Please be patient as the page loads
|
ROD1_HUMAN
|
||||||
| SwissProt Accessions | O95758, Q86YB3, Q86YH9 | Gene names | ROD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of differentiation 1 (Rod1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PTBP1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.975550 (rank : 6) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P26599 | Gene names | PTBP1, PTB | |||
|
Domain Architecture |

|
|||||
| Description | Polypyrimidine tract-binding protein 1 (PTB) (Heterogeneous nuclear ribonucleoprotein I) (hnRNP I) (57 kDa RNA-binding protein PPTB-1). | |||||
|
PTBP1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.988258 (rank : 3) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P17225 | Gene names | Ptbp1, Ptb | |||
|
Domain Architecture |

|
|||||
| Description | Polypyrimidine tract-binding protein 1 (PTB) (Heterogeneous nuclear ribonucleoprotein I) (hnRNP I). | |||||
|
PTBP2_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.982471 (rank : 4) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9UKA9, Q8N0Z1, Q8N160, Q8NFB0, Q8NFB1, Q969N9, Q96Q76 | Gene names | PTBP2, NPTB, PTB, PTBLP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypyrimidine tract-binding protein 2 (Neurally-enriched homolog of PTB) (Neural polypyrimidine tract-binding protein) (PTB-like protein). | |||||
|
PTBP2_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.982377 (rank : 5) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q91Z31, Q7TNW7, Q9CUW2, Q9QYC2, Q9R0V9 | Gene names | Ptbp2, Brptb, Nptb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypyrimidine tract-binding protein 2 (Brain-enriched polypyrimidine tract-binding protein) (Brain-enriched PTB) (RRM-type RNA-binding protein brPTB) (Neural polypyrimidine tract-binding protein). | |||||
|
ROD1_HUMAN
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O95758, Q86YB3, Q86YH9 | Gene names | ROD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of differentiation 1 (Rod1). | |||||
|
ROD1_MOUSE
|
||||||
| θ value | 0 (rank : 6) | NC score | 0.998290 (rank : 2) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8BHD7, Q923C3 | Gene names | Rod1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of differentiation 1 (Rod1). | |||||
|
HNRLL_MOUSE
|
||||||
| θ value | 5.96599e-36 (rank : 7) | NC score | 0.799804 (rank : 8) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q921F4, Q8BIP6, Q99J40 | Gene names | Hnrpll | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heterogeneous nuclear ribonucleoprotein L-like. | |||||
|
HNRLL_HUMAN
|
||||||
| θ value | 2.96089e-35 (rank : 8) | NC score | 0.802813 (rank : 7) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8WVV9, Q659B9, Q8IVH5, Q8IVH6, Q96HR5 | Gene names | HNRPLL, SRRF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heterogeneous nuclear ribonucleoprotein L-like (Stromal RNA-regulating factor) (BLOCK24 protein). | |||||
|
HNRPL_HUMAN
|
||||||
| θ value | 3.86705e-35 (rank : 9) | NC score | 0.766507 (rank : 9) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P14866, Q9H3P3 | Gene names | HNRPL | |||
|
Domain Architecture |
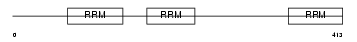
|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein L (hnRNP L). | |||||
|
HNRPL_MOUSE
|
||||||
| θ value | 6.59618e-35 (rank : 10) | NC score | 0.765728 (rank : 10) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8R081, O54789, Q8K0S7 | Gene names | Hnrpl | |||
|
Domain Architecture |
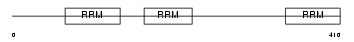
|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein L (hnRNP L). | |||||
|
MATR3_HUMAN
|
||||||
| θ value | 1.09739e-05 (rank : 11) | NC score | 0.308785 (rank : 12) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P43243, Q9UHW0, Q9UQ27 | Gene names | MATR3, KIAA0723 | |||
|
Domain Architecture |

|
|||||
| Description | Matrin-3. | |||||
|
MATR3_MOUSE
|
||||||
| θ value | 1.09739e-05 (rank : 12) | NC score | 0.311313 (rank : 11) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8K310 | Gene names | Matr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Matrin-3. | |||||
|
ELAV4_MOUSE
|
||||||
| θ value | 9.29e-05 (rank : 13) | NC score | 0.155041 (rank : 17) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q61701, O55010 | Gene names | Elavl4, Hud | |||
|
Domain Architecture |
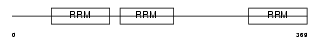
|
|||||
| Description | ELAV-like protein 4 (Paraneoplastic encephalomyelitis antigen HuD) (Hu-antigen D). | |||||
|
ELAV4_HUMAN
|
||||||
| θ value | 0.000158464 (rank : 14) | NC score | 0.153747 (rank : 18) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P26378, Q96J74 | Gene names | ELAVL4, HUD, PNEM | |||
|
Domain Architecture |
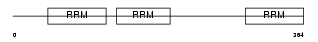
|
|||||
| Description | ELAV-like protein 4 (Paraneoplastic encephalomyelitis antigen HuD) (Hu-antigen D). | |||||
|
ZN638_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 15) | NC score | 0.184435 (rank : 16) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q14966, Q53R34, Q5XJ05, Q68DP3, Q6P2H2, Q7Z3T7, Q8NF92, Q8TCA1, Q9H2G1, Q9NP37 | Gene names | ZNF638, NP220, ZFML | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 638 (Nuclear protein 220) (Zinc-finger matrin-like protein) (Cutaneous T-cell lymphoma-associated antigen se33-1) (CTCL tumor antigen se33-1). | |||||
|
HTSF1_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 16) | NC score | 0.128998 (rank : 29) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O43719, Q59G06, Q99730 | Gene names | HTATSF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 (Tat-SF1). | |||||
|
ZN638_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 17) | NC score | 0.198510 (rank : 15) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q61464, Q6DFV9, Q8C941 | Gene names | Znf638, Np220, Zfml | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 638 (Nuclear protein 220) (Zinc-finger matrin-like protein). | |||||
|
ELAV3_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 18) | NC score | 0.148338 (rank : 20) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q14576, Q16135, Q96CL8, Q96QS9 | Gene names | ELAVL3, HUC, PLE21 | |||
|
Domain Architecture |
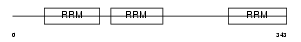
|
|||||
| Description | ELAV-like protein 3 (Hu-antigen C) (HuC) (Paraneoplastic cerebellar degeneration-associated antigen) (Paraneoplastic limbic encephalitis antigen 21). | |||||
|
ELAV3_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 19) | NC score | 0.146292 (rank : 26) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q60900, Q60901 | Gene names | Elavl3, Huc | |||
|
Domain Architecture |
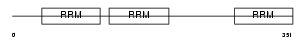
|
|||||
| Description | ELAV-like protein 3 (Hu-antigen C) (HuC). | |||||
|
RBM19_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 20) | NC score | 0.105865 (rank : 37) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9Y4C8, Q9BPY6, Q9UFN5 | Gene names | RBM19, KIAA0682 | |||
|
Domain Architecture |

