Please be patient as the page loads
|
PTBP2_HUMAN
|
||||||
| SwissProt Accessions | Q9UKA9, Q8N0Z1, Q8N160, Q8NFB0, Q8NFB1, Q969N9, Q96Q76 | Gene names | PTBP2, NPTB, PTB, PTBLP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypyrimidine tract-binding protein 2 (Neurally-enriched homolog of PTB) (Neural polypyrimidine tract-binding protein) (PTB-like protein). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PTBP1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.988147 (rank : 3) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P26599 | Gene names | PTBP1, PTB | |||
|
Domain Architecture |

|
|||||
| Description | Polypyrimidine tract-binding protein 1 (PTB) (Heterogeneous nuclear ribonucleoprotein I) (hnRNP I) (57 kDa RNA-binding protein PPTB-1). | |||||
|
PTBP1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.980214 (rank : 6) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P17225 | Gene names | Ptbp1, Ptb | |||
|
Domain Architecture |

|
|||||
| Description | Polypyrimidine tract-binding protein 1 (PTB) (Heterogeneous nuclear ribonucleoprotein I) (hnRNP I). | |||||
|
PTBP2_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | Q9UKA9, Q8N0Z1, Q8N160, Q8NFB0, Q8NFB1, Q969N9, Q96Q76 | Gene names | PTBP2, NPTB, PTB, PTBLP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypyrimidine tract-binding protein 2 (Neurally-enriched homolog of PTB) (Neural polypyrimidine tract-binding protein) (PTB-like protein). | |||||
|
PTBP2_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.999946 (rank : 2) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | Q91Z31, Q7TNW7, Q9CUW2, Q9QYC2, Q9R0V9 | Gene names | Ptbp2, Brptb, Nptb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypyrimidine tract-binding protein 2 (Brain-enriched polypyrimidine tract-binding protein) (Brain-enriched PTB) (RRM-type RNA-binding protein brPTB) (Neural polypyrimidine tract-binding protein). | |||||
|
ROD1_HUMAN
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.982471 (rank : 4) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O95758, Q86YB3, Q86YH9 | Gene names | ROD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of differentiation 1 (Rod1). | |||||
|
ROD1_MOUSE
|
||||||
| θ value | 0 (rank : 6) | NC score | 0.980288 (rank : 5) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8BHD7, Q923C3 | Gene names | Rod1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of differentiation 1 (Rod1). | |||||
|
HNRLL_HUMAN
|
||||||
| θ value | 4.71619e-41 (rank : 7) | NC score | 0.783992 (rank : 7) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8WVV9, Q659B9, Q8IVH5, Q8IVH6, Q96HR5 | Gene names | HNRPLL, SRRF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heterogeneous nuclear ribonucleoprotein L-like (Stromal RNA-regulating factor) (BLOCK24 protein). | |||||
|
HNRLL_MOUSE
|
||||||
| θ value | 6.15952e-41 (rank : 8) | NC score | 0.780440 (rank : 8) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q921F4, Q8BIP6, Q99J40 | Gene names | Hnrpll | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heterogeneous nuclear ribonucleoprotein L-like. | |||||
|
HNRPL_HUMAN
|
||||||
| θ value | 1.6852e-22 (rank : 9) | NC score | 0.734917 (rank : 9) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P14866, Q9H3P3 | Gene names | HNRPL | |||
|
Domain Architecture |
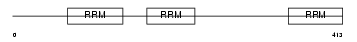
|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein L (hnRNP L). | |||||
|
HNRPL_MOUSE
|
||||||
| θ value | 1.6852e-22 (rank : 10) | NC score | 0.734129 (rank : 10) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8R081, O54789, Q8K0S7 | Gene names | Hnrpl | |||
|
Domain Architecture |
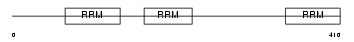
|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein L (hnRNP L). | |||||
|
MATR3_HUMAN
|
||||||
| θ value | 1.69304e-06 (rank : 11) | NC score | 0.311924 (rank : 12) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P43243, Q9UHW0, Q9UQ27 | Gene names | MATR3, KIAA0723 | |||
|
Domain Architecture |

|
|||||
| Description | Matrin-3. | |||||
|
MATR3_MOUSE
|
||||||
| θ value | 1.69304e-06 (rank : 12) | NC score | 0.312859 (rank : 11) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8K310 | Gene names | Matr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Matrin-3. | |||||
|
RU2B_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 13) | NC score | 0.250090 (rank : 13) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P08579, Q9UJD4 | Gene names | SNRPB2 | |||
|
Domain Architecture |
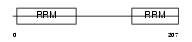
|
|||||
| Description | U2 small nuclear ribonucleoprotein B''. | |||||
|
RU2B_MOUSE
|
||||||
| θ value | 9.29e-05 (rank : 14) | NC score | 0.247556 (rank : 14) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9CQI7, Q9CW35, Q9CZ66 | Gene names | Snrpb2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U2 small nuclear ribonucleoprotein B''. | |||||
|
NONO_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 15) | NC score | 0.199312 (rank : 18) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q15233, O00201, P30807, Q12786, Q9BQC5 | Gene names | NONO, NRB54 | |||
|
Domain Architecture |
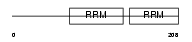
|
|||||
| Description | Non-POU domain-containing octamer-binding protein (NonO protein) (54 kDa nuclear RNA- and DNA-binding protein) (p54(nrb)) (p54nrb) (55 kDa nuclear protein) (NMT55) (DNA-binding p52/p100 complex, 52 kDa subunit). | |||||
|
NONO_MOUSE
|
||||||
| θ value | 0.00020696 (rank : 16) | NC score | 0.200482 (rank : 16) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q99K48, Q63887, Q9CYQ4, Q9DBP2 | Gene names | Nono | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Non-POU domain-containing octamer-binding protein (NonO protein). | |||||
|
ELAV3_HUMAN
|
||||||
| θ value | 0.00035302 (rank : 17) | NC score | 0.183726 (rank : 28) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q14576, Q16135, Q96CL8, Q96QS9 | Gene names | ELAVL3, HUC, PLE21 | |||
|
Domain Architecture |
|
|||||
| Description | ELAV-like protein 3 (Hu-antigen C) (HuC) (Paraneoplastic cerebellar degeneration-associated antigen) (Paraneoplastic limbic encephalitis antigen 21). | |||||
|
ELAV3_MOUSE
|
||||||
| θ value | 0.00035302 (rank : 18) | NC score | 0.180817 (rank : 30) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q60900, Q60901 | Gene names | Elavl3, Huc | |||
|
Domain Architecture |
|
|||||
| Description | ELAV-like protein 3 (Hu-antigen C) (HuC). | |||||
|
RBM4B_HUMAN
|
||||||
| θ value | 0.00035302 (rank : 19) | NC score | 0.185725 (rank : 27) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9BQ04 | Gene names | RBM4B, RBM30 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 4B (RNA-binding motif protein 4B) (RNA-binding protein 30) (RNA-binding motif protein 30). | |||||
|
RBM4B_MOUSE
|
||||||
| θ value | 0.00035302 (rank : 20) | NC score | 0.185774 (rank : 26) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8VE92 | Gene names | Rbm4b, Rbm30 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 4B (RNA-binding motif protein 4B) (RNA-binding protein 30) (RNA-binding motif protein 30). | |||||
|
RBM4_MOUSE
|
||||||
| θ value | 0.00035302 (rank : 21) | NC score | 0.185939 (rank : 24) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8C7Q4, O08752, Q8BN66 | Gene names | Rbm4, Rbm4a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 4 (RNA-binding motif protein 4) (RNA-binding motif protein 4a) (Lark homolog) (mLark). | |||||
|
RBM4_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 22) | NC score | 0.185902 (rank : 25) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9BWF3, O02916, Q6P1P2, Q8WU85 | Gene names | RBM4, RBM4A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 4 (RNA-binding motif protein 4) (RNA-binding motif protein 4a) (Lark homolog) (hLark). | |||||
|
RBM7_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 23) | NC score | 0.201969 (rank : 15) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9Y580 | Gene names | RBM7 | |||
|
Domain Architecture |
|
|||||
| Description | RNA-binding protein 7 (RNA-binding motif protein 7). | |||||
|
ZN638_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 24) | NC score | 0.200433 (rank : 17) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q61464, Q6DFV9, Q8C941 | Gene names | Znf638, Np220, Zfml | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 638 (Nuclear protein 220) (Zinc-finger matrin-like protein). | |||||
|
RBM7_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 25) | NC score | 0.190281 (rank : 20) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9CQT2, Q3UB91, Q7TQE3 | Gene names | Rbm7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 7 (RNA-binding motif protein 7). | |||||
|
ZN638_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 26) | NC score | 0.186860 (rank : 23) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q14966, Q53R34, Q5XJ05, Q68DP3, Q6P2H2, Q7Z3T7, Q8NF92, Q8TCA1, Q9H2G1, Q9NP37 | Gene names | ZNF638, NP220, ZFML | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 638 (Nuclear protein 220) (Zinc-finger matrin-like protein) (Cutaneous T-cell lymphoma-associated antigen se33-1) (CTCL tumor antigen se33-1). | |||||
|
ELAV2_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 27) | NC score | 0.180486 (rank : 31) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q12926, Q13235, Q59G15, Q8NEM4, Q9H1Q8 | Gene names | ELAVL2, HUB | |||
|
Domain Architecture |
|
|||||
| Description | ELAV-like protein 2 (Hu-antigen B) (HuB) (ELAV-like neuronal protein 1) (Nervous system-specific RNA-binding protein Hel-N1). | |||||
|
ELAV2_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 28) | NC score | 0.180850 (rank : 29) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q60899 | Gene names | Elavl2, Hub | |||
|
Domain Architecture |
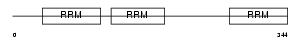
|
|||||
| Description | ELAV-like protein 2 (Hu-antigen B) (HuB) (ELAV-like neuronal protein 1) (Nervous system-specific RNA-binding protein Mel-N1). | |||||
|
ELAV4_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 29) | NC score | 0.188263 (rank : 21) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q61701, O55010 | Gene names | Elavl4, Hud | |||
|
Domain Architecture |
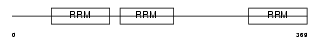
|
|||||
| Description | ELAV-like protein 4 (Paraneoplastic encephalomyelitis antigen HuD) (Hu-antigen D). | |||||
|
ELAV4_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 30) | NC score | 0.186953 (rank : 22) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P26378, Q96J74 | Gene names | ELAVL4, HUD, PNEM | |||
|
Domain Architecture |
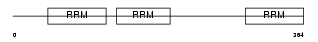
|
|||||
| Description | ELAV-like protein 4 (Paraneoplastic encephalomyelitis antigen HuD) (Hu-antigen D). | |||||
|
DRBP1_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 31) | NC score | 0.141800 (rank : 36) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8IUH3, Q6NYL0, Q8NFC9 | Gene names | DRBP1, DRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Developmentally-regulated RNA-binding protein 1 (RB-1). | |||||
|
RU17_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 32) | NC score | 0.100461 (rank : 62) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P08621, P78493, P78494, Q15364, Q15686, Q15687, Q15689, Q99377, Q9UE45, Q9UE46, Q9UE47, Q9UE48, Q9UFQ6 | Gene names | SNRP70, RPU1, U1AP1 | |||
|
Domain Architecture |

|
|||||
| Description | U1 small nuclear ribonucleoprotein 70 kDa (U1 snRNP 70 kDa) (snRNP70) (U1-70K). | |||||
|
RU17_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 33) | NC score | 0.101078 (rank : 59) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q62376, Q3UIW4 | Gene names | Snrp70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U1 small nuclear ribonucleoprotein 70 kDa (U1 SNRNP 70 kDa) (snRNP70). | |||||
|
ELAV1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 34) | NC score | 0.163700 (rank : 33) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q15717, Q9BTT1 | Gene names | ELAVL1, HUR | |||
|
Domain Architecture |
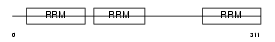
|
|||||
| Description | ELAV-like protein 1 (Hu-antigen R) (HuR). | |||||
|
ELAV1_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 35) | NC score | 0.141316 (rank : 38) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P70372, Q60745 | Gene names | Elavl1, Elra, Hua | |||
|
Domain Architecture |
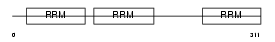
|
|||||
| Description | ELAV-like protein 1 (Hu-antigen R) (HuR) (Elav-like generic protein) (MelG). | |||||
|
SFRS2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 36) | NC score | 0.143425 (rank : 34) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q01130 | Gene names | SFRS2 | |||
|
Domain Architecture |
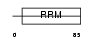
|
|||||
| Description | Splicing factor, arginine/serine-rich 2 (Splicing factor SC35) (SC-35) (Splicing component, 35 kDa) (Protein PR264). | |||||
|
SFRS2_MOUSE
|
||||||
| θ value | 0.279714 (rank : 37) | NC score | 0.143425 (rank : 35) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q62093, Q542L3, Q60701 | Gene names | Sfrs2, Pr264, Sfrs10 | |||
|
Domain Architecture |
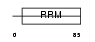
|
|||||
| Description | Splicing factor, arginine/serine-rich 2 (Splicing factor SC35) (SC-35) (Splicing component, 35 kDa) (Protein PR264) (Putative myelin regulatory factor 1) (MRF-1). | |||||
|
ZCRB1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 38) | NC score | 0.197308 (rank : 19) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q8TBF4, Q6PJX0 | Gene names | ZCRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC-type and RNA-binding motif-containing protein 1 (U11/U12 snRNP 31 kDa protein) (U11/U12-31K). | |||||
|
HTSF1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 39) | NC score | 0.117097 (rank : 49) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O43719, Q59G06, Q99730 | Gene names | HTATSF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 (Tat-SF1). | |||||
|
SAFB1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 40) | NC score | 0.098024 (rank : 65) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 493 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q15424, O60406, Q59HH8 | Gene names | SAFB, HAP, HET, SAFB1 | |||
|
Domain Architecture |

