCAP). | |||||
|
PININ_HUMAN
|
||||||
| NC score | 0.049234 (rank : 8) | θ value | 1.81305 (rank : 6) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9H307, O60899, Q53EM7, Q6P5X4, Q7KYL1, Q99738, Q9UHZ9, Q9UQR9 | Gene names | PNN, DRS, MEMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin (140 kDa nuclear and cell adhesion-related phosphoprotein) (Domain-rich serine protein) (DRS-protein) (DRSP) (Melanoma metastasis clone A protein) (Desmosome-associated protein) (SR-like protein) (Nuclear protein SDK3). | |||||
|
IFT74_HUMAN
|
||||||
| NC score | 0.036573 (rank : 9) | θ value | 1.38821 (rank : 5) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q96LB3, Q6PGQ8, Q9H643, Q9H8G7 | Gene names | IFT74, CCDC2, CMG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 74 homolog (Coiled-coil domain-containing protein 2) (Capillary morphogenesis protein 1) (CMG-1). | |||||
|
SMC2_HUMAN
|
||||||
| NC score | 0.032446 (rank : 10) | θ value | 0.47712 (rank : 3) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O95347, Q9P1P2 | Gene names | SMC2L1, CAPE, SMC2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (hCAP-E) (XCAP-E homolog). | |||||
|
XYLT1_HUMAN
|
||||||
| NC score | 0.028920 (rank : 11) | θ value | 5.27518 (rank : 19) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86Y38, Q9H1B6 | Gene names | XYLT1, XT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Xylosyltransferase 1 (EC 2.4.2.26) (Xylosyltransferase I) (XylT-I) (XT-I) (Peptide O-xylosyltransferase 1). | |||||
|
RAD50_MOUSE
|
||||||
| NC score | 0.028195 (rank : 12) | θ value | 1.81305 (rank : 8) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 1366 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P70388, Q6PEB0, Q8C2T7, Q9CU59 | Gene names | Rad50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (mRad50). | |||||
|
XYLT1_MOUSE
|
||||||
| NC score | 0.027772 (rank : 13) | θ value | 6.88961 (rank : 22) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q811B1, Q9EPL1 | Gene names | Xylt1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Xylosyltransferase 1 (EC 2.4.2.26) (Xylosyltransferase I) (Peptide O- xylosyltransferase 1). | |||||
|
NEBU_HUMAN
|
||||||
| NC score | 0.027166 (rank : 14) | θ value | 3.0926 (rank : 12) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P20929, Q15346 | Gene names | NEB | |||
|
Domain Architecture |

|
|||||
| Description | Nebulin. | |||||
|
SRGP2_MOUSE
|
||||||
| NC score | 0.026362 (rank : 15) | θ value | 3.0926 (rank : 14) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q812A2, Q80U09, Q8BKP4, Q8BLD0, Q925I2 | Gene names | Srgap3, Arhgap14, Kiaa0411, Srgap2 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 3 (srGAP3) (srGAP2) (WAVE- associated Rac GTPase-activating protein) (WRP) (Rho-GTPase-activating protein 14). | |||||
|
SRGP2_HUMAN
|
||||||
| NC score | 0.026192 (rank : 16) | θ value | 3.0926 (rank : 13) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O43295, Q8IX13, Q8IZV8 | Gene names | SRGAP3, ARHGAP14, KIAA0411, MEGAP, SRGAP2 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 3 (srGAP3) (srGAP2) (WAVE- associated Rac GTPase-activating protein) (WRP) (Mental disorder- associated GAP) (Rho-GTPase-activating protein 14). | |||||
|
WDR67_HUMAN
|
||||||
| NC score | 0.025558 (rank : 17) | θ value | 3.0926 (rank : 16) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 782 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96DN5, Q3MIR6, Q8TBP9 | Gene names | WDR67 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 67. | |||||
|
TXLNB_MOUSE
|
||||||
| NC score | 0.023408 (rank : 18) | θ value | 6.88961 (rank : 21) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 640 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8VBT1, Q3UVB8, Q8BUK2 | Gene names | Txlnb, Mdp77 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-taxilin (Muscle-derived protein 77). | |||||
|
RNF8_MOUSE
|
||||||
| NC score | 0.021572 (rank : 19) | θ value | 5.27518 (rank : 17) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VC56, Q9JK13 | Gene names | Rnf8 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin ligase protein RNF8 (EC 6.3.2.-) (RING finger protein 8) (AIP37) (ActA-interacting protein 37) (LaXp180). | |||||
|
CR034_MOUSE
|
||||||
| NC score | 0.020595 (rank : 20) | θ value | 8.99809 (rank : 24) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8CDV0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C18orf34 homolog. | |||||
|
CASP_MOUSE
|
||||||
| NC score | 0.020313 (rank : 21) | θ value | 8.99809 (rank : 23) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 1152 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P70403, Q7TQJ4, Q91WN6 | Gene names | Cutl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
CUTL1_MOUSE
|
||||||
| NC score | 0.019380 (rank : 22) | θ value | 8.99809 (rank : 27) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P53564, O08994, P70301, Q571L6, Q91ZD2 | Gene names | Cutl1, Cux, Cux1, Kiaa4047 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP) (Homeobox protein Cux). | |||||
|
KINH_MOUSE
|
||||||
| NC score | 0.017550 (rank : 23) | θ value | 2.36792 (rank : 9) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q61768, O08711, Q61580 | Gene names | Kif5b, Khcs, Kns1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain (Ubiquitous kinesin heavy chain) (UKHC). | |||||
|
CTRO_MOUSE
|
||||||
| NC score | 0.011614 (rank : 24) | θ value | 8.99809 (rank : 26) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 2344 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P49025, O88528, O88937, O88938, Q3UM99, Q8CIJ1 | Gene names | Cit | |||
|
Domain Architecture |

|
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
CTRO_HUMAN
|
||||||
| NC score | 0.011012 (rank : 25) | θ value | 8.99809 (rank : 25) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 2092 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O14578, Q6XUH8, Q86UQ9, Q9UPZ7 | Gene names | CIT, KIAA0949, STK21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
TITIN_HUMAN
|
||||||
| NC score | 0.005485 (rank : 26) | θ value | 5.27518 (rank : 18) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
BORIS_HUMAN
|
||||||
| NC score | 0.001990 (rank : 27) | θ value | 6.88961 (rank : 20) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 743 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NI51, Q5JUG4, Q9BZ30, Q9NQJ3 | Gene names | CTCFL, BORIS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional repressor CTCFL (CCCTC-binding factor) (Brother of the regulator of imprinted sites) (Zinc finger protein CTCF-T) (CTCF paralog). | |||||