|
|||||
| Description | Probable RNA-binding protein 19 (RNA-binding motif protein 19). | |||||
|
ELAV1_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 21) | NC score | 0.131039 (rank : 28) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q15717, Q9BTT1 | Gene names | ELAVL1, HUR | |||
|
Domain Architecture |
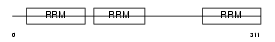
|
|||||
| Description | ELAV-like protein 1 (Hu-antigen R) (HuR). | |||||
|
ELAV2_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 22) | NC score | 0.146839 (rank : 25) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q12926, Q13235, Q59G15, Q8NEM4, Q9H1Q8 | Gene names | ELAVL2, HUB | |||
|
Domain Architecture |
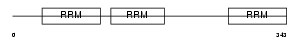
|
|||||
| Description | ELAV-like protein 2 (Hu-antigen B) (HuB) (ELAV-like neuronal protein 1) (Nervous system-specific RNA-binding protein Hel-N1). | |||||
|
ELAV2_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 23) | NC score | 0.147177 (rank : 24) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q60899 | Gene names | Elavl2, Hub | |||
|
Domain Architecture |
|
|||||
| Description | ELAV-like protein 2 (Hu-antigen B) (HuB) (ELAV-like neuronal protein 1) (Nervous system-specific RNA-binding protein Mel-N1). | |||||
|
NONO_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 24) | NC score | 0.147344 (rank : 23) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q15233, O00201, P30807, Q12786, Q9BQC5 | Gene names | NONO, NRB54 | |||
|
Domain Architecture |
|
|||||
| Description | Non-POU domain-containing octamer-binding protein (NonO protein) (54 kDa nuclear RNA- and DNA-binding protein) (p54(nrb)) (p54nrb) (55 kDa nuclear protein) (NMT55) (DNA-binding p52/p100 complex, 52 kDa subunit). | |||||
|
NONO_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 25) | NC score | 0.148755 (rank : 19) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q99K48, Q63887, Q9CYQ4, Q9DBP2 | Gene names | Nono | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Non-POU domain-containing octamer-binding protein (NonO protein). | |||||
|
RU2B_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 26) | NC score | 0.209438 (rank : 13) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P08579, Q9UJD4 | Gene names | SNRPB2 | |||
|
Domain Architecture |
|
|||||
| Description | U2 small nuclear ribonucleoprotein B''. | |||||
|
RU2B_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 27) | NC score | 0.206925 (rank : 14) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9CQI7, Q9CW35, Q9CZ66 | Gene names | Snrpb2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U2 small nuclear ribonucleoprotein B''. | |||||
|
HTSF1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 28) | NC score | 0.110145 (rank : 32) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8BGC0, Q1WWK0, Q9CT41, Q9DAU3 | Gene names | Htatsf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 homolog. | |||||
|
RBM7_HUMAN
|
||||||
| θ value | 0.21417 (rank : 29) | NC score | 0.148315 (rank : 21) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Y580 | Gene names | RBM7 | |||
|
Domain Architecture |
|
|||||
| Description | RNA-binding protein 7 (RNA-binding motif protein 7). | |||||
|
NKX26_MOUSE
|
||||||
| θ value | 0.279714 (rank : 30) | NC score | 0.017701 (rank : 90) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P43688 | Gene names | Nkx2-6, Nkx-2.6, Nkx2f | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Nkx-2.6 (Homeobox protein NK-2 homolog F). | |||||
|
SNRPA_MOUSE
|
||||||
| θ value | 0.279714 (rank : 31) | NC score | 0.117517 (rank : 31) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q62189, Q52LA2 | Gene names | Snrpa, Rnu1a-1 | |||
|
Domain Architecture |

|
|||||
| Description | U1 small nuclear ribonucleoprotein A (U1 snRNP protein A) (U1A protein) (U1-A). | |||||
|
RBM19_MOUSE
|
||||||
| θ value | 0.365318 (rank : 32) | NC score | 0.108144 (rank : 35) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8R3C6, Q8BHR0, Q9CW63 | Gene names | Rbm19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable RNA-binding protein 19 (RNA-binding motif protein 19). | |||||
|
SAFB1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 33) | NC score | 0.082355 (rank : 47) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 493 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q15424, O60406, Q59HH8 | Gene names | SAFB, HAP, HET, SAFB1 | |||
|
Domain Architecture |

|
|||||
| Description | Scaffold attachment factor B (Scaffold attachment factor B1) (SAF-B) (HSP27 estrogen response element-TATA box-binding protein) (HSP27 ERE- TATA-binding protein). | |||||
|
ZCRB1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 34) | NC score | 0.148279 (rank : 22) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8TBF4, Q6PJX0 | Gene names | ZCRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC-type and RNA-binding motif-containing protein 1 (U11/U12 snRNP 31 kDa protein) (U11/U12-31K). | |||||
|
RBM7_MOUSE
|
||||||
| θ value | 0.62314 (rank : 35) | NC score | 0.135437 (rank : 27) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9CQT2, Q3UB91, Q7TQE3 | Gene names | Rbm7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 7 (RNA-binding motif protein 7). | |||||
|
CO4A2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 36) | NC score | 0.011751 (rank : 93) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P08122, Q61375 | Gene names | Col4a2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(IV) chain precursor. | |||||
|
ZCRB1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 37) | NC score | 0.121036 (rank : 30) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9CZ96, Q3TDL3 | Gene names | Zcrb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC-type and RNA-binding motif-containing protein 1 (MADP-1). | |||||
|
MUC5B_HUMAN
|
||||||
| θ value | 1.38821 (rank : 38) | NC score | 0.011191 (rank : 94) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
PM14_HUMAN
|
||||||
| θ value | 1.38821 (rank : 39) | NC score | 0.109602 (rank : 33) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Y3B4 | Gene names | SF3B14 | |||
|
Domain Architecture |
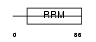
|
|||||
| Description | Pre-mRNA branch site protein p14 (SF3B 14 kDa subunit). | |||||
|
PM14_MOUSE
|
||||||
| θ value | 1.38821 (rank : 40) | NC score | 0.109602 (rank : 34) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P59708, Q3U9T9 | Gene names | Sf3b14 | |||
|
Domain Architecture |
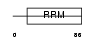
|
|||||
| Description | Pre-mRNA branch site protein p14 (SF3B 14 kDa subunit). | |||||
|
COJA1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 41) | NC score | 0.014211 (rank : 91) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q14993, Q00559, Q05850, Q12885, Q13676, Q5JUF0, Q5T424, Q9H572, Q9NPZ2, Q9NQP2 | Gene names | COL19A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XIX) chain precursor (Collagen alpha-1(Y) chain). | |||||
|
ELAV1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 42) | NC score | 0.107808 (rank : 36) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P70372, Q60745 | Gene names | Elavl1, Elra, Hua | |||
|
Domain Architecture |
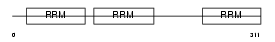
|
|||||
| Description | ELAV-like protein 1 (Hu-antigen R) (HuR) (Elav-like generic protein) (MelG). | |||||
|
RXRA_HUMAN
|
||||||
| θ value | 2.36792 (rank : 43) | NC score | 0.003870 (rank : 98) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P19793, Q2V504 | Gene names | RXRA, NR2B1 | |||
|
Domain Architecture |
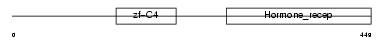
|
|||||
| Description | Retinoic acid receptor RXR-alpha (Retinoid X receptor alpha). | |||||
|
P3C2G_MOUSE
|
||||||
| θ value | 3.0926 (rank : 44) | NC score | 0.021423 (rank : 87) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O70167, O70168 | Gene names | Pik3c2g | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing gamma polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-gamma) (PtdIns-3-kinase C2 gamma) (PI3K-C2gamma). | |||||
|
RBMS1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 45) | NC score | 0.074676 (rank : 49) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q91W59, Q6PEU6, Q6PHC2, Q9WTK4 | Gene names | Rbms1, Mssp, Mssp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding motif, single-stranded-interacting protein 1 (Single- stranded DNA-binding protein MSSP-1). | |||||
|
IF39_MOUSE
|
||||||
| θ value | 4.03905 (rank : 46) | NC score | 0.046381 (rank : 83) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8JZQ9, Q922K2 | Gene names | Eif3s9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 9 (eIF-3 eta) (eIF3 p116). | |||||
|
SFRS2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 47) | NC score | 0.095314 (rank : 43) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q01130 | Gene names | SFRS2 | |||
|
Domain Architecture |
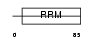
|
|||||
| Description | Splicing factor, arginine/serine-rich 2 (Splicing factor SC35) (SC-35) (Splicing component, 35 kDa) (Protein PR264). | |||||
|
SFRS2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 48) | NC score | 0.095314 (rank : 44) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q62093, Q542L3, Q60701 | Gene names | Sfrs2, Pr264, Sfrs10 | |||
|
Domain Architecture |
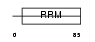
|
|||||
| Description | Splicing factor, arginine/serine-rich 2 (Splicing factor SC35) (SC-35) (Splicing component, 35 kDa) (Protein PR264) (Putative myelin regulatory factor 1) (MRF-1). | |||||
|
ACINU_HUMAN
|
||||||
| θ value | 5.27518 (rank : 49) | NC score | 0.018256 (rank : 89) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UKV3, O75158, Q9UG91, Q9UKV1, Q9UKV2 | Gene names | ACIN1, ACINUS, KIAA0670 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
ACINU_MOUSE
|
||||||
| θ value | 5.27518 (rank : 50) | NC score | 0.018424 (rank : 88) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9JIX8, Q9CSN7, Q9CSR9, Q9CSX7, Q9R046, Q9R047 | Gene names | Acin1, Acinus | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
CO8A2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 51) | NC score | 0.009288 (rank : 96) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P25318, Q68ED0, Q6KAQ4, Q6P1C4 | Gene names | Col8a2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-2(VIII) chain precursor (Endothelial collagen). | |||||
|
RBMS1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 52) | NC score | 0.069495 (rank : 51) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P29558, Q15433, Q8WV20 | Gene names | RBMS1, MSSP, MSSP1, SCR2 | |||
|
Domain Architecture |
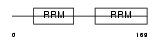
|
|||||
| Description | RNA-binding motif, single-stranded-interacting protein 1 (Single- stranded DNA-binding protein MSSP-1) (Suppressor of CDC2 with RNA- binding motif 2). | |||||
|
CO4A2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 53) | NC score | 0.009733 (rank : 95) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 454 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P08572 | Gene names | COL4A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(IV) chain precursor. | |||||
|
IF39_HUMAN
|
||||||
| θ value | 6.88961 (rank : 54) | NC score | 0.033275 (rank : 86) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P55884, Q9UMF9 | Gene names | EIF3S9 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 9 (eIF-3 eta) (eIF3 p116) (eIF3 p110) (eIF3b) (Prt1 homolog) (hPrt1). | |||||
|
MINT_HUMAN
|
||||||
| θ value | 6.88961 (rank : 55) | NC score | 0.041563 (rank : 84) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
SYNG_HUMAN
|
||||||
| θ value | 6.88961 (rank : 56) | NC score | 0.012996 (rank : 92) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UMZ2, Q6ZT17 | Gene names | AP1GBP1, SYNG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP1 subunit gamma-binding protein 1 (Gamma-synergin). | |||||
|
ATX1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 57) | NC score | 0.006982 (rank : 97) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P54254 | Gene names | Atxn1, Sca1 | |||
|
Domain Architecture |