|
|||||
| Description | Scaffold attachment factor B (Scaffold attachment factor B1) (SAF-B) (HSP27 estrogen response element-TATA box-binding protein) (HSP27 ERE- TATA-binding protein). | |||||
|
DRBP1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 41) | NC score | 0.132675 (rank : 39) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8BHN5, Q8C1Z5 | Gene names | Drbp1, Drb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Developmentally-regulated RNA-binding protein 1 (RB-1). | |||||
|
NUCL_MOUSE
|
||||||
| θ value | 0.813845 (rank : 42) | NC score | 0.090354 (rank : 82) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P09405, Q548M9, Q61991, Q99K50 | Gene names | Ncl, Nuc | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolin (Protein C23). | |||||
|
RBMX2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 43) | NC score | 0.125786 (rank : 42) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q8R0F5, Q3TI42 | Gene names | Rbmx2 | |||
|
Domain Architecture |
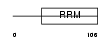
|
|||||
| Description | RNA-binding motif protein, X-linked 2. | |||||
|
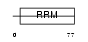
SFRS7_HUMAN
|
||||||
| θ value | 0.813845 (rank : 44) | NC score | 0.114002 (rank : 50) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q16629 | Gene names | SFRS7 | |||
|
Domain Architecture |
|
|||||
| Description | Splicing factor, arginine/serine-rich 7 (Splicing factor 9G8). | |||||
|
RBM19_MOUSE
|
||||||
| θ value | 1.06291 (rank : 45) | NC score | 0.131757 (rank : 40) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q8R3C6, Q8BHR0, Q9CW63 | Gene names | Rbm19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable RNA-binding protein 19 (RNA-binding motif protein 19). | |||||
|
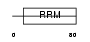
HNRPC_MOUSE
|
||||||
| θ value | 1.38821 (rank : 46) | NC score | 0.077800 (rank : 109) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9Z204, Q3TLB5, Q501Q3, Q8C2G5, Q99KE2, Q9CQT3, Q9CY83 | Gene names | Hnrpc, Hnrnpc | |||
|
Domain Architecture |
|
|||||
| Description | Heterogeneous nuclear ribonucleoproteins C1/C2 (hnRNP C1 / hnRNP C2). | |||||
|
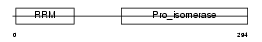
PPIE_HUMAN
|
||||||
| θ value | 1.38821 (rank : 47) | NC score | 0.066015 (rank : 137) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9UNP9, O43634, O43635, Q5TGA0, Q9UIZ5 | Gene names | PPIE, CYP33 | |||
|
Domain Architecture |
|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase E (EC 5.2.1.8) (PPIase E) (Rotamase E) (Cyclophilin E) (Cyclophilin 33). | |||||
|
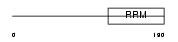
TRA2A_HUMAN
|
||||||
| θ value | 1.38821 (rank : 48) | NC score | 0.097771 (rank : 67) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q13595 | Gene names | TRA2A | |||
|
Domain Architecture |
|
|||||
| Description | Transformer-2 protein homolog (TRA-2 alpha). | |||||
|
TRA2A_MOUSE
|
||||||
| θ value | 1.38821 (rank : 49) | NC score | 0.097984 (rank : 66) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q6PFR5 | Gene names | Tra2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transformer-2 protein homolog (TRA-2 alpha). | |||||
|
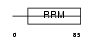
RALY_MOUSE
|
||||||
| θ value | 1.81305 (rank : 50) | NC score | 0.085030 (rank : 94) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q64012, Q99K76, Q9CXH8, Q9QZX6 | Gene names | Raly, Merc | |||
|
Domain Architecture |
|
|||||
| Description | RNA-binding protein Raly (hnRNP associated with lethal yellow protein) (Maternally expressed hnRNP C-related protein). | |||||
|
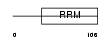
RBMX2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 51) | NC score | 0.118885 (rank : 47) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9Y388, Q5JY82, Q9Y3I8 | Gene names | RBMX2 | |||
|
Domain Architecture |
|
|||||
| Description | RNA-binding motif protein, X-linked 2. | |||||
|
RBY1A_HUMAN
|
||||||
| θ value | 1.81305 (rank : 52) | NC score | 0.106625 (rank : 51) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q15414, Q15377, Q8NHR0 | Gene names | RBMY1A1, RBM1, YRRM1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding motif protein, Y chromosome, family 1 member A1 (RNA- binding motif protein 1). | |||||
|
ZCRB1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 53) | NC score | 0.171090 (rank : 32) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9CZ96, Q3TDL3 | Gene names | Zcrb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC-type and RNA-binding motif-containing protein 1 (MADP-1). | |||||
|
HTSF1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 54) | NC score | 0.097694 (rank : 68) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8BGC0, Q1WWK0, Q9CT41, Q9DAU3 | Gene names | Htatsf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 homolog. | |||||
|
MK67I_HUMAN
|
||||||
| θ value | 2.36792 (rank : 55) | NC score | 0.086518 (rank : 89) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9BYG3, Q8TB66, Q96ED4 | Gene names | MKI67IP, NIFK, NOPP34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MKI67 FHA domain-interacting nucleolar phosphoprotein (Nucleolar protein interacting with the FHA domain of pKI-67) (hNIFK) (Nucleolar phosphoprotein Nopp34). | |||||
|
NEP_MOUSE
|
||||||
| θ value | 2.36792 (rank : 56) | NC score | 0.013005 (rank : 195) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61391, Q6NXX5, Q8K251 | Gene names | Mme | |||
|
Domain Architecture |

|
|||||
| Description | Neprilysin (EC 3.4.24.11) (Neutral endopeptidase) (NEP) (Enkephalinase) (Neutral endopeptidase 24.11) (Atriopeptidase) (CD10 antigen). | |||||
|
P3C2G_MOUSE
|
||||||
| θ value | 2.36792 (rank : 57) | NC score | 0.021797 (rank : 188) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O70167, O70168 | Gene names | Pik3c2g | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing gamma polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-gamma) (PtdIns-3-kinase C2 gamma) (PI3K-C2gamma). | |||||
|
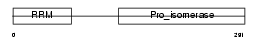
PPIE_MOUSE
|
||||||
| θ value | 2.36792 (rank : 58) | NC score | 0.064187 (rank : 142) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9QZH3, Q9D8C0 | Gene names | Ppie, Cyp33 | |||
|
Domain Architecture |
|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase E (EC 5.2.1.8) (PPIase E) (Rotamase E) (Cyclophilin E) (Cyclophilin 33). | |||||
|
SNRPA_HUMAN
|
||||||
| θ value | 2.36792 (rank : 59) | NC score | 0.122640 (rank : 46) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P09012 | Gene names | SNRPA | |||
|
Domain Architecture |

|
|||||
| Description | U1 small nuclear ribonucleoprotein A (U1 snRNP protein A) (U1A protein) (U1-A). | |||||
|
SNRPA_MOUSE
|
||||||
| θ value | 2.36792 (rank : 60) | NC score | 0.141609 (rank : 37) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q62189, Q52LA2 | Gene names | Snrpa, Rnu1a-1 | |||
|
Domain Architecture |

|
|||||
| Description | U1 small nuclear ribonucleoprotein A (U1 snRNP protein A) (U1A protein) (U1-A). | |||||
|
PAR14_MOUSE
|
||||||
| θ value | 3.0926 (rank : 61) | NC score | 0.015035 (rank : 194) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q2EMV9, Q571C6, Q8BN35, Q8VDT6 | Gene names | Parp14, Kiaa1268 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly [ADP-ribose] polymerase 14 (EC 2.4.2.30) (PARP-14) (Collaborator of STAT6) (CoaSt6). | |||||
|
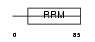
RALY_HUMAN
|
||||||
| θ value | 3.0926 (rank : 62) | NC score | 0.078760 (rank : 106) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9UKM9, Q14621, Q5QPL8, Q9BQX6, Q9UJE3 | Gene names | RALY, P542 | |||
|
Domain Architecture |
|
|||||
| Description | RNA-binding protein Raly (hnRNP associated with lethal yellow homolog) (Autoantigen p542). | |||||
|
SFRS3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 63) | NC score | 0.100766 (rank : 60) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P84103, O08831, P23152 | Gene names | SFRS3, SRP20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 3 (Pre-mRNA-splicing factor SRP20). | |||||
|
SFRS3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 64) | NC score | 0.100766 (rank : 61) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P84104, O08831, P23152, Q564E9 | Gene names | Sfrs3, Srp20, X16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 3 (Pre-mRNA-splicing factor SRP20) (X16 protein). | |||||
|
RBM19_HUMAN
|
||||||
| θ value | 4.03905 (rank : 65) | NC score | 0.128465 (rank : 41) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9Y4C8, Q9BPY6, Q9UFN5 | Gene names | RBM19, KIAA0682 | |||
|
Domain Architecture |

|
|||||
| Description | Probable RNA-binding protein 19 (RNA-binding motif protein 19). | |||||
|
SFPQ_HUMAN
|
||||||
| θ value | 4.03905 (rank : 66) | NC score | 0.104379 (rank : 54) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
SFPQ_MOUSE
|
||||||
| θ value | 4.03905 (rank : 67) | NC score | 0.104193 (rank : 55) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8VIJ6, Q9ERW2 | Gene names | Sfpq | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit). | |||||
|
CA103_HUMAN
|
||||||
| θ value | 5.27518 (rank : 68) | NC score | 0.019285 (rank : 192) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5T3J3, Q86XS4, Q8N3B6, Q96HT4, Q9NUM5, Q9NV32 | Gene names | C1orf103, RIF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf103 (Receptor-interacting factor 1). | |||||
|
RBM23_HUMAN
|
||||||
| θ value | 5.27518 (rank : 69) | NC score | 0.102668 (rank : 56) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q86U06, Q8ND16, Q8TB88, Q8WY40, Q9BUJ1, Q9NVV7 | Gene names | RBM23, RNPC4 | |||
|
Domain Architecture |
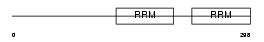
|
|||||
| Description | Probable RNA-binding protein 23 (RNA-binding motif protein 23) (RNA- binding region-containing protein 4) (Splicing factor SF2). | |||||
|
SPT16_MOUSE
|
||||||
| θ value | 5.27518 (rank : 70) | NC score | 0.021600 (rank : 189) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q920B9, Q3TZ48, Q3UFH0, Q921H4 | Gene names | Supt16h, Fact140, Factp140 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FACT complex subunit SPT16 (Facilitates chromatin transcription complex subunit SPT16) (FACT 140 kDa subunit) (FACTp140) (Chromatin- specific transcription elongation factor 140 kDa subunit). | |||||
|
SYTC_HUMAN
|
||||||
| θ value | 5.27518 (rank : 71) | NC score | 0.022281 (rank : 187) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P26639, Q96FP5, Q9BWA6 | Gene names | TARS | |||
|
Domain Architecture |

|
|||||
| Description | Threonyl-tRNA synthetase, cytoplasmic (EC 6.1.1.3) (Threonine--tRNA ligase) (ThrRS). | |||||
|
TRA2B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 72) | NC score | 0.105227 (rank : 52) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P62995, O15449, Q15815, Q64283 | Gene names | SFRS10, TRA2B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine/serine-rich-splicing factor 10 (Transformer-2-beta) (HTRA2- beta) (Transformer 2 protein homolog). | |||||
|
TRA2B_MOUSE
|
||||||
| θ value | 5.27518 (rank : 73) | NC score | 0.105227 (rank : 53) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P62996, O15449, Q15815, Q64283 | Gene names | Sfrs10, Silg41, Tra2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine/serine-rich-splicing factor 10 (Transformer-2-beta) (HTRA2- beta) (Transformer 2 protein homolog) (Silica-induced gene 41 protein) (SIG-41). | |||||
|
EAN57_HUMAN
|
||||||
| θ value | 6.88961 (rank : 74) | NC score | 0.018688 (rank : 193) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43247, Q6ICF2, Q9Y4V8 | Gene names | EAN57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein EAN57. | |||||
|
RBMS1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 75) | NC score | 0.095666 (rank : 69) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P29558, Q15433, Q8WV20 | Gene names | RBMS1, MSSP, MSSP1, SCR2 | |||
|
Domain Architecture |
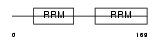
|
|||||
| Description | RNA-binding motif, single-stranded-interacting protein 1 (Single- stranded DNA-binding protein MSSP-1) (Suppressor of CDC2 with RNA- binding motif 2). | |||||
|
RBMS1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 76) | NC score | 0.099788 (rank : 63) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q91W59, Q6PEU6, Q6PHC2, Q9WTK4 | Gene names | Rbms1, Mssp, Mssp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding motif, single-stranded-interacting protein 1 (Single- stranded DNA-binding protein MSSP-1). | |||||
|
SFRS7_MOUSE
|
||||||
| θ value | 6.88961 (rank : 77) | NC score | 0.102231 (rank : 57) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8BL97, Q8BMC6, Q8BUR2, Q8R2N4, Q8R3E9, Q91YS1 | Gene names | Sfrs7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 7. | |||||
|
SPT16_HUMAN
|
||||||
| θ value | 6.88961 (rank : 78) | NC score | 0.019830 (rank : 191) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y5B9, Q6GMT8, Q6P2F1, Q6PJM1, Q9NRX0 | Gene names | SUPT16H, FACT140, FACTP140 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FACT complex subunit SPT16 (Facilitates chromatin transcription complex subunit SPT16) (hSPT16) (FACT 140 kDa subunit) (FACTp140) (Chromatin-specific transcription elongation factor 140 kDa subunit). | |||||
|
SYTC_MOUSE
|
||||||
| θ value | 6.88961 (rank : 79) | NC score | 0.020442 (rank : 190) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D0R2, Q3TL42, Q7TMQ2, Q8BMI6, Q9CX03 | Gene names | Tars | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Threonyl-tRNA synthetase, cytoplasmic (EC 6.1.1.3) (Threonine--tRNA ligase) (ThrRS). | |||||
|
ACF_HUMAN
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | 0.085586 (rank : 92) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9NQ94, Q5SZQ0, Q9NQ93, Q9NQX8, Q9NQX9, Q9NXC9, Q9NZD3 | Gene names | ACF, ASP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | APOBEC1 complementation factor (APOBEC1-stimulating protein). | |||||
|
ACF_MOUSE
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | 0.090771 (rank : 81) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q5YD48, Q5FW55, Q5YD47 | Gene names | Acf, Asp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | APOBEC1 complementation factor (APOBEC1-stimulating protein). | |||||
|
HNRPC_HUMAN
|
||||||
| θ value | 8.99809 (rank : 82) | NC score | 0.067700 (rank : 130) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P07910, P22628, Q53EX2, Q59FD3, Q5FWE8, Q86SF8, Q86U45, Q96HK7, Q96HM4, Q96IY5, Q9BTS3 | Gene names | HNRPC | |||
|
Domain Architecture |
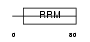
|
|||||
| Description | Heterogeneous nuclear ribonucleoproteins C1/C2 (hnRNP C1 / hnRNP C2). | |||||
|
IF39_MOUSE
|
||||||
| θ value | 8.99809 (rank : 83) | NC score | 0.043686 (rank : 185) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8JZQ9, Q922K2 | Gene names | Eif3s9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 9 (eIF-3 eta) (eIF3 p116). | |||||
|
PM14_HUMAN
|
||||||
| θ value | 8.99809 (rank : 84) | NC score | 0.124052 (rank : 43) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9Y3B4 | Gene names | SF3B14 | |||
|
Domain Architecture |
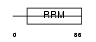
|
|||||
| Description | Pre-mRNA branch site protein p14 (SF3B 14 kDa subunit). | |||||
|
PM14_MOUSE
|
||||||
| θ value | 8.99809 (rank : 85) | NC score | 0.124052 (rank : 44) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P59708, Q3U9T9 | Gene names | Sf3b14 | |||
|
Domain Architecture |
|
|||||
| Description | Pre-mRNA branch site protein p14 (SF3B 14 kDa subunit). | |||||
|
RBM14_HUMAN
|
||||||
| θ value | 8.99809 (rank : 86) | NC score | 0.118765 (rank : 48) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q96PK6, O75932, Q53GV1, Q68DQ9, Q96PK5 | Gene names | RBM14, SIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 14 (RNA-binding motif protein 14) (RRM-containing coactivator activator/modulator) (Synaptotagmin-interacting protein) (SYT-interacting protein). | |||||
|
RBM14_MOUSE
|
||||||
| θ value | 8.99809 (rank : 87) | NC score | 0.123693 (rank : 45) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8C2Q3, Q3TJB6, Q91Z21, Q9DBI6 | Gene names | Rbm14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 14 (RNA-binding motif protein 14). | |||||
|
SFR12_HUMAN
|
||||||
| θ value | 8.99809 (rank : 88) | NC score | 0.025740 (rank : 186) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8WXA9, Q86X37 | Gene names | SFRS12, SRRP86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86) (Splicing regulatory protein 508) (SRrp508). | |||||
|
UBF1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 89) | NC score | 0.011332 (rank : 196) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P17480 | Gene names | UBTF, UBF, UBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1) (Autoantigen NOR-90). | |||||
|
A2BP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.071601 (rank : 119) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9NWB1, Q9NS20 | Gene names | A2BP1, A2BP | |||
|
Domain Architecture |
|
|||||
| Description | Ataxin-2-binding protein 1. | |||||
|
A2BP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.071089 (rank : 123) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9JJ43, Q9JJB5 | Gene names | A2bp1, A2bp | |||
|
Domain Architecture |
|
|||||
| Description | Ataxin-2-binding protein 1. | |||||
|
BOLL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.057324 (rank : 169) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8N9W6, Q969U3 | Gene names | BOLL, BOULE | |||
|
Domain Architecture |
|
|||||
| Description | Protein boule-like. | |||||
|
BOLL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.058124 (rank : 167) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q924M5, Q9D4V2 | Gene names | Boll, Boule | |||
|
Domain Architecture |
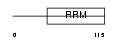
|
|||||
| Description | Protein boule-like. | |||||
|
CIRBP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.082876 (rank : 97) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q14011 | Gene names | CIRBP, A18HNRNP, CIRP | |||
|
Domain Architecture |
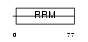
|
|||||
| Description | Cold-inducible RNA-binding protein (Glycine-rich RNA-binding protein CIRP) (A18 hnRNP). | |||||
|
CIRBP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.082660 (rank : 98) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P60824, O09069, O09148, Q3V1A6, Q61413 | Gene names | Cirbp, Cirp | |||
|
Domain Architecture |
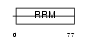
|
|||||
| Description | Cold-inducible RNA-binding protein (Glycine-rich RNA-binding protein CIRP) (A18 hnRNP). | |||||
|
CSTF2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.073272 (rank : 113) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P33240, Q5H951, Q6LA74, Q8N502 | Gene names | CSTF2 | |||
|
Domain Architecture |
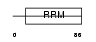
|
|||||
| Description | Cleavage stimulation factor 64 kDa subunit (CSTF 64 kDa subunit) (CF-1 64 kDa subunit) (CstF-64). | |||||
|
CSTF2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.074134 (rank : 110) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8BIQ5, Q8K1Y6, Q9ERC2 | Gene names | Cstf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage stimulation factor 64 kDa subunit (CSTF 64 kDa subunit) (CF-1 64 kDa subunit) (CstF-64). | |||||
|
CSTFT_HUMAN
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.072422 (rank : 115) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9H0L4, O75174, Q53HK6, Q7LGE8, Q8N6T1 | Gene names | CSTF2T, KIAA0689 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage stimulation factor 64 kDa subunit, tau variant (CSTF 64 kDa subunit, tau variant) (CF-1 64 kDa subunit, tau variant) (TauCstF-64). | |||||
|
CSTFT_MOUSE
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.071802 (rank : 117) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8C7E9, Q6A016, Q8BHH7, Q8C3W7, Q8R2Y1, Q9EPU3 | Gene names | Cstf2t, Kiaa0689 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage stimulation factor 64 kDa subunit, tau variant (CSTF 64 kDa subunit, tau variant) (CF-1 64 kDa subunit, tau variant) (TauCstF-64). | |||||
|
CUGB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.090058 (rank : 83) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q92879, Q9NP83, Q9NR06 | Gene names | CUGBP1, BRUNOL2, CUGBP, NAB50 | |||
|
Domain Architecture |

|
|||||
| Description | CUG triplet repeat RNA-binding protein 1 (CUG-BP1) (RNA-binding protein BRUNOL-2) (Deadenylation factor CUG-BP) (50 kDa Nuclear polyadenylated RNA-binding protein) (EDEN-BP). | |||||
|
CUGB1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.089912 (rank : 84) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P28659, Q9CXE5, Q9EPJ8, Q9JI37 | Gene names | Cugbp1, Brunol2, Cugbp | |||
|
Domain Architecture |

|
|||||
| Description | CUG triplet repeat RNA-binding protein 1 (CUG-BP1) (RNA-binding protein BRUNOL-2) (Deadenylation factor CUG-BP) (Deadenylation factor EDEN-BP) (Brain protein F41). | |||||
|
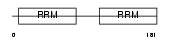
DAZP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.069569 (rank : 126) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q96EP5, Q96MJ3, Q9NRR9 | Gene names | DAZAP1 | |||
|
Domain Architecture |
|
|||||
| Description | DAZ-associated protein 1 (Deleted in azoospermia-associated protein 1). | |||||
|
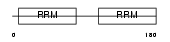
DAZP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.071053 (rank : 124) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9JII5 | Gene names | Dazap1 | |||
|
Domain Architecture |
|
|||||
| Description | DAZ-associated protein 1 (Deleted in azoospermia-associated protein 1). | |||||
|
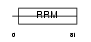
FUSIP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.095508 (rank : 70) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | O75494, O60572, Q96G09, Q96P17 | Gene names | FUSIP1, TASR | |||
|
Domain Architecture |
|
|||||
| Description | FUS-interacting serine-arginine-rich protein 1 (TLS-associated protein with Ser-Arg repeats) (TLS-associated protein with SR repeats) (TASR) (TLS-associated serine-arginine protein) (TLS-associated SR protein) (40 kDa SR-repressor protein) (SRrp40) (Splicing factor SRp38). | |||||
|
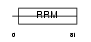
FUSIP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.095508 (rank : 71) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9R0U0, O70307, O88468, Q3TGW7, Q8CFZ1, Q9R0T9 | Gene names | Fusip1, Nssr | |||
|
Domain Architecture |
|
|||||
| Description | FUS-interacting serine-arginine-rich protein 1 (TLS-associated protein with Ser-Arg repeats) (TLS-associated protein with SR repeats) (TASR) (TLS-associated serine-arginine protein) (TLS-associated SR protein) (Neural-specific SR protein) (Neural-salient serine/arginine-rich protein). | |||||
|
HNRCL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.050583 (rank : 183) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O60812 | Gene names | HNRPCL1 | |||
|
Domain Architecture |
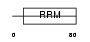
|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein C-like 1 (hnRNP core protein C-like 1). | |||||
|
HNRPD_HUMAN
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.063823 (rank : 143) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q14103, Q01858, Q14100, Q14101, Q14102 | Gene names | HNRPD, AUF1 | |||
|
Domain Architecture |
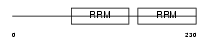
|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein D0 (hnRNP D0) (AU-rich element RNA-binding protein 1). | |||||
|
HNRPD_MOUSE
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.063537 (rank : 145) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q60668, Q60667, Q80ZJ0, Q91X94 | Gene names | Hnrpd, Auf1 | |||
|
Domain Architecture |
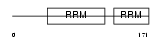
|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein D0 (hnRNP D0) (AU-rich element RNA-binding protein 1). | |||||
|
HNRPG_HUMAN
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.090935 (rank : 79) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P38159, Q5JQ67, Q969R3 | Gene names | RBMX, HNRPG, RBMXP1 | |||
|
Domain Architecture |
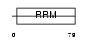
|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein G (hnRNP G) (RNA-binding motif protein, X chromosome) (Glycoprotein p43). | |||||
|
HNRPG_MOUSE
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.087388 (rank : 87) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | O35479 | Gene names | Rbmx, Hnrnpg, Hnrpg, Rbmxp1, Rbmxrt | |||
|
Domain Architecture |
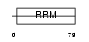
|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein G (hnRNP G) (RNA-binding motif protein, X chromosome). | |||||
|
HNRPM_HUMAN
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.053094 (rank : 179) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P52272, Q15584, Q8WZ44, Q96H56, Q9BWL9, Q9Y492 | Gene names | HNRPM | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein M (hnRNP M). | |||||
|
HNRPM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.059523 (rank : 163) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9D0E1, Q6P1B2, Q99JQ0 | Gene names | Hnrpm | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heterogeneous nuclear ribonucleoprotein M (hnRNP M). | |||||
|
HNRPQ_HUMAN
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.090817 (rank : 80) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O60506, Q53H05, Q5TCG2, Q8IW78, Q8N599, Q96LC1, Q96LC2, Q9Y583 | Gene names | SYNCRIP, HNRPQ, NSAP1 | |||
|
Domain Architecture |
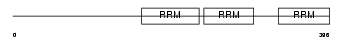
|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein Q (hnRNP Q) (hnRNP-Q) (Synaptotagmin-binding, cytoplasmic RNA-interacting protein) (Glycine- and tyrosine-rich RNA-binding protein) (GRY-RBP) (NS1-associated protein 1). | |||||
|
HNRPQ_MOUSE
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.087833 (rank : 86) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q7TMK9, O88991, Q80YV6, Q8BGP1, Q8C5K6, Q8CGC2, Q91ZR0, Q99KU9, Q9QYF4 | Gene names | Syncrip, Hnrpq, Nsap1, Nsap1l | |||
|
Domain Architecture |
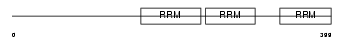
|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein Q (hnRNP Q) (hnRNP-Q) (Synaptotagmin-binding, cytoplasmic RNA-interacting protein) (Glycine- and tyrosine-rich RNA-binding protein) (GRY-RBP) (NS1-associated protein 1) (pp68). | |||||
|
HNRPR_HUMAN
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.082001 (rank : 103) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O43390 | Gene names | HNRPR | |||
|
Domain Architecture |
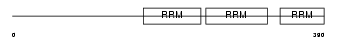
|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein R (hnRNP R). | |||||
|
IF2B2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.061958 (rank : 154) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Y6M1 | Gene names | IGF2BP2, IMP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulin-like growth factor 2 mRNA-binding protein 2 (IGF-II mRNA- binding protein 2) (IMP-2) (Hepatocellular carcinoma autoantigen p62). | |||||
|
IF2B2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.052961 (rank : 180) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q5SF07, Q3TCU4, Q7TQF9 | Gene names | Igf2bp2, Imp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulin-like growth factor 2 mRNA-binding protein 2 (IGF-II mRNA- binding protein 2) (IMP-2). | |||||
|
IF34_HUMAN
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.071755 (rank : 118) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O75821, O14801, Q969U5 | Gene names | EIF3S4 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 4 (eIF-3 delta) (eIF3 p44) (eIF-3 RNA-binding subunit) (eIF3 p42) (eIF3g). | |||||
|
IF34_MOUSE
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.072030 (rank : 116) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9Z1D1, Q9R079 | Gene names | Eif3s4, Eif3p42 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 4 (eIF-3 delta) (eIF3 p44) (eIF-3 RNA-binding subunit) (eIF3 p42) (Eif3p42) (eIF3g). | |||||
|
MINT_HUMAN
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.055656 (rank : 173) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
MINT_MOUSE
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.053973 (rank : 174) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
MSI1H_HUMAN
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.063701 (rank : 144) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O43347, Q96PU0, Q96PU1, Q96PU2, Q96PU3 | Gene names | MSI1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein Musashi homolog 1 (Musashi-1). | |||||
|
MSI1H_MOUSE
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.062740 (rank : 149) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q61474, Q8BNC7 | Gene names | Msi1, Msi1h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein Musashi homolog 1 (Musashi-1). | |||||
|
MSI2H_HUMAN
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.062259 (rank : 151) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q96DH6, Q7Z6M7, Q8N9T4 | Gene names | MSI2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein Musashi homolog 2 (Musashi-2). | |||||
|
MSI2H_MOUSE
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.062802 (rank : 148) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q920Q6, Q8BQ90, Q920Q7 | Gene names | Msi2, Msi2h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein Musashi homolog 2 (Musashi-2). | |||||
|
MYEF2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.053585 (rank : 175) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9P2K5, Q6NUM5, Q7L388, Q7Z4B7, Q9H922, Q9NUQ1 | Gene names | MYEF2, KIAA1341 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin expression factor 2 (MyEF-2) (MST156). | |||||
|
MYEF2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.057325 (rank : 168) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8C854, Q60690, Q6P930, Q6ZPT3, Q8QZZ1 | Gene names | Myef2, Kiaa1341, Mef2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin expression factor 2 (MyEF-2) (MEF-2). | |||||
|
NCBP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.071389 (rank : 120) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P52298, Q14924, Q2TS50 | Gene names | NCBP2, CBP20, PIG55 | |||
|
Domain Architecture |
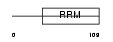
|
|||||
| Description | Nuclear cap-binding protein subunit 2 (20 kDa nuclear cap-binding protein) (NCBP 20 kDa subunit) (CBP20) (NCBP-interacting protein 1) (NIP1) (Cell proliferation-inducing gene 55 protein). | |||||
|
NCBP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.071123 (rank : 122) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9CQ49, Q3UI86 | Gene names | Ncbp2 | |||
|
Domain Architecture |
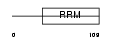
|
|||||
| Description | Nuclear cap-binding protein subunit 2 (20 kDa nuclear cap-binding protein) (NCBP 20 kDa subunit) (CBP20). | |||||
|
NUCL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.079818 (rank : 104) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P19338, Q53SK1, Q8NB06 | Gene names | NCL | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolin (Protein C23). | |||||
|
PABP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.091866 (rank : 77) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P11940, Q15097, Q93004 | Gene names | PABPC1, PAB1, PABP1, PABPC2 | |||
|
Domain Architecture |