|
|||||
| Description | Ataxin-1 (Spinocerebellar ataxia type 1 protein homolog). | |||||
|
MINT_MOUSE
|
||||||
| θ value | 8.99809 (rank : 58) | NC score | 0.038692 (rank : 85) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
SFRS3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 59) | NC score | 0.052107 (rank : 78) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P84103, O08831, P23152 | Gene names | SFRS3, SRP20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 3 (Pre-mRNA-splicing factor SRP20). | |||||
|
SFRS3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 60) | NC score | 0.052107 (rank : 79) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P84104, O08831, P23152, Q564E9 | Gene names | Sfrs3, Srp20, X16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 3 (Pre-mRNA-splicing factor SRP20) (X16 protein). | |||||
|
ACF_MOUSE
|
||||||
| θ value | θ > 10 (rank : 61) | NC score | 0.057337 (rank : 60) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q5YD48, Q5FW55, Q5YD47 | Gene names | Acf, Asp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | APOBEC1 complementation factor (APOBEC1-stimulating protein). | |||||
|
DRBP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 62) | NC score | 0.094843 (rank : 45) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8IUH3, Q6NYL0, Q8NFC9 | Gene names | DRBP1, DRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Developmentally-regulated RNA-binding protein 1 (RB-1). | |||||
|
DRBP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 63) | NC score | 0.085569 (rank : 46) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BHN5, Q8C1Z5 | Gene names | Drbp1, Drb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Developmentally-regulated RNA-binding protein 1 (RB-1). | |||||
|
FUSIP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 64) | NC score | 0.050246 (rank : 81) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O75494, O60572, Q96G09, Q96P17 | Gene names | FUSIP1, TASR | |||
|
Domain Architecture |
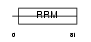
|
|||||
| Description | FUS-interacting serine-arginine-rich protein 1 (TLS-associated protein with Ser-Arg repeats) (TLS-associated protein with SR repeats) (TASR) (TLS-associated serine-arginine protein) (TLS-associated SR protein) (40 kDa SR-repressor protein) (SRrp40) (Splicing factor SRp38). | |||||
|
FUSIP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.050246 (rank : 82) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9R0U0, O70307, O88468, Q3TGW7, Q8CFZ1, Q9R0T9 | Gene names | Fusip1, Nssr | |||
|
Domain Architecture |
|
|||||
| Description | FUS-interacting serine-arginine-rich protein 1 (TLS-associated protein with Ser-Arg repeats) (TLS-associated protein with SR repeats) (TASR) (TLS-associated serine-arginine protein) (TLS-associated SR protein) (Neural-specific SR protein) (Neural-salient serine/arginine-rich protein). | |||||
|
HNRPQ_HUMAN
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.055002 (rank : 69) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O60506, Q53H05, Q5TCG2, Q8IW78, Q8N599, Q96LC1, Q96LC2, Q9Y583 | Gene names | SYNCRIP, HNRPQ, NSAP1 | |||
|
Domain Architecture |
|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein Q (hnRNP Q) (hnRNP-Q) (Synaptotagmin-binding, cytoplasmic RNA-interacting protein) (Glycine- and tyrosine-rich RNA-binding protein) (GRY-RBP) (NS1-associated protein 1). | |||||
|
HNRPQ_MOUSE
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.051879 (rank : 80) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q7TMK9, O88991, Q80YV6, Q8BGP1, Q8C5K6, Q8CGC2, Q91ZR0, Q99KU9, Q9QYF4 | Gene names | Syncrip, Hnrpq, Nsap1, Nsap1l | |||
|
Domain Architecture |
|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein Q (hnRNP Q) (hnRNP-Q) (Synaptotagmin-binding, cytoplasmic RNA-interacting protein) (Glycine- and tyrosine-rich RNA-binding protein) (GRY-RBP) (NS1-associated protein 1) (pp68). | |||||
|
MK67I_HUMAN
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.066313 (rank : 52) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9BYG3, Q8TB66, Q96ED4 | Gene names | MKI67IP, NIFK, NOPP34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MKI67 FHA domain-interacting nucleolar phosphoprotein (Nucleolar protein interacting with the FHA domain of pKI-67) (hNIFK) (Nucleolar phosphoprotein Nopp34). | |||||
|
NUCL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.055732 (rank : 65) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P09405, Q548M9, Q61991, Q99K50 | Gene names | Ncl, Nuc | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolin (Protein C23). | |||||
|
PABP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.056387 (rank : 64) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P11940, Q15097, Q93004 | Gene names | PABPC1, PAB1, PABP1, PABPC2 | |||
|
Domain Architecture |

|
|||||
| Description | Polyadenylate-binding protein 1 (Poly(A)-binding protein 1) (PABP 1). | |||||
|
PABP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.057071 (rank : 62) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P29341 | Gene names | Pabpc1, Pabp1 | |||
|
Domain Architecture |

|
|||||
| Description | Polyadenylate-binding protein 1 (Poly(A)-binding protein 1) (PABP 1). | |||||
|
PABP3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.052974 (rank : 74) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9H361, Q8NHV0, Q9H086 | Gene names | PABPC3, PABP3, PABPL3 | |||
|
Domain Architecture |