|
|||||
| Description | Polyadenylate-binding protein 1 (Poly(A)-binding protein 1) (PABP 1). | |||||
|
PABP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.092624 (rank : 76) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P29341 | Gene names | Pabpc1, Pabp1 | |||
|
Domain Architecture |

|
|||||
| Description | Polyadenylate-binding protein 1 (Poly(A)-binding protein 1) (PABP 1). | |||||
|
PABP3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.088687 (rank : 85) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9H361, Q8NHV0, Q9H086 | Gene names | PABPC3, PABP3, PABPL3 | |||
|
Domain Architecture |

|
|||||
| Description | Polyadenylate-binding protein 3 (Poly(A)-binding protein 3) (PABP 3) (Testis-specific poly(A)-binding protein). | |||||
|
PABP4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.087272 (rank : 88) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q13310, Q6P0N3 | Gene names | PABPC4, APP1, PABP4 | |||
|
Domain Architecture |

|
|||||
| Description | Polyadenylate-binding protein 4 (Poly(A)-binding protein 4) (PABP 4) (Inducible poly(A)-binding protein) (iPABP) (Activated-platelet protein 1) (APP-1). | |||||
|
PABP5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.099759 (rank : 64) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q96DU9, Q6P529, Q9UFE5 | Gene names | PABPC5, PABP5 | |||
|
Domain Architecture |
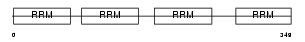
|
|||||
| Description | Polyadenylate-binding protein 5 (Poly(A)-binding protein 5) (PABP 5). | |||||
|
PPIL4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.056527 (rank : 172) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8WUA2, Q7Z3Q5 | Gene names | PPIL4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peptidyl-prolyl cis-trans isomerase-like 4 (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-like protein PPIL4). | |||||
|
PPIL4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.056630 (rank : 170) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9CXG3, Q3TJX1, Q68FC7, Q9CT22 | Gene names | Ppil4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peptidyl-prolyl cis-trans isomerase-like 4 (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-like protein PPIL4). | |||||
|
RAVR1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.072756 (rank : 114) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8IY67, Q8IY60, Q8TF24 | Gene names | RAVER1, KIAA1978 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribonucleoprotein PTB-binding 1 (Protein raver-1). | |||||
|
RAVR1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.071179 (rank : 121) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9CW46, Q5DTT6, Q811K0, Q8C3Z1, Q8CA14 | Gene names | Raver1, Kiaa1978 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribonucleoprotein PTB-binding 1 (Protein raver-1). | |||||
|
RAVR2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.073888 (rank : 111) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9HCJ3, Q6P141, Q9NPV7 | Gene names | RAVER2, KIAA1579 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribonucleoprotein PTB-binding 2 (Protein raver-2). | |||||
|
RAVR2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.066222 (rank : 136) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q7TPD6 | Gene names | Raver2, Kiaa1579 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribonucleoprotein PTB-binding 2 (Protein raver-2). | |||||
|
RB15B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.062298 (rank : 150) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8NDT2, Q6QE19, Q9BV96 | Gene names | RBM15B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative RNA-binding protein 15B (RNA-binding motif protein 15B). | |||||
|
RB15B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.060982 (rank : 157) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q6PHZ5, Q8C6G2 | Gene names | Rbm15b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative RNA-binding protein 15B (RNA-binding motif protein 15B). | |||||
|
RBM11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.094098 (rank : 73) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P57052 | Gene names | RBM11 | |||
|
Domain Architecture |
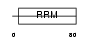
|
|||||
| Description | Putative RNA-binding protein 11 (RNA-binding motif protein 11). | |||||
|
RBM15_HUMAN
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.051820 (rank : 182) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 309 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q96T37, Q5D058, Q96PE4, Q96SC5, Q96SC6, Q96SC9, Q96SD0, Q96T38, Q9BRA5, Q9H6R8, Q9H9Y0 | Gene names | RBM15, OTT | |||
|
Domain Architecture |

|
|||||
| Description | Putative RNA-binding protein 15 (RNA-binding motif protein 15) (One- twenty two protein). | |||||
|
RBM28_HUMAN
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.085762 (rank : 91) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9NW13, Q96CV3 | Gene names | RBM28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 28 (RNA-binding motif protein 28). | |||||
|
RBM28_MOUSE
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.093049 (rank : 75) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q8CGC6, Q8BG25, Q8VEJ8, Q9CS22, Q9CSE6 | Gene names | Rbm28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 28 (RNA-binding motif protein 28). | |||||
|
RBM34_HUMAN
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.065757 (rank : 138) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P42696, Q8N2Z8, Q9H5A1 | Gene names | RBM34, KIAA0117 | |||
|
Domain Architecture |
|
|||||
| Description | RNA-binding protein 34 (RNA-binding motif protein 34). | |||||
|
RBM34_MOUSE
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.064269 (rank : 141) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8C5L7 | Gene names | Rbm34, D8Ertd233e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 34 (RNA-binding motif protein 34). | |||||
|
RBM3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.078639 (rank : 107) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P98179 | Gene names | RBM3, RNPL | |||
|
Domain Architecture |
|
|||||
| Description | Putative RNA-binding protein 3 (RNA-binding motif protein 3) (RNPL). | |||||
|
RBM3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.079812 (rank : 105) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O89086 | Gene names | Rbm3 | |||
|
Domain Architecture |
|
|||||
| Description | Putative RNA-binding protein 3 (RNA-binding motif protein 3). | |||||
|
RBM8A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.083350 (rank : 95) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9Y5S9, Q6FHD1, Q9GZX8, Q9NZI4 | Gene names | RBM8A, RBM8 | |||
|
Domain Architecture |
|
|||||
| Description | RNA-binding protein 8A (RNA-binding motif protein 8A) (Ribonucleoprotein RBM8A) (RNA-binding protein Y14) (Binder of OVCA1- 1) (BOV-1). | |||||
|
RBM8A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.083350 (rank : 96) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9CWZ3, Q9D6U5 | Gene names | Rbm8a, Rbm8 | |||
|
Domain Architecture |
|
|||||
| Description | RNA-binding protein 8A (RNA-binding motif protein 8A) (Ribonucleoprotein RBM8A). | |||||
|
RBM9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.078165 (rank : 108) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | O43251, Q5TF71, Q6IC09, Q8TD00, Q8WYB1, Q96DZ6, Q96NL7, Q9UGW4, Q9UH33 | Gene names | RBM9, HRNBP2, RTA | |||
|
Domain Architecture |
|
|||||
| Description | RNA-binding protein 9 (RNA-binding motif protein 9) (Hexaribonucleotide-binding protein 2) (Repressor of tamoxifen transcriptional activity). | |||||
|
RBMS2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.082543 (rank : 99) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q15434 | Gene names | RBMS2, SCR3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding motif, single-stranded-interacting protein 2 (Suppressor of CDC2 with RNA-binding motif 3). | |||||
|
RBMS2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.082007 (rank : 102) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8VC70 | Gene names | Rbms2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding motif, single-stranded-interacting protein 2. | |||||
|
RBPMS_MOUSE
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.050432 (rank : 184) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9WVB0 | Gene names | Rbpms, Hermes | |||
|
Domain Architecture |
|
|||||
| Description | RNA-binding protein with multiple splicing (RBP-MS) (HEart, RRM Expressed Sequence) (Hermes). | |||||
|
REFP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.052958 (rank : 181) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9JJW6, Q8C869 | Gene names | Refbp2, Ref2 | |||
|
Domain Architecture |
|
|||||
| Description | RNA and export factor-binding protein 2. | |||||
|
RNPC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.068260 (rank : 128) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9H0Z9, Q15350, Q15351, Q9BYK3, Q9BYK4 | Gene names | RNPC1, SEB4 | |||
|
Domain Architecture |
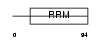
|
|||||
| Description | RNA-binding region-containing protein 1 (HSRNASEB) (ssDNA-binding protein SEB4) (CLL-associated antigen KW-5). | |||||
|
RNPC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.065198 (rank : 140) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q62176, Q922Z2 | Gene names | Rnpc1, Seb4, Seb4l | |||
|
Domain Architecture |
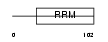
|
|||||
| Description | RNA-binding region-containing protein 1 (ssDNA-binding protein SEB4). | |||||
|
RNPC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.082219 (rank : 100) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q14498, Q14499 | Gene names | RNPC2, HCC1 | |||
|
Domain Architecture |
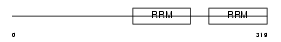
|
|||||
| Description | RNA-binding region-containing protein 2 (Hepatocellular carcinoma protein 1) (Splicing factor HCC1). | |||||
|
RNPC2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.082210 (rank : 101) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q8VH51, Q99KV0 | Gene names | Rnpc2, Caper | |||
|
Domain Architecture |
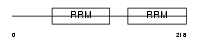
|
|||||
| Description | RNA-binding region-containing protein 2 (Coactivator of activating protein 1 and estrogen receptors) (Coactivator of AP-1 and ERs) (Transcription coactivator CAPER). | |||||
|
RNPS1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.067378 (rank : 131) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q15287, O75308, Q8WY42, Q9NYG3 | Gene names | RNPS1, LDC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein with serine-rich domain 1 (SR-related protein LDC2). | |||||
|
RNPS1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.067378 (rank : 132) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q99M28, Q3TMJ1, Q922H8 | Gene names | Rnps1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein with serine-rich domain 1. | |||||
|
ROA0_HUMAN
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.053354 (rank : 176) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q13151 | Gene names | HNRPA0 | |||
|
Domain Architecture |
|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein A0 (hnRNP A0). | |||||
|
ROA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.059965 (rank : 162) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P09651, Q6PJZ7 | Gene names | HNRPA1 | |||
|
Domain Architecture |
|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein A1 (Helix-destabilizing protein) (Single-strand RNA-binding protein) (hnRNP core protein A1). | |||||
|
ROA1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.060182 (rank : 161) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P49312, P97312, Q3V269 | Gene names | Hnrpa1, Fli-2, Tis | |||
|
Domain Architecture |
|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein A1 (Helix-destabilizing protein) (Single-strand-binding protein) (hnRNP core protein A1) (HDP- 1) (Topoisomerase-inhibitor suppressed). | |||||
|
ROA2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.060257 (rank : 159) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P22626, P22627 | Gene names | HNRPA2B1 | |||
|
Domain Architecture |
|
|||||
| Description | Heterogeneous nuclear ribonucleoproteins A2/B1 (hnRNP A2 / hnRNP B1). | |||||
|
ROA2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.060188 (rank : 160) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O88569 | Gene names | Hnrpa2b1 | |||
|
Domain Architecture |
|
|||||
| Description | Heterogeneous nuclear ribonucleoproteins A2/B1 (hnRNP A2 / hnRNP B1). | |||||
|
ROA3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.053116 (rank : 177) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P51991, Q6URK5 | Gene names | HNRPA3 | |||
|
Domain Architecture |
|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein A3 (hnRNP A3). | |||||
|
ROA3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.053106 (rank : 178) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8BG05, Q8BHF8 | Gene names | Hnrpa3, Hnrnpa3 | |||
|
Domain Architecture |
|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein A3 (hnRNP A3). | |||||
|
ROAA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.056551 (rank : 171) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q99729, Q04150, Q8N7U3, Q9BQ99 | Gene names | HNRPAB, ABBP1 | |||
|
Domain Architecture |
|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein A/B (hnRNP A/B) (APOBEC-1- binding protein 1) (ABBP-1). | |||||
|
ROAA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.059206 (rank : 165) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q99020, Q8BPE0 | Gene names | Hnrpab, Cbf-a, Cgbfa | |||
|
Domain Architecture |
|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein A/B (hnRNP A/B) (CArG-binding factor-A) (CBF-A). | |||||
|
SART3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.061917 (rank : 155) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 301 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q15020, Q58F06, Q8IUS1, Q96J95 | Gene names | SART3, KIAA0156, TIP110 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Squamous cell carcinoma antigen recognized by T-cells 3 (SART-3) (hSART-3) (Tat-interacting protein of 110 kDa) (Tip110). | |||||
|
SART3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.062049 (rank : 152) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9JLI8, Q6ZQI2, Q8BPK9, Q8C3B7, Q8CFU9 | Gene names | Sart3, Kiaa0156 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Squamous cell carcinoma antigen recognized by T-cells 3 (SART-3) (mSART-3) (Tumor-rejection antigen SART3). | |||||
|
SF3B4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.094343 (rank : 72) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q15427 | Gene names | SF3B4, SAP49 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 4 (Spliceosome-associated protein 49) (SAP 49) (SF3b50) (Pre-mRNA-splicing factor SF3b 49 kDa subunit). | |||||
|
SFRS1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.067075 (rank : 133) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q07955, Q13809 | Gene names | SFRS1, ASF, SF2, SF2P33 | |||
|
Domain Architecture |
|
|||||
| Description | Splicing factor, arginine/serine-rich 1 (pre-mRNA-splicing factor SF2, P33 subunit) (Alternative-splicing factor 1) (ASF-1). | |||||
|
SFRS1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.067075 (rank : 134) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q6PDM2, Q3UCH2, Q5SXC5, Q8BJV3, Q8C1H9 | Gene names | Sfrs1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 1. | |||||
|
SFRS4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.058233 (rank : 166) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q08170, Q5VXP1, Q9BUA4, Q9UEB5 | Gene names | SFRS4, SRP75 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4 (Pre-mRNA-splicing factor SRP75) (SRP001LB). | |||||
|
SFRS4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.059384 (rank : 164) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8VE97, Q9JJC3 | Gene names | Sfrs4 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4. | |||||
|
SFRS5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 181) | NC score | 0.061519 (rank : 156) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q13243, O14797, Q16662 | Gene names | SFRS5, HRS, SRP40 | |||
|
Domain Architecture |
|
|||||
| Description | Splicing factor, arginine/serine-rich 5 (Pre-mRNA-splicing factor SRP40) (Delayed-early protein HRS). | |||||
|
SFRS5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 182) | NC score | 0.063317 (rank : 147) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O35326 | Gene names | Sfrs5, Hrs | |||
|
Domain Architecture |
|
|||||
| Description | Splicing factor, arginine/serine-rich 5 (Pre-mRNA-splicing factor SRP40) (Delayed-early protein HRS). | |||||
|
SFRS6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 183) | NC score | 0.066278 (rank : 135) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q13247, Q13244, Q13245, Q96J06, Q9UJB8, Q9Y3N7 | Gene names | SFRS6, SRP55 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 6 (Pre-mRNA-splicing factor SRP55). | |||||
|
SFRS9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 184) | NC score | 0.073341 (rank : 112) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q13242 | Gene names | SFRS9, SRP30C | |||
|
Domain Architecture |
|
|||||
| Description | Splicing factor, arginine/serine-rich 9 (Pre-mRNA-splicing factor SRp30C). | |||||
|
SFRS9_MOUSE
|
||||||
| θ value | θ > 10 (rank : 185) | NC score | 0.070719 (rank : 125) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9D0B0, Q9CRN3 | Gene names | Sfrs9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 9. | |||||
|
SRR35_HUMAN
|
||||||
| θ value | θ > 10 (rank : 186) | NC score | 0.101089 (rank : 58) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q8WXF0, Q8WW25 | Gene names | SRRP35 | |||
|
Domain Architecture |
|
|||||
| Description | 35 kDa SR repressor protein (SRrp35). | |||||
|
TADBP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 187) | NC score | 0.065667 (rank : 139) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q13148, Q96DJ0 | Gene names | TARDBP, TDP43 | |||
|
Domain Architecture |
|
|||||
| Description | TAR DNA-binding protein 43 (TDP-43). | |||||
|
TADBP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 188) | NC score | 0.061980 (rank : 153) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q921F2 | Gene names | Tardbp, Tdp43 | |||
|
Domain Architecture |
|
|||||
| Description | TAR DNA-binding protein 43 (TDP-43). | |||||
|
THOC4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 189) | NC score | 0.060875 (rank : 158) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q86V81, O43672 | Gene names | THOC4, ALY, BEF | |||
|
Domain Architecture |
|
|||||
| Description | THO complex subunit 4 (Tho4) (Ally of AML-1 and LEF-1) (Transcriptional coactivator Aly/REF) (bZIP-enhancing factor BEF). | |||||
|
THOC4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 190) | NC score | 0.063321 (rank : 146) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O08583, Q8CBM4, Q9JJW7 | Gene names | Thoc4, Aly, Ref1, Refbp1 | |||
|
Domain Architecture |
|
|||||
| Description | THO complex subunit 4 (Tho4) (RNA and export factor-binding protein 1) (REF1-I) (Ally of AML-1 and LEF-1) (Aly/REF). | |||||
|
TIA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 191) | NC score | 0.093460 (rank : 74) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P31483 | Gene names | TIA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolysin TIA-1 isoform p40 (RNA-binding protein TIA-1) (p40-TIA-1) [Contains: Nucleolysin TIA-1 isoform p15 (p15-TIA-1)]. | |||||
|
TIA1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 192) | NC score | 0.091537 (rank : 78) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P52912 | Gene names | Tia1, Tia | |||
|
Domain Architecture |
|
|||||
| Description | Nucleolysin TIA-1 (RNA-binding protein TIA-1). | |||||
|
TIAR_HUMAN
|
||||||
| θ value | θ > 10 (rank : 193) | NC score | 0.086277 (rank : 90) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q01085 | Gene names | TIAL1 | |||
|
Domain Architecture |
|
|||||
| Description | Nucleolysin TIAR (TIA-1-related protein). | |||||
|
TIAR_MOUSE
|
||||||
| θ value | θ > 10 (rank : 194) | NC score | 0.085450 (rank : 93) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P70318 | Gene names | Tial1 | |||
|
Domain Architecture |
|
|||||
| Description | Nucleolysin TIAR (TIA-1-related protein). | |||||
|
U2AF2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 195) | NC score | 0.068260 (rank : 129) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P26368 | Gene names | U2AF2, U2AF65 | |||
|
Domain Architecture |
|
|||||
| Description | Splicing factor U2AF 65 kDa subunit (U2 auxiliary factor 65 kDa subunit) (U2 snRNP auxiliary factor large subunit) (hU2AF(65)). | |||||
|
U2AF2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 196) | NC score | 0.068524 (rank : 127) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P26369 | Gene names | U2af2, U2af65 | |||
|
Domain Architecture |
|
|||||
| Description | Splicing factor U2AF 65 kDa subunit (U2 auxiliary factor 65 kDa subunit) (U2 snRNP auxiliary factor large subunit). | |||||
|
PTBP2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | Q9UKA9, Q8N0Z1, Q8N160, Q8NFB0, Q8NFB1, Q969N9, Q96Q76 | Gene names | PTBP2, NPTB, PTB, PTBLP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypyrimidine tract-binding protein 2 (Neurally-enriched homolog of PTB) (Neural polypyrimidine tract-binding protein) (PTB-like protein). | |||||
|
PTBP2_MOUSE
|
||||||
| NC score | 0.999946 (rank : 2) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | Q91Z31, Q7TNW7, Q9CUW2, Q9QYC2, Q9R0V9 | Gene names | Ptbp2, Brptb, Nptb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypyrimidine tract-binding protein 2 (Brain-enriched polypyrimidine tract-binding protein) (Brain-enriched PTB) (RRM-type RNA-binding protein brPTB) (Neural polypyrimidine tract-binding protein). | |||||
|
PTBP1_HUMAN
|
||||||
| NC score | 0.988147 (rank : 3) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P26599 | Gene names | PTBP1, PTB | |||
|
Domain Architecture |

|
|||||
| Description | Polypyrimidine tract-binding protein 1 (PTB) (Heterogeneous nuclear ribonucleoprotein I) (hnRNP I) (57 kDa RNA-binding protein PPTB-1). | |||||
|
ROD1_HUMAN
|
||||||
| NC score | 0.982471 (rank : 4) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O95758, Q86YB3, Q86YH9 | Gene names | ROD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of differentiation 1 (Rod1). | |||||
|
ROD1_MOUSE
|
||||||
| NC score | 0.980288 (rank : 5) | θ value | 0 (rank : 6) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8BHD7, Q923C3 | Gene names | Rod1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of differentiation 1 (Rod1). | |||||
|
PTBP1_MOUSE
|
||||||
| NC score | 0.980214 (rank : 6) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P17225 | Gene names | Ptbp1, Ptb | |||
|
Domain Architecture |

|
|||||
| Description | Polypyrimidine tract-binding protein 1 (PTB) (Heterogeneous nuclear ribonucleoprotein I) (hnRNP I). | |||||
|
HNRLL_HUMAN
|
||||||
| NC score | 0.783992 (rank : 7) | θ value | 4.71619e-41 (rank : 7) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8WVV9, Q659B9, Q8IVH5, Q8IVH6, Q96HR5 | Gene names | HNRPLL, SRRF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heterogeneous nuclear ribonucleoprotein L-like (Stromal RNA-regulating factor) (BLOCK24 protein). | |||||
|
HNRLL_MOUSE
|
||||||
| NC score | 0.780440 (rank : 8) | θ value | 6.15952e-41 (rank : 8) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q921F4, Q8BIP6, Q99J40 | Gene names | Hnrpll | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heterogeneous nuclear ribonucleoprotein L-like. | |||||
|
HNRPL_HUMAN
|
||||||
| NC score | 0.734917 (rank : 9) | θ value | 1.6852e-22 (rank : 9) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P14866, Q9H3P3 | Gene names | HNRPL | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein L (hnRNP L). | |||||
|
HNRPL_MOUSE
|
||||||
| NC score | 0.734129 (rank : 10) | θ value | 1.6852e-22 (rank : 10) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8R081, O54789, Q8K0S7 | Gene names | Hnrpl | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein L (hnRNP L). | |||||
|
MATR3_MOUSE
|
||||||
| NC score | 0.312859 (rank : 11) | θ value | 1.69304e-06 (rank : 12) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8K310 | Gene names | Matr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Matrin-3. | |||||
|
MATR3_HUMAN
|
||||||
| NC score | 0.311924 (rank : 12) | θ value | 1.69304e-06 (rank : 11) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P43243, Q9UHW0, Q9UQ27 | Gene names | MATR3, KIAA0723 | |||
|
Domain Architecture |

|
|||||
| Description | Matrin-3. | |||||
|
RU2B_HUMAN
|
||||||
| NC score | 0.250090 (rank : 13) | θ value | 9.29e-05 (rank : 13) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P08579, Q9UJD4 | Gene names | SNRPB2 | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein B''. | |||||
|
RU2B_MOUSE
|
||||||
| NC score | 0.247556 (rank : 14) | θ value | 9.29e-05 (rank : 14) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9CQI7, Q9CW35, Q9CZ66 | Gene names | Snrpb2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U2 small nuclear ribonucleoprotein B''. | |||||
|
RBM7_HUMAN
|
||||||
| NC score | 0.201969 (rank : 15) | θ value | 0.00298849 (rank : 23) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9Y580 | Gene names | RBM7 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 7 (RNA-binding motif protein 7). | |||||
|
NONO_MOUSE
|
||||||
| NC score | 0.200482 (rank : 16) | θ value | 0.00020696 (rank : 16) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q99K48, Q63887, Q9CYQ4, Q9DBP2 | Gene names | Nono | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Non-POU domain-containing octamer-binding protein (NonO protein). | |||||
|
ZN638_MOUSE
|
||||||
| NC score | 0.200433 (rank : 17) | θ value | 0.00509761 (rank : 24) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q61464, Q6DFV9, Q8C941 | Gene names | Znf638, Np220, Zfml | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 638 (Nuclear protein 220) (Zinc-finger matrin-like protein). | |||||
|
NONO_HUMAN
|
||||||
| NC score | 0.199312 (rank : 18) | θ value | 0.00020696 (rank : 15) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q15233, O00201, P30807, Q12786, Q9BQC5 | Gene names | NONO, NRB54 | |||
|
Domain Architecture |

|
|||||
| Description | Non-POU domain-containing octamer-binding protein (NonO protein) (54 kDa nuclear RNA- and DNA-binding protein) (p54(nrb)) (p54nrb) (55 kDa nuclear protein) (NMT55) (DNA-binding p52/p100 complex, 52 kDa subunit). | |||||
|
ZCRB1_HUMAN
|
||||||
| NC score | 0.197308 (rank : 19) | θ value | 0.279714 (rank : 38) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q8TBF4, Q6PJX0 | Gene names | ZCRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC-type and RNA-binding motif-containing protein 1 (U11/U12 snRNP 31 kDa protein) (U11/U12-31K). | |||||
|
RBM7_MOUSE
|
||||||
| NC score | 0.190281 (rank : 20) | θ value | 0.00869519 (rank : 25) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9CQT2, Q3UB91, Q7TQE3 | Gene names | Rbm7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 7 (RNA-binding motif protein 7). | |||||
|
ELAV4_MOUSE
|
||||||
| NC score | 0.188263 (rank : 21) | θ value | 0.0252991 (rank : 29) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q61701, O55010 | Gene names | Elavl4, Hud | |||
|
Domain Architecture |

|
|||||
| Description | ELAV-like protein 4 (Paraneoplastic encephalomyelitis antigen HuD) (Hu-antigen D). | |||||
|
ELAV4_HUMAN
|
||||||
| NC score | 0.186953 (rank : 22) | θ value | 0.0330416 (rank : 30) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P26378, Q96J74 | Gene names | ELAVL4, HUD, PNEM | |||
|
Domain Architecture |

|
|||||
| Description | ELAV-like protein 4 (Paraneoplastic encephalomyelitis antigen HuD) (Hu-antigen D). | |||||
|
ZN638_HUMAN
|
||||||
| NC score | 0.186860 (rank : 23) | θ value | 0.00869519 (rank : 26) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q14966, Q53R34, Q5XJ05, Q68DP3, Q6P2H2, Q7Z3T7, Q8NF92, Q8TCA1, Q9H2G1, Q9NP37 | Gene names | ZNF638, NP220, ZFML | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 638 (Nuclear protein 220) (Zinc-finger matrin-like protein) (Cutaneous T-cell lymphoma-associated antigen se33-1) (CTCL tumor antigen se33-1). | |||||
|
RBM4_MOUSE
|
||||||
| NC score | 0.185939 (rank : 24) | θ value | 0.00035302 (rank : 21) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8C7Q4, O08752, Q8BN66 | Gene names | Rbm4, Rbm4a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 4 (RNA-binding motif protein 4) (RNA-binding motif protein 4a) (Lark homolog) (mLark). | |||||
|
RBM4_HUMAN
|
||||||
| NC score | 0.185902 (rank : 25) | θ value | 0.000461057 (rank : 22) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9BWF3, O02916, Q6P1P2, Q8WU85 | Gene names | RBM4, RBM4A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 4 (RNA-binding motif protein 4) (RNA-binding motif protein 4a) (Lark homolog) (hLark). | |||||
|
RBM4B_MOUSE
|
||||||
| NC score | 0.185774 (rank : 26) | θ value | 0.00035302 (rank : 20) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8VE92 | Gene names | Rbm4b, Rbm30 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 4B (RNA-binding motif protein 4B) (RNA-binding protein 30) (RNA-binding motif protein 30). | |||||
|
RBM4B_HUMAN
|
||||||
| NC score | 0.185725 (rank : 27) | θ value | 0.00035302 (rank : 19) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9BQ04 | Gene names | RBM4B, RBM30 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 4B (RNA-binding motif protein 4B) (RNA-binding protein 30) (RNA-binding motif protein 30). | |||||
|
ELAV3_HUMAN
|
||||||
| NC score | 0.183726 (rank : 28) | θ value | 0.00035302 (rank : 17) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q14576, Q16135, Q96CL8, Q96QS9 | Gene names | ELAVL3, HUC, PLE21 | |||
|
Domain Architecture |