|
|||||
| Description | Polyadenylate-binding protein 3 (Poly(A)-binding protein 3) (PABP 3) (Testis-specific poly(A)-binding protein). | |||||
|
PABP4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.053080 (rank : 73) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q13310, Q6P0N3 | Gene names | PABPC4, APP1, PABP4 | |||
|
Domain Architecture |

|
|||||
| Description | Polyadenylate-binding protein 4 (Poly(A)-binding protein 4) (PABP 4) (Inducible poly(A)-binding protein) (iPABP) (Activated-platelet protein 1) (APP-1). | |||||
|
PABP5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.063529 (rank : 54) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q96DU9, Q6P529, Q9UFE5 | Gene names | PABPC5, PABP5 | |||
|
Domain Architecture |
|
|||||
| Description | Polyadenylate-binding protein 5 (Poly(A)-binding protein 5) (PABP 5). | |||||
|
RBM11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.055326 (rank : 67) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P57052 | Gene names | RBM11 | |||
|
Domain Architecture |
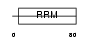
|
|||||
| Description | Putative RNA-binding protein 11 (RNA-binding motif protein 11). | |||||
|
RBM14_HUMAN
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.052114 (rank : 77) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q96PK6, O75932, Q53GV1, Q68DQ9, Q96PK5 | Gene names | RBM14, SIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 14 (RNA-binding motif protein 14) (RRM-containing coactivator activator/modulator) (Synaptotagmin-interacting protein) (SYT-interacting protein). | |||||
|
RBM14_MOUSE
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.057335 (rank : 61) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8C2Q3, Q3TJB6, Q91Z21, Q9DBI6 | Gene names | Rbm14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 14 (RNA-binding motif protein 14). | |||||
|
RBM23_HUMAN
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.065392 (rank : 53) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q86U06, Q8ND16, Q8TB88, Q8WY40, Q9BUJ1, Q9NVV7 | Gene names | RBM23, RNPC4 | |||
|
Domain Architecture |
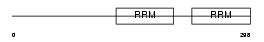
|
|||||
| Description | Probable RNA-binding protein 23 (RNA-binding motif protein 23) (RNA- binding region-containing protein 4) (Splicing factor SF2). | |||||
|
RBM28_MOUSE
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.053471 (rank : 70) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8CGC6, Q8BG25, Q8VEJ8, Q9CS22, Q9CSE6 | Gene names | Rbm28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 28 (RNA-binding motif protein 28). | |||||
|
RBM4B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.095962 (rank : 40) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9BQ04 | Gene names | RBM4B, RBM30 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 4B (RNA-binding motif protein 4B) (RNA-binding protein 30) (RNA-binding motif protein 30). | |||||
|
RBM4B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.095991 (rank : 39) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8VE92 | Gene names | Rbm4b, Rbm30 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 4B (RNA-binding motif protein 4B) (RNA-binding protein 30) (RNA-binding motif protein 30). | |||||
|
RBM4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.095685 (rank : 42) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9BWF3, O02916, Q6P1P2, Q8WU85 | Gene names | RBM4, RBM4A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 4 (RNA-binding motif protein 4) (RNA-binding motif protein 4a) (Lark homolog) (hLark). | |||||
|
RBM4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.095803 (rank : 41) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8C7Q4, O08752, Q8BN66 | Gene names | Rbm4, Rbm4a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 4 (RNA-binding motif protein 4) (RNA-binding motif protein 4a) (Lark homolog) (mLark). | |||||
|
RBMS2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.057948 (rank : 58) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q15434 | Gene names | RBMS2, SCR3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding motif, single-stranded-interacting protein 2 (Suppressor of CDC2 with RNA-binding motif 3). | |||||
|
RBMS2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.057434 (rank : 59) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8VC70 | Gene names | Rbms2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding motif, single-stranded-interacting protein 2. | |||||
|
RBMX2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.073454 (rank : 50) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9Y388, Q5JY82, Q9Y3I8 | Gene names | RBMX2 | |||
|
Domain Architecture |
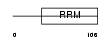
|
|||||
| Description | RNA-binding motif protein, X-linked 2. | |||||
|
RBMX2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.078134 (rank : 48) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8R0F5, Q3TI42 | Gene names | Rbmx2 | |||
|
Domain Architecture |
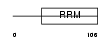
|
|||||
| Description | RNA-binding motif protein, X-linked 2. | |||||
|
RBY1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.056771 (rank : 63) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q15414, Q15377, Q8NHR0 | Gene names | RBMY1A1, RBM1, YRRM1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding motif protein, Y chromosome, family 1 member A1 (RNA- binding motif protein 1). | |||||
|
SF3B4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.058077 (rank : 57) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q15427 | Gene names | SF3B4, SAP49 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 4 (Spliceosome-associated protein 49) (SAP 49) (SF3b50) (Pre-mRNA-splicing factor SF3b 49 kDa subunit). | |||||
|
SFPQ_HUMAN
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.063110 (rank : 56) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
SFPQ_MOUSE
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.063231 (rank : 55) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8VIJ6, Q9ERW2 | Gene names | Sfpq | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit). | |||||
|
SFRS7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.052702 (rank : 75) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q16629 | Gene names | SFRS7 | |||
|
Domain Architecture |
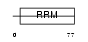
|
|||||
| Description | Splicing factor, arginine/serine-rich 7 (Splicing factor 9G8). | |||||
|
SNRPA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.096340 (rank : 38) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P09012 | Gene names | SNRPA | |||
|
Domain Architecture |

|
|||||
| Description | U1 small nuclear ribonucleoprotein A (U1 snRNP protein A) (U1A protein) (U1-A). | |||||
|
SRR35_HUMAN
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.055622 (rank : 66) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8WXF0, Q8WW25 | Gene names | SRRP35 | |||
|
Domain Architecture |
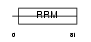
|
|||||
| Description | 35 kDa SR repressor protein (SRrp35). | |||||
|
TIA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.055154 (rank : 68) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P31483 | Gene names | TIA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolysin TIA-1 isoform p40 (RNA-binding protein TIA-1) (p40-TIA-1) [Contains: Nucleolysin TIA-1 isoform p15 (p15-TIA-1)]. | |||||
|
TIA1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.052442 (rank : 76) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P52912 | Gene names | Tia1, Tia | |||
|
Domain Architecture |
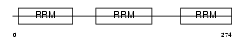
|
|||||
| Description | Nucleolysin TIA-1 (RNA-binding protein TIA-1). | |||||
|
TRA2B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.053145 (rank : 71) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P62995, O15449, Q15815, Q64283 | Gene names | SFRS10, TRA2B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine/serine-rich-splicing factor 10 (Transformer-2-beta) (HTRA2- beta) (Transformer 2 protein homolog). | |||||
|
TRA2B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.053145 (rank : 72) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P62996, O15449, Q15815, Q64283 | Gene names | Sfrs10, Silg41, Tra2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine/serine-rich-splicing factor 10 (Transformer-2-beta) (HTRA2- beta) (Transformer 2 protein homolog) (Silica-induced gene 41 protein) (SIG-41). | |||||
|
ROD1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O95758, Q86YB3, Q86YH9 | Gene names | ROD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of differentiation 1 (Rod1). | |||||
|
ROD1_MOUSE
|
||||||
| NC score | 0.998290 (rank : 2) | θ value | 0 (rank : 6) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8BHD7, Q923C3 | Gene names | Rod1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of differentiation 1 (Rod1). | |||||
|
PTBP1_MOUSE
|
||||||
| NC score | 0.988258 (rank : 3) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P17225 | Gene names | Ptbp1, Ptb | |||
|
Domain Architecture |

|
|||||
| Description | Polypyrimidine tract-binding protein 1 (PTB) (Heterogeneous nuclear ribonucleoprotein I) (hnRNP I). | |||||
|
PTBP2_HUMAN
|
||||||
| NC score | 0.982471 (rank : 4) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9UKA9, Q8N0Z1, Q8N160, Q8NFB0, Q8NFB1, Q969N9, Q96Q76 | Gene names | PTBP2, NPTB, PTB, PTBLP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypyrimidine tract-binding protein 2 (Neurally-enriched homolog of PTB) (Neural polypyrimidine tract-binding protein) (PTB-like protein). | |||||
|
PTBP2_MOUSE
|
||||||
| NC score | 0.982377 (rank : 5) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q91Z31, Q7TNW7, Q9CUW2, Q9QYC2, Q9R0V9 | Gene names | Ptbp2, Brptb, Nptb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypyrimidine tract-binding protein 2 (Brain-enriched polypyrimidine tract-binding protein) (Brain-enriched PTB) (RRM-type RNA-binding protein brPTB) (Neural polypyrimidine tract-binding protein). | |||||
|
PTBP1_HUMAN
|
||||||
| NC score | 0.975550 (rank : 6) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P26599 | Gene names | PTBP1, PTB | |||
|
Domain Architecture |

|
|||||
| Description | Polypyrimidine tract-binding protein 1 (PTB) (Heterogeneous nuclear ribonucleoprotein I) (hnRNP I) (57 kDa RNA-binding protein PPTB-1). | |||||
|
HNRLL_HUMAN
|
||||||
| NC score | 0.802813 (rank : 7) | θ value | 2.96089e-35 (rank : 8) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8WVV9, Q659B9, Q8IVH5, Q8IVH6, Q96HR5 | Gene names | HNRPLL, SRRF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heterogeneous nuclear ribonucleoprotein L-like (Stromal RNA-regulating factor) (BLOCK24 protein). | |||||
|
HNRLL_MOUSE
|
||||||
| NC score | 0.799804 (rank : 8) | θ value | 5.96599e-36 (rank : 7) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q921F4, Q8BIP6, Q99J40 | Gene names | Hnrpll | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heterogeneous nuclear ribonucleoprotein L-like. | |||||
|
HNRPL_HUMAN
|
||||||
| NC score | 0.766507 (rank : 9) | θ value | 3.86705e-35 (rank : 9) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P14866, Q9H3P3 | Gene names | HNRPL | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein L (hnRNP L). | |||||
|
HNRPL_MOUSE
|
||||||
| NC score | 0.765728 (rank : 10) | θ value | 6.59618e-35 (rank : 10) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8R081, O54789, Q8K0S7 | Gene names | Hnrpl | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein L (hnRNP L). | |||||
|
MATR3_MOUSE
|
||||||
| NC score | 0.311313 (rank : 11) | θ value | 1.09739e-05 (rank : 12) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8K310 | Gene names | Matr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Matrin-3. | |||||
|
MATR3_HUMAN
|
||||||
| NC score | 0.308785 (rank : 12) | θ value | 1.09739e-05 (rank : 11) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P43243, Q9UHW0, Q9UQ27 | Gene names | MATR3, KIAA0723 | |||
|
Domain Architecture |

|
|||||
| Description | Matrin-3. | |||||
|
RU2B_HUMAN
|
||||||
| NC score | 0.209438 (rank : 13) | θ value | 0.0736092 (rank : 26) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P08579, Q9UJD4 | Gene names | SNRPB2 | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein B''. | |||||
|
RU2B_MOUSE
|
||||||
| NC score | 0.206925 (rank : 14) | θ value | 0.0736092 (rank : 27) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9CQI7, Q9CW35, Q9CZ66 | Gene names | Snrpb2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U2 small nuclear ribonucleoprotein B''. | |||||
|
ZN638_MOUSE
|
||||||
| NC score | 0.198510 (rank : 15) | θ value | 0.0148317 (rank : 17) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q61464, Q6DFV9, Q8C941 | Gene names | Znf638, Np220, Zfml | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 638 (Nuclear protein 220) (Zinc-finger matrin-like protein). | |||||
|
ZN638_HUMAN
|
||||||
| NC score | 0.184435 (rank : 16) | θ value | 0.00509761 (rank : 15) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q14966, Q53R34, Q5XJ05, Q68DP3, Q6P2H2, Q7Z3T7, Q8NF92, Q8TCA1, Q9H2G1, Q9NP37 | Gene names | ZNF638, NP220, ZFML | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 638 (Nuclear protein 220) (Zinc-finger matrin-like protein) (Cutaneous T-cell lymphoma-associated antigen se33-1) (CTCL tumor antigen se33-1). | |||||
|
ELAV4_MOUSE
|
||||||
| NC score | 0.155041 (rank : 17) | θ value | 9.29e-05 (rank : 13) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q61701, O55010 | Gene names | Elavl4, Hud | |||
|
Domain Architecture |

|
|||||
| Description | ELAV-like protein 4 (Paraneoplastic encephalomyelitis antigen HuD) (Hu-antigen D). | |||||
|
ELAV4_HUMAN
|
||||||
| NC score | 0.153747 (rank : 18) | θ value | 0.000158464 (rank : 14) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P26378, Q96J74 | Gene names | ELAVL4, HUD, PNEM | |||
|
Domain Architecture |

|
|||||
| Description | ELAV-like protein 4 (Paraneoplastic encephalomyelitis antigen HuD) (Hu-antigen D). | |||||
|
NONO_MOUSE
|
||||||
| NC score | 0.148755 (rank : 19) | θ value | 0.0563607 (rank : 25) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q99K48, Q63887, Q9CYQ4, Q9DBP2 | Gene names | Nono | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Non-POU domain-containing octamer-binding protein (NonO protein). | |||||
|
ELAV3_HUMAN
|
||||||
| NC score | 0.148338 (rank : 20) | θ value | 0.0252991 (rank : 18) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q14576, Q16135, Q96CL8, Q96QS9 | Gene names | ELAVL3, HUC, PLE21 | |||
|
Domain Architecture |

|
|||||
| Description | ELAV-like protein 3 (Hu-antigen C) (HuC) (Paraneoplastic cerebellar degeneration-associated antigen) (Paraneoplastic limbic encephalitis antigen 21). | |||||
|
RBM7_HUMAN
|
||||||
| NC score | 0.148315 (rank : 21) | θ value | 0.21417 (rank : 29) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Y580 | Gene names | RBM7 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 7 (RNA-binding motif protein 7). | |||||
|
ZCRB1_HUMAN
|
||||||
| NC score | 0.148279 (rank : 22) | θ value | 0.47712 (rank : 34) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8TBF4, Q6PJX0 | Gene names | ZCRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC-type and RNA-binding motif-containing protein 1 (U11/U12 snRNP 31 kDa protein) (U11/U12-31K). | |||||
|
NONO_HUMAN
|
||||||
| NC score | 0.147344 (rank : 23) | θ value | 0.0563607 (rank : 24) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q15233, O00201, P30807, Q12786, Q9BQC5 | Gene names | NONO, NRB54 | |||
|
Domain Architecture |

|
|||||
| Description | Non-POU domain-containing octamer-binding protein (NonO protein) (54 kDa nuclear RNA- and DNA-binding protein) (p54(nrb)) (p54nrb) (55 kDa nuclear protein) (NMT55) (DNA-binding p52/p100 complex, 52 kDa subunit). | |||||
|
ELAV2_MOUSE
|
||||||
| NC score | 0.147177 (rank : 24) | θ value | 0.0563607 (rank : 23) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q60899 | Gene names | Elavl2, Hub | |||
|
Domain Architecture |

|
|||||
| Description | ELAV-like protein 2 (Hu-antigen B) (HuB) (ELAV-like neuronal protein 1) (Nervous system-specific RNA-binding protein Mel-N1). | |||||
|
ELAV2_HUMAN
|
||||||
| NC score | 0.146839 (rank : 25) | θ value | 0.0431538 (rank : 22) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q12926, Q13235, Q59G15, Q8NEM4, Q9H1Q8 | Gene names | ELAVL2, HUB | |||
|
Domain Architecture |