|
|||||
| Description | ELAV-like protein 3 (Hu-antigen C) (HuC) (Paraneoplastic cerebellar degeneration-associated antigen) (Paraneoplastic limbic encephalitis antigen 21). | |||||
|
ELAV2_MOUSE
|
||||||
| NC score | 0.180850 (rank : 29) | θ value | 0.0113563 (rank : 28) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q60899 | Gene names | Elavl2, Hub | |||
|
Domain Architecture |

|
|||||
| Description | ELAV-like protein 2 (Hu-antigen B) (HuB) (ELAV-like neuronal protein 1) (Nervous system-specific RNA-binding protein Mel-N1). | |||||
|
ELAV3_MOUSE
|
||||||
| NC score | 0.180817 (rank : 30) | θ value | 0.00035302 (rank : 18) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q60900, Q60901 | Gene names | Elavl3, Huc | |||
|
Domain Architecture |

|
|||||
| Description | ELAV-like protein 3 (Hu-antigen C) (HuC). | |||||
|
ELAV2_HUMAN
|
||||||
| NC score | 0.180486 (rank : 31) | θ value | 0.0113563 (rank : 27) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q12926, Q13235, Q59G15, Q8NEM4, Q9H1Q8 | Gene names | ELAVL2, HUB | |||
|
Domain Architecture |

|
|||||
| Description | ELAV-like protein 2 (Hu-antigen B) (HuB) (ELAV-like neuronal protein 1) (Nervous system-specific RNA-binding protein Hel-N1). | |||||
|
ZCRB1_MOUSE
|
||||||
| NC score | 0.171090 (rank : 32) | θ value | 1.81305 (rank : 53) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9CZ96, Q3TDL3 | Gene names | Zcrb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC-type and RNA-binding motif-containing protein 1 (MADP-1). | |||||
|
ELAV1_HUMAN
|
||||||
| NC score | 0.163700 (rank : 33) | θ value | 0.0961366 (rank : 34) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q15717, Q9BTT1 | Gene names | ELAVL1, HUR | |||
|
Domain Architecture |

|
|||||
| Description | ELAV-like protein 1 (Hu-antigen R) (HuR). | |||||
|
SFRS2_HUMAN
|
||||||
| NC score | 0.143425 (rank : 34) | θ value | 0.279714 (rank : 36) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q01130 | Gene names | SFRS2 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 2 (Splicing factor SC35) (SC-35) (Splicing component, 35 kDa) (Protein PR264). | |||||
|
SFRS2_MOUSE
|
||||||
| NC score | 0.143425 (rank : 35) | θ value | 0.279714 (rank : 37) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q62093, Q542L3, Q60701 | Gene names | Sfrs2, Pr264, Sfrs10 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 2 (Splicing factor SC35) (SC-35) (Splicing component, 35 kDa) (Protein PR264) (Putative myelin regulatory factor 1) (MRF-1). | |||||
|
DRBP1_HUMAN
|
||||||
| NC score | 0.141800 (rank : 36) | θ value | 0.0736092 (rank : 31) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8IUH3, Q6NYL0, Q8NFC9 | Gene names | DRBP1, DRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Developmentally-regulated RNA-binding protein 1 (RB-1). | |||||
|
SNRPA_MOUSE
|
||||||
| NC score | 0.141609 (rank : 37) | θ value | 2.36792 (rank : 60) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q62189, Q52LA2 | Gene names | Snrpa, Rnu1a-1 | |||
|
Domain Architecture |

|
|||||
| Description | U1 small nuclear ribonucleoprotein A (U1 snRNP protein A) (U1A protein) (U1-A). | |||||
|
ELAV1_MOUSE
|
||||||
| NC score | 0.141316 (rank : 38) | θ value | 0.0961366 (rank : 35) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P70372, Q60745 | Gene names | Elavl1, Elra, Hua | |||
|
Domain Architecture |

|
|||||
| Description | ELAV-like protein 1 (Hu-antigen R) (HuR) (Elav-like generic protein) (MelG). | |||||
|
DRBP1_MOUSE
|
||||||
| NC score | 0.132675 (rank : 39) | θ value | 0.813845 (rank : 41) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8BHN5, Q8C1Z5 | Gene names | Drbp1, Drb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Developmentally-regulated RNA-binding protein 1 (RB-1). | |||||
|
RBM19_MOUSE
|
||||||
| NC score | 0.131757 (rank : 40) | θ value | 1.06291 (rank : 45) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q8R3C6, Q8BHR0, Q9CW63 | Gene names | Rbm19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable RNA-binding protein 19 (RNA-binding motif protein 19). | |||||
|
RBM19_HUMAN
|
||||||
| NC score | 0.128465 (rank : 41) | θ value | 4.03905 (rank : 65) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9Y4C8, Q9BPY6, Q9UFN5 | Gene names | RBM19, KIAA0682 | |||
|
Domain Architecture |

|
|||||
| Description | Probable RNA-binding protein 19 (RNA-binding motif protein 19). | |||||
|
RBMX2_MOUSE
|
||||||
| NC score | 0.125786 (rank : 42) | θ value | 0.813845 (rank : 43) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q8R0F5, Q3TI42 | Gene names | Rbmx2 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding motif protein, X-linked 2. | |||||
|
PM14_HUMAN
|
||||||
| NC score | 0.124052 (rank : 43) | θ value | 8.99809 (rank : 84) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9Y3B4 | Gene names | SF3B14 | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA branch site protein p14 (SF3B 14 kDa subunit). | |||||
|
PM14_MOUSE
|
||||||
| NC score | 0.124052 (rank : 44) | θ value | 8.99809 (rank : 85) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P59708, Q3U9T9 | Gene names | Sf3b14 | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA branch site protein p14 (SF3B 14 kDa subunit). | |||||
|
RBM14_MOUSE
|
||||||
| NC score | 0.123693 (rank : 45) | θ value | 8.99809 (rank : 87) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8C2Q3, Q3TJB6, Q91Z21, Q9DBI6 | Gene names | Rbm14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 14 (RNA-binding motif protein 14). | |||||
|
SNRPA_HUMAN
|
||||||
| NC score | 0.122640 (rank : 46) | θ value | 2.36792 (rank : 59) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P09012 | Gene names | SNRPA | |||
|
Domain Architecture |

|
|||||
| Description | U1 small nuclear ribonucleoprotein A (U1 snRNP protein A) (U1A protein) (U1-A). | |||||
|
RBMX2_HUMAN
|
||||||
| NC score | 0.118885 (rank : 47) | θ value | 1.81305 (rank : 51) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9Y388, Q5JY82, Q9Y3I8 | Gene names | RBMX2 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding motif protein, X-linked 2. | |||||
|
RBM14_HUMAN
|
||||||
| NC score | 0.118765 (rank : 48) | θ value | 8.99809 (rank : 86) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q96PK6, O75932, Q53GV1, Q68DQ9, Q96PK5 | Gene names | RBM14, SIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 14 (RNA-binding motif protein 14) (RRM-containing coactivator activator/modulator) (Synaptotagmin-interacting protein) (SYT-interacting protein). | |||||
|
HTSF1_HUMAN
|
||||||
| NC score | 0.117097 (rank : 49) | θ value | 0.62314 (rank : 39) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O43719, Q59G06, Q99730 | Gene names | HTATSF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 (Tat-SF1). | |||||
|
SFRS7_HUMAN
|
||||||
| NC score | 0.114002 (rank : 50) | θ value | 0.813845 (rank : 44) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q16629 | Gene names | SFRS7 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 7 (Splicing factor 9G8). | |||||
|
RBY1A_HUMAN
|
||||||
| NC score | 0.106625 (rank : 51) | θ value | 1.81305 (rank : 52) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q15414, Q15377, Q8NHR0 | Gene names | RBMY1A1, RBM1, YRRM1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding motif protein, Y chromosome, family 1 member A1 (RNA- binding motif protein 1). | |||||
|
TRA2B_HUMAN
|
||||||
| NC score | 0.105227 (rank : 52) | θ value | 5.27518 (rank : 72) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P62995, O15449, Q15815, Q64283 | Gene names | SFRS10, TRA2B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine/serine-rich-splicing factor 10 (Transformer-2-beta) (HTRA2- beta) (Transformer 2 protein homolog). | |||||
|
TRA2B_MOUSE
|
||||||
| NC score | 0.105227 (rank : 53) | θ value | 5.27518 (rank : 73) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P62996, O15449, Q15815, Q64283 | Gene names | Sfrs10, Silg41, Tra2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine/serine-rich-splicing factor 10 (Transformer-2-beta) (HTRA2- beta) (Transformer 2 protein homolog) (Silica-induced gene 41 protein) (SIG-41). | |||||
|
SFPQ_HUMAN
|
||||||
| NC score | 0.104379 (rank : 54) | θ value | 4.03905 (rank : 66) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
SFPQ_MOUSE
|
||||||
| NC score | 0.104193 (rank : 55) | θ value | 4.03905 (rank : 67) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8VIJ6, Q9ERW2 | Gene names | Sfpq | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit). | |||||
|
RBM23_HUMAN
|
||||||
| NC score | 0.102668 (rank : 56) | θ value | 5.27518 (rank : 69) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q86U06, Q8ND16, Q8TB88, Q8WY40, Q9BUJ1, Q9NVV7 | Gene names | RBM23, RNPC4 | |||
|
Domain Architecture |

|
|||||
| Description | Probable RNA-binding protein 23 (RNA-binding motif protein 23) (RNA- binding region-containing protein 4) (Splicing factor SF2). | |||||
|
SFRS7_MOUSE
|
||||||
| NC score | 0.102231 (rank : 57) | θ value | 6.88961 (rank : 77) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8BL97, Q8BMC6, Q8BUR2, Q8R2N4, Q8R3E9, Q91YS1 | Gene names | Sfrs7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 7. | |||||
|
SRR35_HUMAN
|
||||||
| NC score | 0.101089 (rank : 58) | θ value | θ > 10 (rank : 186) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q8WXF0, Q8WW25 | Gene names | SRRP35 | |||
|
Domain Architecture |

|
|||||
| Description | 35 kDa SR repressor protein (SRrp35). | |||||
|
RU17_MOUSE
|
||||||
| NC score | 0.101078 (rank : 59) | θ value | 0.0736092 (rank : 33) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q62376, Q3UIW4 | Gene names | Snrp70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U1 small nuclear ribonucleoprotein 70 kDa (U1 SNRNP 70 kDa) (snRNP70). | |||||
|
SFRS3_HUMAN
|
||||||
| NC score | 0.100766 (rank : 60) | θ value | 3.0926 (rank : 63) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P84103, O08831, P23152 | Gene names | SFRS3, SRP20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 3 (Pre-mRNA-splicing factor SRP20). | |||||
|
SFRS3_MOUSE
|
||||||
| NC score | 0.100766 (rank : 61) | θ value | 3.0926 (rank : 64) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P84104, O08831, P23152, Q564E9 | Gene names | Sfrs3, Srp20, X16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 3 (Pre-mRNA-splicing factor SRP20) (X16 protein). | |||||
|
RU17_HUMAN
|
||||||
| NC score | 0.100461 (rank : 62) | θ value | 0.0736092 (rank : 32) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P08621, P78493, P78494, Q15364, Q15686, Q15687, Q15689, Q99377, Q9UE45, Q9UE46, Q9UE47, Q9UE48, Q9UFQ6 | Gene names | SNRP70, RPU1, U1AP1 | |||
|
Domain Architecture |

|
|||||
| Description | U1 small nuclear ribonucleoprotein 70 kDa (U1 snRNP 70 kDa) (snRNP70) (U1-70K). | |||||
|
RBMS1_MOUSE
|
||||||
| NC score | 0.099788 (rank : 63) | θ value | 6.88961 (rank : 76) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q91W59, Q6PEU6, Q6PHC2, Q9WTK4 | Gene names | Rbms1, Mssp, Mssp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding motif, single-stranded-interacting protein 1 (Single- stranded DNA-binding protein MSSP-1). | |||||
|
PABP5_HUMAN
|
||||||
| NC score | 0.099759 (rank : 64) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q96DU9, Q6P529, Q9UFE5 | Gene names | PABPC5, PABP5 | |||
|
Domain Architecture |

|
|||||
| Description | Polyadenylate-binding protein 5 (Poly(A)-binding protein 5) (PABP 5). | |||||
|
SAFB1_HUMAN
|
||||||
| NC score | 0.098024 (rank : 65) | θ value | 0.62314 (rank : 40) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 493 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q15424, O60406, Q59HH8 | Gene names | SAFB, HAP, HET, SAFB1 | |||
|
Domain Architecture |

|
|||||
| Description | Scaffold attachment factor B (Scaffold attachment factor B1) (SAF-B) (HSP27 estrogen response element-TATA box-binding protein) (HSP27 ERE- TATA-binding protein). | |||||
|
TRA2A_MOUSE
|
||||||
| NC score | 0.097984 (rank : 66) | θ value | 1.38821 (rank : 49) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q6PFR5 | Gene names | Tra2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transformer-2 protein homolog (TRA-2 alpha). | |||||
|
TRA2A_HUMAN
|
||||||
| NC score | 0.097771 (rank : 67) | θ value | 1.38821 (rank : 48) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q13595 | Gene names | TRA2A | |||
|
Domain Architecture |