|
|||||
| Description | ELAV-like protein 2 (Hu-antigen B) (HuB) (ELAV-like neuronal protein 1) (Nervous system-specific RNA-binding protein Hel-N1). | |||||
|
ELAV3_MOUSE
|
||||||
| NC score | 0.146292 (rank : 26) | θ value | 0.0252991 (rank : 19) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q60900, Q60901 | Gene names | Elavl3, Huc | |||
|
Domain Architecture |

|
|||||
| Description | ELAV-like protein 3 (Hu-antigen C) (HuC). | |||||
|
RBM7_MOUSE
|
||||||
| NC score | 0.135437 (rank : 27) | θ value | 0.62314 (rank : 35) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9CQT2, Q3UB91, Q7TQE3 | Gene names | Rbm7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 7 (RNA-binding motif protein 7). | |||||
|
ELAV1_HUMAN
|
||||||
| NC score | 0.131039 (rank : 28) | θ value | 0.0431538 (rank : 21) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q15717, Q9BTT1 | Gene names | ELAVL1, HUR | |||
|
Domain Architecture |

|
|||||
| Description | ELAV-like protein 1 (Hu-antigen R) (HuR). | |||||
|
HTSF1_HUMAN
|
||||||
| NC score | 0.128998 (rank : 29) | θ value | 0.0113563 (rank : 16) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O43719, Q59G06, Q99730 | Gene names | HTATSF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 (Tat-SF1). | |||||
|
ZCRB1_MOUSE
|
||||||
| NC score | 0.121036 (rank : 30) | θ value | 1.06291 (rank : 37) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9CZ96, Q3TDL3 | Gene names | Zcrb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC-type and RNA-binding motif-containing protein 1 (MADP-1). | |||||
|
SNRPA_MOUSE
|
||||||
| NC score | 0.117517 (rank : 31) | θ value | 0.279714 (rank : 31) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q62189, Q52LA2 | Gene names | Snrpa, Rnu1a-1 | |||
|
Domain Architecture |

|
|||||
| Description | U1 small nuclear ribonucleoprotein A (U1 snRNP protein A) (U1A protein) (U1-A). | |||||
|
HTSF1_MOUSE
|
||||||
| NC score | 0.110145 (rank : 32) | θ value | 0.21417 (rank : 28) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8BGC0, Q1WWK0, Q9CT41, Q9DAU3 | Gene names | Htatsf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 homolog. | |||||
|
PM14_HUMAN
|
||||||
| NC score | 0.109602 (rank : 33) | θ value | 1.38821 (rank : 39) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Y3B4 | Gene names | SF3B14 | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA branch site protein p14 (SF3B 14 kDa subunit). | |||||
|
PM14_MOUSE
|
||||||
| NC score | 0.109602 (rank : 34) | θ value | 1.38821 (rank : 40) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P59708, Q3U9T9 | Gene names | Sf3b14 | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA branch site protein p14 (SF3B 14 kDa subunit). | |||||
|
RBM19_MOUSE
|
||||||
| NC score | 0.108144 (rank : 35) | θ value | 0.365318 (rank : 32) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8R3C6, Q8BHR0, Q9CW63 | Gene names | Rbm19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable RNA-binding protein 19 (RNA-binding motif protein 19). | |||||
|
ELAV1_MOUSE
|
||||||
| NC score | 0.107808 (rank : 36) | θ value | 1.81305 (rank : 42) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P70372, Q60745 | Gene names | Elavl1, Elra, Hua | |||
|
Domain Architecture |

|
|||||
| Description | ELAV-like protein 1 (Hu-antigen R) (HuR) (Elav-like generic protein) (MelG). | |||||
|
RBM19_HUMAN
|
||||||
| NC score | 0.105865 (rank : 37) | θ value | 0.0330416 (rank : 20) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9Y4C8, Q9BPY6, Q9UFN5 | Gene names | RBM19, KIAA0682 | |||
|
Domain Architecture |

|
|||||
| Description | Probable RNA-binding protein 19 (RNA-binding motif protein 19). | |||||
|
SNRPA_HUMAN
|
||||||
| NC score | 0.096340 (rank : 38) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P09012 | Gene names | SNRPA | |||
|
Domain Architecture |

|
|||||
| Description | U1 small nuclear ribonucleoprotein A (U1 snRNP protein A) (U1A protein) (U1-A). | |||||
|
RBM4B_MOUSE
|
||||||
| NC score | 0.095991 (rank : 39) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8VE92 | Gene names | Rbm4b, Rbm30 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 4B (RNA-binding motif protein 4B) (RNA-binding protein 30) (RNA-binding motif protein 30). | |||||
|
RBM4B_HUMAN
|
||||||
| NC score | 0.095962 (rank : 40) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9BQ04 | Gene names | RBM4B, RBM30 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 4B (RNA-binding motif protein 4B) (RNA-binding protein 30) (RNA-binding motif protein 30). | |||||
|
RBM4_MOUSE
|
||||||
| NC score | 0.095803 (rank : 41) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8C7Q4, O08752, Q8BN66 | Gene names | Rbm4, Rbm4a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 4 (RNA-binding motif protein 4) (RNA-binding motif protein 4a) (Lark homolog) (mLark). | |||||
|
RBM4_HUMAN
|
||||||
| NC score | 0.095685 (rank : 42) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9BWF3, O02916, Q6P1P2, Q8WU85 | Gene names | RBM4, RBM4A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 4 (RNA-binding motif protein 4) (RNA-binding motif protein 4a) (Lark homolog) (hLark). | |||||
|
SFRS2_HUMAN
|
||||||
| NC score | 0.095314 (rank : 43) | θ value | 4.03905 (rank : 47) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q01130 | Gene names | SFRS2 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 2 (Splicing factor SC35) (SC-35) (Splicing component, 35 kDa) (Protein PR264). | |||||
|
SFRS2_MOUSE
|
||||||
| NC score | 0.095314 (rank : 44) | θ value | 4.03905 (rank : 48) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q62093, Q542L3, Q60701 | Gene names | Sfrs2, Pr264, Sfrs10 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 2 (Splicing factor SC35) (SC-35) (Splicing component, 35 kDa) (Protein PR264) (Putative myelin regulatory factor 1) (MRF-1). | |||||
|
DRBP1_HUMAN
|
||||||
| NC score | 0.094843 (rank : 45) | θ value | θ > 10 (rank : 62) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8IUH3, Q6NYL0, Q8NFC9 | Gene names | DRBP1, DRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Developmentally-regulated RNA-binding protein 1 (RB-1). | |||||
|
DRBP1_MOUSE
|
||||||
| NC score | 0.085569 (rank : 46) | θ value | θ > 10 (rank : 63) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BHN5, Q8C1Z5 | Gene names | Drbp1, Drb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Developmentally-regulated RNA-binding protein 1 (RB-1). | |||||
|
SAFB1_HUMAN
|
||||||
| NC score | 0.082355 (rank : 47) | θ value | 0.365318 (rank : 33) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 493 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q15424, O60406, Q59HH8 | Gene names | SAFB, HAP, HET, SAFB1 | |||
|
Domain Architecture |