|
|||||
| Description | Transformer-2 protein homolog (TRA-2 alpha). | |||||
|
HTSF1_MOUSE
|
||||||
| NC score | 0.097694 (rank : 68) | θ value | 2.36792 (rank : 54) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8BGC0, Q1WWK0, Q9CT41, Q9DAU3 | Gene names | Htatsf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 homolog. | |||||
|
RBMS1_HUMAN
|
||||||
| NC score | 0.095666 (rank : 69) | θ value | 6.88961 (rank : 75) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P29558, Q15433, Q8WV20 | Gene names | RBMS1, MSSP, MSSP1, SCR2 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding motif, single-stranded-interacting protein 1 (Single- stranded DNA-binding protein MSSP-1) (Suppressor of CDC2 with RNA- binding motif 2). | |||||
|
FUSIP_HUMAN
|
||||||
| NC score | 0.095508 (rank : 70) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | O75494, O60572, Q96G09, Q96P17 | Gene names | FUSIP1, TASR | |||
|
Domain Architecture |

|
|||||
| Description | FUS-interacting serine-arginine-rich protein 1 (TLS-associated protein with Ser-Arg repeats) (TLS-associated protein with SR repeats) (TASR) (TLS-associated serine-arginine protein) (TLS-associated SR protein) (40 kDa SR-repressor protein) (SRrp40) (Splicing factor SRp38). | |||||
|
FUSIP_MOUSE
|
||||||
| NC score | 0.095508 (rank : 71) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9R0U0, O70307, O88468, Q3TGW7, Q8CFZ1, Q9R0T9 | Gene names | Fusip1, Nssr | |||
|
Domain Architecture |

|
|||||
| Description | FUS-interacting serine-arginine-rich protein 1 (TLS-associated protein with Ser-Arg repeats) (TLS-associated protein with SR repeats) (TASR) (TLS-associated serine-arginine protein) (TLS-associated SR protein) (Neural-specific SR protein) (Neural-salient serine/arginine-rich protein). | |||||
|
SF3B4_HUMAN
|
||||||
| NC score | 0.094343 (rank : 72) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q15427 | Gene names | SF3B4, SAP49 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 4 (Spliceosome-associated protein 49) (SAP 49) (SF3b50) (Pre-mRNA-splicing factor SF3b 49 kDa subunit). | |||||
|
RBM11_HUMAN
|
||||||
| NC score | 0.094098 (rank : 73) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P57052 | Gene names | RBM11 | |||
|
Domain Architecture |

|
|||||
| Description | Putative RNA-binding protein 11 (RNA-binding motif protein 11). | |||||
|
TIA1_HUMAN
|
||||||
| NC score | 0.093460 (rank : 74) | θ value | θ > 10 (rank : 191) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P31483 | Gene names | TIA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolysin TIA-1 isoform p40 (RNA-binding protein TIA-1) (p40-TIA-1) [Contains: Nucleolysin TIA-1 isoform p15 (p15-TIA-1)]. | |||||
|
RBM28_MOUSE
|
||||||
| NC score | 0.093049 (rank : 75) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q8CGC6, Q8BG25, Q8VEJ8, Q9CS22, Q9CSE6 | Gene names | Rbm28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 28 (RNA-binding motif protein 28). | |||||
|
PABP1_MOUSE
|
||||||
| NC score | 0.092624 (rank : 76) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P29341 | Gene names | Pabpc1, Pabp1 | |||
|
Domain Architecture |

|
|||||
| Description | Polyadenylate-binding protein 1 (Poly(A)-binding protein 1) (PABP 1). | |||||
|
PABP1_HUMAN
|
||||||
| NC score | 0.091866 (rank : 77) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P11940, Q15097, Q93004 | Gene names | PABPC1, PAB1, PABP1, PABPC2 | |||
|
Domain Architecture |

|
|||||
| Description | Polyadenylate-binding protein 1 (Poly(A)-binding protein 1) (PABP 1). | |||||
|
TIA1_MOUSE
|
||||||
| NC score | 0.091537 (rank : 78) | θ value | θ > 10 (rank : 192) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P52912 | Gene names | Tia1, Tia | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolysin TIA-1 (RNA-binding protein TIA-1). | |||||
|
HNRPG_HUMAN
|
||||||
| NC score | 0.090935 (rank : 79) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P38159, Q5JQ67, Q969R3 | Gene names | RBMX, HNRPG, RBMXP1 | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein G (hnRNP G) (RNA-binding motif protein, X chromosome) (Glycoprotein p43). | |||||
|
HNRPQ_HUMAN
|
||||||
| NC score | 0.090817 (rank : 80) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O60506, Q53H05, Q5TCG2, Q8IW78, Q8N599, Q96LC1, Q96LC2, Q9Y583 | Gene names | SYNCRIP, HNRPQ, NSAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein Q (hnRNP Q) (hnRNP-Q) (Synaptotagmin-binding, cytoplasmic RNA-interacting protein) (Glycine- and tyrosine-rich RNA-binding protein) (GRY-RBP) (NS1-associated protein 1). | |||||
|
ACF_MOUSE
|
||||||
| NC score | 0.090771 (rank : 81) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q5YD48, Q5FW55, Q5YD47 | Gene names | Acf, Asp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | APOBEC1 complementation factor (APOBEC1-stimulating protein). | |||||
|
NUCL_MOUSE
|
||||||
| NC score | 0.090354 (rank : 82) | θ value | 0.813845 (rank : 42) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P09405, Q548M9, Q61991, Q99K50 | Gene names | Ncl, Nuc | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolin (Protein C23). | |||||
|
CUGB1_HUMAN
|
||||||
| NC score | 0.090058 (rank : 83) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q92879, Q9NP83, Q9NR06 | Gene names | CUGBP1, BRUNOL2, CUGBP, NAB50 | |||
|
Domain Architecture |

|
|||||
| Description | CUG triplet repeat RNA-binding protein 1 (CUG-BP1) (RNA-binding protein BRUNOL-2) (Deadenylation factor CUG-BP) (50 kDa Nuclear polyadenylated RNA-binding protein) (EDEN-BP). | |||||
|
CUGB1_MOUSE
|
||||||
| NC score | 0.089912 (rank : 84) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P28659, Q9CXE5, Q9EPJ8, Q9JI37 | Gene names | Cugbp1, Brunol2, Cugbp | |||
|
Domain Architecture |

|
|||||
| Description | CUG triplet repeat RNA-binding protein 1 (CUG-BP1) (RNA-binding protein BRUNOL-2) (Deadenylation factor CUG-BP) (Deadenylation factor EDEN-BP) (Brain protein F41). | |||||
|
PABP3_HUMAN
|
||||||
| NC score | 0.088687 (rank : 85) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9H361, Q8NHV0, Q9H086 | Gene names | PABPC3, PABP3, PABPL3 | |||
|
Domain Architecture |

|
|||||
| Description | Polyadenylate-binding protein 3 (Poly(A)-binding protein 3) (PABP 3) (Testis-specific poly(A)-binding protein). | |||||
|
HNRPQ_MOUSE
|
||||||
| NC score | 0.087833 (rank : 86) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q7TMK9, O88991, Q80YV6, Q8BGP1, Q8C5K6, Q8CGC2, Q91ZR0, Q99KU9, Q9QYF4 | Gene names | Syncrip, Hnrpq, Nsap1, Nsap1l | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein Q (hnRNP Q) (hnRNP-Q) (Synaptotagmin-binding, cytoplasmic RNA-interacting protein) (Glycine- and tyrosine-rich RNA-binding protein) (GRY-RBP) (NS1-associated protein 1) (pp68). | |||||
|
HNRPG_MOUSE
|
||||||
| NC score | 0.087388 (rank : 87) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | O35479 | Gene names | Rbmx, Hnrnpg, Hnrpg, Rbmxp1, Rbmxrt | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein G (hnRNP G) (RNA-binding motif protein, X chromosome). | |||||
|
PABP4_HUMAN
|
||||||
| NC score | 0.087272 (rank : 88) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q13310, Q6P0N3 | Gene names | PABPC4, APP1, PABP4 | |||
|
Domain Architecture |

|
|||||
| Description | Polyadenylate-binding protein 4 (Poly(A)-binding protein 4) (PABP 4) (Inducible poly(A)-binding protein) (iPABP) (Activated-platelet protein 1) (APP-1). | |||||
|
MK67I_HUMAN
|
||||||
| NC score | 0.086518 (rank : 89) | θ value | 2.36792 (rank : 55) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9BYG3, Q8TB66, Q96ED4 | Gene names | MKI67IP, NIFK, NOPP34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MKI67 FHA domain-interacting nucleolar phosphoprotein (Nucleolar protein interacting with the FHA domain of pKI-67) (hNIFK) (Nucleolar phosphoprotein Nopp34). | |||||
|
TIAR_HUMAN
|
||||||
| NC score | 0.086277 (rank : 90) | θ value | θ > 10 (rank : 193) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q01085 | Gene names | TIAL1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolysin TIAR (TIA-1-related protein). | |||||
|
RBM28_HUMAN
|
||||||
| NC score | 0.085762 (rank : 91) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9NW13, Q96CV3 | Gene names | RBM28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 28 (RNA-binding motif protein 28). | |||||
|
ACF_HUMAN
|
||||||
| NC score | 0.085586 (rank : 92) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9NQ94, Q5SZQ0, Q9NQ93, Q9NQX8, Q9NQX9, Q9NXC9, Q9NZD3 | Gene names | ACF, ASP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | APOBEC1 complementation factor (APOBEC1-stimulating protein). | |||||
|
TIAR_MOUSE
|
||||||
| NC score | 0.085450 (rank : 93) | θ value | θ > 10 (rank : 194) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P70318 | Gene names | Tial1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolysin TIAR (TIA-1-related protein). | |||||
|
RALY_MOUSE
|
||||||
| NC score | 0.085030 (rank : 94) | θ value | 1.81305 (rank : 50) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q64012, Q99K76, Q9CXH8, Q9QZX6 | Gene names | Raly, Merc | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein Raly (hnRNP associated with lethal yellow protein) (Maternally expressed hnRNP C-related protein). | |||||
|
RBM8A_HUMAN
|
||||||
| NC score | 0.083350 (rank : 95) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9Y5S9, Q6FHD1, Q9GZX8, Q9NZI4 | Gene names | RBM8A, RBM8 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 8A (RNA-binding motif protein 8A) (Ribonucleoprotein RBM8A) (RNA-binding protein Y14) (Binder of OVCA1- 1) (BOV-1). | |||||
|
RBM8A_MOUSE
|
||||||
| NC score | 0.083350 (rank : 96) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9CWZ3, Q9D6U5 | Gene names | Rbm8a, Rbm8 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 8A (RNA-binding motif protein 8A) (Ribonucleoprotein RBM8A). | |||||
|
CIRBP_HUMAN
|
||||||
| NC score | 0.082876 (rank : 97) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q14011 | Gene names | CIRBP, A18HNRNP, CIRP | |||
|
Domain Architecture |

|
|||||
| Description | Cold-inducible RNA-binding protein (Glycine-rich RNA-binding protein CIRP) (A18 hnRNP). | |||||
|
CIRBP_MOUSE
|
||||||
| NC score | 0.082660 (rank : 98) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P60824, O09069, O09148, Q3V1A6, Q61413 | Gene names | Cirbp, Cirp | |||
|
Domain Architecture |

|
|||||
| Description | Cold-inducible RNA-binding protein (Glycine-rich RNA-binding protein CIRP) (A18 hnRNP). | |||||
|
RBMS2_HUMAN
|
||||||
| NC score | 0.082543 (rank : 99) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q15434 | Gene names | RBMS2, SCR3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding motif, single-stranded-interacting protein 2 (Suppressor of CDC2 with RNA-binding motif 3). | |||||
|
RNPC2_HUMAN
|
||||||
| NC score | 0.082219 (rank : 100) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q14498, Q14499 | Gene names | RNPC2, HCC1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding region-containing protein 2 (Hepatocellular carcinoma protein 1) (Splicing factor HCC1). | |||||
|
RNPC2_MOUSE
|
||||||
| NC score | 0.082210 (rank : 101) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q8VH51, Q99KV0 | Gene names | Rnpc2, Caper | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding region-containing protein 2 (Coactivator of activating protein 1 and estrogen receptors) (Coactivator of AP-1 and ERs) (Transcription coactivator CAPER). | |||||
|
RBMS2_MOUSE
|
||||||
| NC score | 0.082007 (rank : 102) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8VC70 | Gene names | Rbms2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding motif, single-stranded-interacting protein 2. | |||||
|
HNRPR_HUMAN
|
||||||
| NC score | 0.082001 (rank : 103) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O43390 | Gene names | HNRPR | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein R (hnRNP R). | |||||
|
NUCL_HUMAN
|
||||||
| NC score | 0.079818 (rank : 104) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P19338, Q53SK1, Q8NB06 | Gene names | NCL | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolin (Protein C23). | |||||
|
RBM3_MOUSE
|
||||||
| NC score | 0.079812 (rank : 105) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O89086 | Gene names | Rbm3 | |||
|
Domain Architecture |

|
|||||
| Description | Putative RNA-binding protein 3 (RNA-binding motif protein 3). | |||||
|
RALY_HUMAN
|
||||||
| NC score | 0.078760 (rank : 106) | θ value | 3.0926 (rank : 62) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9UKM9, Q14621, Q5QPL8, Q9BQX6, Q9UJE3 | Gene names | RALY, P542 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein Raly (hnRNP associated with lethal yellow homolog) (Autoantigen p542). | |||||
|
RBM3_HUMAN
|
||||||
| NC score | 0.078639 (rank : 107) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P98179 | Gene names | RBM3, RNPL | |||
|
Domain Architecture |