|
|||||
| Description | Scaffold attachment factor B (Scaffold attachment factor B1) (SAF-B) (HSP27 estrogen response element-TATA box-binding protein) (HSP27 ERE- TATA-binding protein). | |||||
|
RBMX2_MOUSE
|
||||||
| NC score | 0.078134 (rank : 48) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8R0F5, Q3TI42 | Gene names | Rbmx2 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding motif protein, X-linked 2. | |||||
|
RBMS1_MOUSE
|
||||||
| NC score | 0.074676 (rank : 49) | θ value | 3.0926 (rank : 45) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q91W59, Q6PEU6, Q6PHC2, Q9WTK4 | Gene names | Rbms1, Mssp, Mssp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding motif, single-stranded-interacting protein 1 (Single- stranded DNA-binding protein MSSP-1). | |||||
|
RBMX2_HUMAN
|
||||||
| NC score | 0.073454 (rank : 50) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9Y388, Q5JY82, Q9Y3I8 | Gene names | RBMX2 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding motif protein, X-linked 2. | |||||
|
RBMS1_HUMAN
|
||||||
| NC score | 0.069495 (rank : 51) | θ value | 5.27518 (rank : 52) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P29558, Q15433, Q8WV20 | Gene names | RBMS1, MSSP, MSSP1, SCR2 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding motif, single-stranded-interacting protein 1 (Single- stranded DNA-binding protein MSSP-1) (Suppressor of CDC2 with RNA- binding motif 2). | |||||
|
MK67I_HUMAN
|
||||||
| NC score | 0.066313 (rank : 52) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9BYG3, Q8TB66, Q96ED4 | Gene names | MKI67IP, NIFK, NOPP34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MKI67 FHA domain-interacting nucleolar phosphoprotein (Nucleolar protein interacting with the FHA domain of pKI-67) (hNIFK) (Nucleolar phosphoprotein Nopp34). | |||||
|
RBM23_HUMAN
|
||||||
| NC score | 0.065392 (rank : 53) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q86U06, Q8ND16, Q8TB88, Q8WY40, Q9BUJ1, Q9NVV7 | Gene names | RBM23, RNPC4 | |||
|
Domain Architecture |

|
|||||
| Description | Probable RNA-binding protein 23 (RNA-binding motif protein 23) (RNA- binding region-containing protein 4) (Splicing factor SF2). | |||||
|
PABP5_HUMAN
|
||||||
| NC score | 0.063529 (rank : 54) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q96DU9, Q6P529, Q9UFE5 | Gene names | PABPC5, PABP5 | |||
|
Domain Architecture |

|
|||||
| Description | Polyadenylate-binding protein 5 (Poly(A)-binding protein 5) (PABP 5). | |||||
|
SFPQ_MOUSE
|
||||||
| NC score | 0.063231 (rank : 55) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8VIJ6, Q9ERW2 | Gene names | Sfpq | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit). | |||||
|
SFPQ_HUMAN
|
||||||
| NC score | 0.063110 (rank : 56) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
SF3B4_HUMAN
|
||||||
| NC score | 0.058077 (rank : 57) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q15427 | Gene names | SF3B4, SAP49 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 4 (Spliceosome-associated protein 49) (SAP 49) (SF3b50) (Pre-mRNA-splicing factor SF3b 49 kDa subunit). | |||||
|
RBMS2_HUMAN
|
||||||
| NC score | 0.057948 (rank : 58) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q15434 | Gene names | RBMS2, SCR3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding motif, single-stranded-interacting protein 2 (Suppressor of CDC2 with RNA-binding motif 3). | |||||
|
RBMS2_MOUSE
|
||||||
| NC score | 0.057434 (rank : 59) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8VC70 | Gene names | Rbms2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding motif, single-stranded-interacting protein 2. | |||||
|
ACF_MOUSE
|
||||||
| NC score | 0.057337 (rank : 60) | θ value | θ > 10 (rank : 61) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q5YD48, Q5FW55, Q5YD47 | Gene names | Acf, Asp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | APOBEC1 complementation factor (APOBEC1-stimulating protein). | |||||
|
RBM14_MOUSE
|
||||||
| NC score | 0.057335 (rank : 61) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8C2Q3, Q3TJB6, Q91Z21, Q9DBI6 | Gene names | Rbm14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 14 (RNA-binding motif protein 14). | |||||
|
PABP1_MOUSE
|
||||||
| NC score | 0.057071 (rank : 62) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P29341 | Gene names | Pabpc1, Pabp1 | |||
|
Domain Architecture |

|
|||||
| Description | Polyadenylate-binding protein 1 (Poly(A)-binding protein 1) (PABP 1). | |||||
|
RBY1A_HUMAN
|
||||||
| NC score | 0.056771 (rank : 63) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q15414, Q15377, Q8NHR0 | Gene names | RBMY1A1, RBM1, YRRM1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding motif protein, Y chromosome, family 1 member A1 (RNA- binding motif protein 1). | |||||
|
PABP1_HUMAN
|
||||||
| NC score | 0.056387 (rank : 64) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P11940, Q15097, Q93004 | Gene names | PABPC1, PAB1, PABP1, PABPC2 | |||
|
Domain Architecture |

|
|||||
| Description | Polyadenylate-binding protein 1 (Poly(A)-binding protein 1) (PABP 1). | |||||
|
NUCL_MOUSE
|
||||||
| NC score | 0.055732 (rank : 65) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P09405, Q548M9, Q61991, Q99K50 | Gene names | Ncl, Nuc | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolin (Protein C23). | |||||
|
SRR35_HUMAN
|
||||||
| NC score | 0.055622 (rank : 66) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8WXF0, Q8WW25 | Gene names | SRRP35 | |||
|
Domain Architecture |

|
|||||
| Description | 35 kDa SR repressor protein (SRrp35). | |||||
|
RBM11_HUMAN
|
||||||
| NC score | 0.055326 (rank : 67) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P57052 | Gene names | RBM11 | |||
|
Domain Architecture |

|
|||||
| Description | Putative RNA-binding protein 11 (RNA-binding motif protein 11). | |||||
|
TIA1_HUMAN
|
||||||
| NC score | 0.055154 (rank : 68) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P31483 | Gene names | TIA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolysin TIA-1 isoform p40 (RNA-binding protein TIA-1) (p40-TIA-1) [Contains: Nucleolysin TIA-1 isoform p15 (p15-TIA-1)]. | |||||
|
HNRPQ_HUMAN
|
||||||
| NC score | 0.055002 (rank : 69) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O60506, Q53H05, Q5TCG2, Q8IW78, Q8N599, Q96LC1, Q96LC2, Q9Y583 | Gene names | SYNCRIP, HNRPQ, NSAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein Q (hnRNP Q) (hnRNP-Q) (Synaptotagmin-binding, cytoplasmic RNA-interacting protein) (Glycine- and tyrosine-rich RNA-binding protein) (GRY-RBP) (NS1-associated protein 1). | |||||
|
RBM28_MOUSE
|
||||||
| NC score | 0.053471 (rank : 70) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8CGC6, Q8BG25, Q8VEJ8, Q9CS22, Q9CSE6 | Gene names | Rbm28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 28 (RNA-binding motif protein 28). | |||||
|
TRA2B_HUMAN
|
||||||
| NC score | 0.053145 (rank : 71) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P62995, O15449, Q15815, Q64283 | Gene names | SFRS10, TRA2B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine/serine-rich-splicing factor 10 (Transformer-2-beta) (HTRA2- beta) (Transformer 2 protein homolog). | |||||
|
TRA2B_MOUSE
|
||||||
| NC score | 0.053145 (rank : 72) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P62996, O15449, Q15815, Q64283 | Gene names | Sfrs10, Silg41, Tra2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine/serine-rich-splicing factor 10 (Transformer-2-beta) (HTRA2- beta) (Transformer 2 protein homolog) (Silica-induced gene 41 protein) (SIG-41). | |||||
|
PABP4_HUMAN
|
||||||
| NC score | 0.053080 (rank : 73) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q13310, Q6P0N3 | Gene names | PABPC4, APP1, PABP4 | |||
|
Domain Architecture |

|
|||||
| Description | Polyadenylate-binding protein 4 (Poly(A)-binding protein 4) (PABP 4) (Inducible poly(A)-binding protein) (iPABP) (Activated-platelet protein 1) (APP-1). | |||||
|
PABP3_HUMAN
|
||||||
| NC score | 0.052974 (rank : 74) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9H361, Q8NHV0, Q9H086 | Gene names | PABPC3, PABP3, PABPL3 | |||
|
Domain Architecture |

|
|||||
| Description | Polyadenylate-binding protein 3 (Poly(A)-binding protein 3) (PABP 3) (Testis-specific poly(A)-binding protein). | |||||
|
SFRS7_HUMAN
|
||||||
| NC score | 0.052702 (rank : 75) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q16629 | Gene names | SFRS7 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 7 (Splicing factor 9G8). | |||||
|
TIA1_MOUSE
|
||||||
| NC score | 0.052442 (rank : 76) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P52912 | Gene names | Tia1, Tia | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolysin TIA-1 (RNA-binding protein TIA-1). | |||||
|
RBM14_HUMAN
|
||||||
| NC score | 0.052114 (rank : 77) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q96PK6, O75932, Q53GV1, Q68DQ9, Q96PK5 | Gene names | RBM14, SIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 14 (RNA-binding motif protein 14) (RRM-containing coactivator activator/modulator) (Synaptotagmin-interacting protein) (SYT-interacting protein). | |||||
|
SFRS3_HUMAN
|
||||||
| NC score | 0.052107 (rank : 78) | θ value | 8.99809 (rank : 59) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P84103, O08831, P23152 | Gene names | SFRS3, SRP20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 3 (Pre-mRNA-splicing factor SRP20). | |||||
|
SFRS3_MOUSE
|
||||||
| NC score | 0.052107 (rank : 79) | θ value | 8.99809 (rank : 60) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P84104, O08831, P23152, Q564E9 | Gene names | Sfrs3, Srp20, X16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 3 (Pre-mRNA-splicing factor SRP20) (X16 protein). | |||||
|
HNRPQ_MOUSE
|
||||||
| NC score | 0.051879 (rank : 80) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q7TMK9, O88991, Q80YV6, Q8BGP1, Q8C5K6, Q8CGC2, Q91ZR0, Q99KU9, Q9QYF4 | Gene names | Syncrip, Hnrpq, Nsap1, Nsap1l | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein Q (hnRNP Q) (hnRNP-Q) (Synaptotagmin-binding, cytoplasmic RNA-interacting protein) (Glycine- and tyrosine-rich RNA-binding protein) (GRY-RBP) (NS1-associated protein 1) (pp68). | |||||
|
FUSIP_HUMAN
|
||||||
| NC score | 0.050246 (rank : 81) | θ value | θ > 10 (rank : 64) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O75494, O60572, Q96G09, Q96P17 | Gene names | FUSIP1, TASR | |||
|
Domain Architecture |

|
|||||
| Description | FUS-interacting serine-arginine-rich protein 1 (TLS-associated protein with Ser-Arg repeats) (TLS-associated protein with SR repeats) (TASR) (TLS-associated serine-arginine protein) (TLS-associated SR protein) (40 kDa SR-repressor protein) (SRrp40) (Splicing factor SRp38). | |||||
|
FUSIP_MOUSE
|
||||||
| NC score | 0.050246 (rank : 82) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9R0U0, O70307, O88468, Q3TGW7, Q8CFZ1, Q9R0T9 | Gene names | Fusip1, Nssr | |||
|
Domain Architecture |

|
|||||
| Description | FUS-interacting serine-arginine-rich protein 1 (TLS-associated protein with Ser-Arg repeats) (TLS-associated protein with SR repeats) (TASR) (TLS-associated serine-arginine protein) (TLS-associated SR protein) (Neural-specific SR protein) (Neural-salient serine/arginine-rich protein). | |||||
|
IF39_MOUSE
|
||||||
| NC score | 0.046381 (rank : 83) | θ value | 4.03905 (rank : 46) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8JZQ9, Q922K2 | Gene names | Eif3s9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 9 (eIF-3 eta) (eIF3 p116). | |||||
|
MINT_HUMAN
|
||||||
| NC score | 0.041563 (rank : 84) | θ value | 6.88961 (rank : 55) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
MINT_MOUSE
|
||||||
| NC score | 0.038692 (rank : 85) | θ value | 8.99809 (rank : 58) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
IF39_HUMAN
|
||||||
| NC score | 0.033275 (rank : 86) | θ value | 6.88961 (rank : 54) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P55884, Q9UMF9 | Gene names | EIF3S9 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 9 (eIF-3 eta) (eIF3 p116) (eIF3 p110) (eIF3b) (Prt1 homolog) (hPrt1). | |||||
|
P3C2G_MOUSE
|
||||||
| NC score | 0.021423 (rank : 87) | θ value | 3.0926 (rank : 44) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O70167, O70168 | Gene names | Pik3c2g | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing gamma polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-gamma) (PtdIns-3-kinase C2 gamma) (PI3K-C2gamma). | |||||
|
ACINU_MOUSE
|
||||||
| NC score | 0.018424 (rank : 88) | θ value | 5.27518 (rank : 50) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9JIX8, Q9CSN7, Q9CSR9, Q9CSX7, Q9R046, Q9R047 | Gene names | Acin1, Acinus | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
ACINU_HUMAN
|
||||||
| NC score | 0.018256 (rank : 89) | θ value | 5.27518 (rank : 49) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UKV3, O75158, Q9UG91, Q9UKV1, Q9UKV2 | Gene names | ACIN1, ACINUS, KIAA0670 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
NKX26_MOUSE
|
||||||
| NC score | 0.017701 (rank : 90) | θ value | 0.279714 (rank : 30) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P43688 | Gene names | Nkx2-6, Nkx-2.6, Nkx2f | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Nkx-2.6 (Homeobox protein NK-2 homolog F). | |||||
|
COJA1_HUMAN
|
||||||
| NC score | 0.014211 (rank : 91) | θ value | 1.81305 (rank : 41) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q14993, Q00559, Q05850, Q12885, Q13676, Q5JUF0, Q5T424, Q9H572, Q9NPZ2, Q9NQP2 | Gene names | COL19A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XIX) chain precursor (Collagen alpha-1(Y) chain). | |||||
|
SYNG_HUMAN
|
||||||
| NC score | 0.012996 (rank : 92) | θ value | 6.88961 (rank : 56) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UMZ2, Q6ZT17 | Gene names | AP1GBP1, SYNG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP1 subunit gamma-binding protein 1 (Gamma-synergin). | |||||
|
CO4A2_MOUSE
|
||||||
| NC score | 0.011751 (rank : 93) | θ value | 1.06291 (rank : 36) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P08122, Q61375 | Gene names | Col4a2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(IV) chain precursor. | |||||
|
MUC5B_HUMAN
|
||||||
| NC score | 0.011191 (rank : 94) | θ value | 1.38821 (rank : 38) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
CO4A2_HUMAN
|
||||||
| NC score | 0.009733 (rank : 95) | θ value | 6.88961 (rank : 53) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 454 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P08572 | Gene names | COL4A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(IV) chain precursor. | |||||
|
CO8A2_MOUSE
|
||||||
| NC score | 0.009288 (rank : 96) | θ value | 5.27518 (rank : 51) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P25318, Q68ED0, Q6KAQ4, Q6P1C4 | Gene names | Col8a2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-2(VIII) chain precursor (Endothelial collagen). | |||||
|
ATX1_MOUSE
|
||||||
| NC score | 0.006982 (rank : 97) | θ value | 8.99809 (rank : 57) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P54254 | Gene names | Atxn1, Sca1 | |||
|
Domain Architecture |

|
|||||
| Description | Ataxin-1 (Spinocerebellar ataxia type 1 protein homolog). | |||||
|
RXRA_HUMAN
|
||||||
| NC score | 0.003870 (rank : 98) | θ value | 2.36792 (rank : 43) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P19793, Q2V504 | Gene names | RXRA, NR2B1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoic acid receptor RXR-alpha (Retinoid X receptor alpha). | |||||