|
|||||
| Description | Putative RNA-binding protein 3 (RNA-binding motif protein 3) (RNPL). | |||||
|
RBM9_HUMAN
|
||||||
| NC score | 0.078165 (rank : 108) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | O43251, Q5TF71, Q6IC09, Q8TD00, Q8WYB1, Q96DZ6, Q96NL7, Q9UGW4, Q9UH33 | Gene names | RBM9, HRNBP2, RTA | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 9 (RNA-binding motif protein 9) (Hexaribonucleotide-binding protein 2) (Repressor of tamoxifen transcriptional activity). | |||||
|
HNRPC_MOUSE
|
||||||
| NC score | 0.077800 (rank : 109) | θ value | 1.38821 (rank : 46) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9Z204, Q3TLB5, Q501Q3, Q8C2G5, Q99KE2, Q9CQT3, Q9CY83 | Gene names | Hnrpc, Hnrnpc | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoproteins C1/C2 (hnRNP C1 / hnRNP C2). | |||||
|
CSTF2_MOUSE
|
||||||
| NC score | 0.074134 (rank : 110) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8BIQ5, Q8K1Y6, Q9ERC2 | Gene names | Cstf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage stimulation factor 64 kDa subunit (CSTF 64 kDa subunit) (CF-1 64 kDa subunit) (CstF-64). | |||||
|
RAVR2_HUMAN
|
||||||
| NC score | 0.073888 (rank : 111) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9HCJ3, Q6P141, Q9NPV7 | Gene names | RAVER2, KIAA1579 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribonucleoprotein PTB-binding 2 (Protein raver-2). | |||||
|
SFRS9_HUMAN
|
||||||
| NC score | 0.073341 (rank : 112) | θ value | θ > 10 (rank : 184) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q13242 | Gene names | SFRS9, SRP30C | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 9 (Pre-mRNA-splicing factor SRp30C). | |||||
|
CSTF2_HUMAN
|
||||||
| NC score | 0.073272 (rank : 113) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P33240, Q5H951, Q6LA74, Q8N502 | Gene names | CSTF2 | |||
|
Domain Architecture |

|
|||||
| Description | Cleavage stimulation factor 64 kDa subunit (CSTF 64 kDa subunit) (CF-1 64 kDa subunit) (CstF-64). | |||||
|
RAVR1_HUMAN
|
||||||
| NC score | 0.072756 (rank : 114) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8IY67, Q8IY60, Q8TF24 | Gene names | RAVER1, KIAA1978 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribonucleoprotein PTB-binding 1 (Protein raver-1). | |||||
|
CSTFT_HUMAN
|
||||||
| NC score | 0.072422 (rank : 115) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9H0L4, O75174, Q53HK6, Q7LGE8, Q8N6T1 | Gene names | CSTF2T, KIAA0689 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage stimulation factor 64 kDa subunit, tau variant (CSTF 64 kDa subunit, tau variant) (CF-1 64 kDa subunit, tau variant) (TauCstF-64). | |||||
|
IF34_MOUSE
|
||||||
| NC score | 0.072030 (rank : 116) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9Z1D1, Q9R079 | Gene names | Eif3s4, Eif3p42 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 4 (eIF-3 delta) (eIF3 p44) (eIF-3 RNA-binding subunit) (eIF3 p42) (Eif3p42) (eIF3g). | |||||
|
CSTFT_MOUSE
|
||||||
| NC score | 0.071802 (rank : 117) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8C7E9, Q6A016, Q8BHH7, Q8C3W7, Q8R2Y1, Q9EPU3 | Gene names | Cstf2t, Kiaa0689 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage stimulation factor 64 kDa subunit, tau variant (CSTF 64 kDa subunit, tau variant) (CF-1 64 kDa subunit, tau variant) (TauCstF-64). | |||||
|
IF34_HUMAN
|
||||||
| NC score | 0.071755 (rank : 118) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O75821, O14801, Q969U5 | Gene names | EIF3S4 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 4 (eIF-3 delta) (eIF3 p44) (eIF-3 RNA-binding subunit) (eIF3 p42) (eIF3g). | |||||
|
A2BP1_HUMAN
|
||||||
| NC score | 0.071601 (rank : 119) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9NWB1, Q9NS20 | Gene names | A2BP1, A2BP | |||
|
Domain Architecture |

|
|||||
| Description | Ataxin-2-binding protein 1. | |||||
|
NCBP2_HUMAN
|
||||||
| NC score | 0.071389 (rank : 120) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P52298, Q14924, Q2TS50 | Gene names | NCBP2, CBP20, PIG55 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear cap-binding protein subunit 2 (20 kDa nuclear cap-binding protein) (NCBP 20 kDa subunit) (CBP20) (NCBP-interacting protein 1) (NIP1) (Cell proliferation-inducing gene 55 protein). | |||||
|
RAVR1_MOUSE
|
||||||
| NC score | 0.071179 (rank : 121) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9CW46, Q5DTT6, Q811K0, Q8C3Z1, Q8CA14 | Gene names | Raver1, Kiaa1978 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribonucleoprotein PTB-binding 1 (Protein raver-1). | |||||
|
NCBP2_MOUSE
|
||||||
| NC score | 0.071123 (rank : 122) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9CQ49, Q3UI86 | Gene names | Ncbp2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear cap-binding protein subunit 2 (20 kDa nuclear cap-binding protein) (NCBP 20 kDa subunit) (CBP20). | |||||
|
A2BP1_MOUSE
|
||||||
| NC score | 0.071089 (rank : 123) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9JJ43, Q9JJB5 | Gene names | A2bp1, A2bp | |||
|
Domain Architecture |

|
|||||
| Description | Ataxin-2-binding protein 1. | |||||
|
DAZP1_MOUSE
|
||||||
| NC score | 0.071053 (rank : 124) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9JII5 | Gene names | Dazap1 | |||
|
Domain Architecture |

|
|||||
| Description | DAZ-associated protein 1 (Deleted in azoospermia-associated protein 1). | |||||
|
SFRS9_MOUSE
|
||||||
| NC score | 0.070719 (rank : 125) | θ value | θ > 10 (rank : 185) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9D0B0, Q9CRN3 | Gene names | Sfrs9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 9. | |||||
|
DAZP1_HUMAN
|
||||||
| NC score | 0.069569 (rank : 126) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q96EP5, Q96MJ3, Q9NRR9 | Gene names | DAZAP1 | |||
|
Domain Architecture |

|
|||||
| Description | DAZ-associated protein 1 (Deleted in azoospermia-associated protein 1). | |||||
|
U2AF2_MOUSE
|
||||||
| NC score | 0.068524 (rank : 127) | θ value | θ > 10 (rank : 196) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P26369 | Gene names | U2af2, U2af65 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor U2AF 65 kDa subunit (U2 auxiliary factor 65 kDa subunit) (U2 snRNP auxiliary factor large subunit). | |||||
|
RNPC1_HUMAN
|
||||||
| NC score | 0.068260 (rank : 128) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9H0Z9, Q15350, Q15351, Q9BYK3, Q9BYK4 | Gene names | RNPC1, SEB4 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding region-containing protein 1 (HSRNASEB) (ssDNA-binding protein SEB4) (CLL-associated antigen KW-5). | |||||
|
U2AF2_HUMAN
|
||||||
| NC score | 0.068260 (rank : 129) | θ value | θ > 10 (rank : 195) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P26368 | Gene names | U2AF2, U2AF65 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor U2AF 65 kDa subunit (U2 auxiliary factor 65 kDa subunit) (U2 snRNP auxiliary factor large subunit) (hU2AF(65)). | |||||
|
HNRPC_HUMAN
|
||||||
| NC score | 0.067700 (rank : 130) | θ value | 8.99809 (rank : 82) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P07910, P22628, Q53EX2, Q59FD3, Q5FWE8, Q86SF8, Q86U45, Q96HK7, Q96HM4, Q96IY5, Q9BTS3 | Gene names | HNRPC | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoproteins C1/C2 (hnRNP C1 / hnRNP C2). | |||||
|
RNPS1_HUMAN
|
||||||
| NC score | 0.067378 (rank : 131) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q15287, O75308, Q8WY42, Q9NYG3 | Gene names | RNPS1, LDC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein with serine-rich domain 1 (SR-related protein LDC2). | |||||
|
RNPS1_MOUSE
|
||||||
| NC score | 0.067378 (rank : 132) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q99M28, Q3TMJ1, Q922H8 | Gene names | Rnps1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein with serine-rich domain 1. | |||||
|
SFRS1_HUMAN
|
||||||
| NC score | 0.067075 (rank : 133) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q07955, Q13809 | Gene names | SFRS1, ASF, SF2, SF2P33 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 1 (pre-mRNA-splicing factor SF2, P33 subunit) (Alternative-splicing factor 1) (ASF-1). | |||||
|
SFRS1_MOUSE
|
||||||
| NC score | 0.067075 (rank : 134) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q6PDM2, Q3UCH2, Q5SXC5, Q8BJV3, Q8C1H9 | Gene names | Sfrs1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 1. | |||||
|
SFRS6_HUMAN
|
||||||
| NC score | 0.066278 (rank : 135) | θ value | θ > 10 (rank : 183) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q13247, Q13244, Q13245, Q96J06, Q9UJB8, Q9Y3N7 | Gene names | SFRS6, SRP55 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 6 (Pre-mRNA-splicing factor SRP55). | |||||
|
RAVR2_MOUSE
|
||||||
| NC score | 0.066222 (rank : 136) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q7TPD6 | Gene names | Raver2, Kiaa1579 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribonucleoprotein PTB-binding 2 (Protein raver-2). | |||||
|
PPIE_HUMAN
|
||||||
| NC score | 0.066015 (rank : 137) | θ value | 1.38821 (rank : 47) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9UNP9, O43634, O43635, Q5TGA0, Q9UIZ5 | Gene names | PPIE, CYP33 | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase E (EC 5.2.1.8) (PPIase E) (Rotamase E) (Cyclophilin E) (Cyclophilin 33). | |||||
|
RBM34_HUMAN
|
||||||
| NC score | 0.065757 (rank : 138) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P42696, Q8N2Z8, Q9H5A1 | Gene names | RBM34, KIAA0117 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 34 (RNA-binding motif protein 34). | |||||
|
TADBP_HUMAN
|
||||||
| NC score | 0.065667 (rank : 139) | θ value | θ > 10 (rank : 187) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q13148, Q96DJ0 | Gene names | TARDBP, TDP43 | |||
|
Domain Architecture |

|
|||||
| Description | TAR DNA-binding protein 43 (TDP-43). | |||||
|
RNPC1_MOUSE
|
||||||
| NC score | 0.065198 (rank : 140) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q62176, Q922Z2 | Gene names | Rnpc1, Seb4, Seb4l | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding region-containing protein 1 (ssDNA-binding protein SEB4). | |||||
|
RBM34_MOUSE
|
||||||
| NC score | 0.064269 (rank : 141) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8C5L7 | Gene names | Rbm34, D8Ertd233e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 34 (RNA-binding motif protein 34). | |||||
|
PPIE_MOUSE
|
||||||
| NC score | 0.064187 (rank : 142) | θ value | 2.36792 (rank : 58) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9QZH3, Q9D8C0 | Gene names | Ppie, Cyp33 | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase E (EC 5.2.1.8) (PPIase E) (Rotamase E) (Cyclophilin E) (Cyclophilin 33). | |||||
|
HNRPD_HUMAN
|
||||||
| NC score | 0.063823 (rank : 143) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q14103, Q01858, Q14100, Q14101, Q14102 | Gene names | HNRPD, AUF1 | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein D0 (hnRNP D0) (AU-rich element RNA-binding protein 1). | |||||
|
MSI1H_HUMAN
|
||||||
| NC score | 0.063701 (rank : 144) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O43347, Q96PU0, Q96PU1, Q96PU2, Q96PU3 | Gene names | MSI1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein Musashi homolog 1 (Musashi-1). | |||||
|
HNRPD_MOUSE
|
||||||
| NC score | 0.063537 (rank : 145) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q60668, Q60667, Q80ZJ0, Q91X94 | Gene names | Hnrpd, Auf1 | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein D0 (hnRNP D0) (AU-rich element RNA-binding protein 1). | |||||
|
THOC4_MOUSE
|
||||||
| NC score | 0.063321 (rank : 146) | θ value | θ > 10 (rank : 190) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O08583, Q8CBM4, Q9JJW7 | Gene names | Thoc4, Aly, Ref1, Refbp1 | |||
|
Domain Architecture |

|
|||||
| Description | THO complex subunit 4 (Tho4) (RNA and export factor-binding protein 1) (REF1-I) (Ally of AML-1 and LEF-1) (Aly/REF). | |||||
|
SFRS5_MOUSE
|
||||||
| NC score | 0.063317 (rank : 147) | θ value | θ > 10 (rank : 182) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O35326 | Gene names | Sfrs5, Hrs | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 5 (Pre-mRNA-splicing factor SRP40) (Delayed-early protein HRS). | |||||
|
MSI2H_MOUSE
|
||||||
| NC score | 0.062802 (rank : 148) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q920Q6, Q8BQ90, Q920Q7 | Gene names | Msi2, Msi2h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein Musashi homolog 2 (Musashi-2). | |||||
|
MSI1H_MOUSE
|
||||||
| NC score | 0.062740 (rank : 149) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q61474, Q8BNC7 | Gene names | Msi1, Msi1h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein Musashi homolog 1 (Musashi-1). | |||||
|
RB15B_HUMAN
|
||||||
| NC score | 0.062298 (rank : 150) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8NDT2, Q6QE19, Q9BV96 | Gene names | RBM15B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative RNA-binding protein 15B (RNA-binding motif protein 15B). | |||||
|
MSI2H_HUMAN
|
||||||
| NC score | 0.062259 (rank : 151) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q96DH6, Q7Z6M7, Q8N9T4 | Gene names | MSI2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein Musashi homolog 2 (Musashi-2). | |||||
|
SART3_MOUSE
|
||||||
| NC score | 0.062049 (rank : 152) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9JLI8, Q6ZQI2, Q8BPK9, Q8C3B7, Q8CFU9 | Gene names | Sart3, Kiaa0156 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Squamous cell carcinoma antigen recognized by T-cells 3 (SART-3) (mSART-3) (Tumor-rejection antigen SART3). | |||||
|
TADBP_MOUSE
|
||||||
| NC score | 0.061980 (rank : 153) | θ value | θ > 10 (rank : 188) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q921F2 | Gene names | Tardbp, Tdp43 | |||
|
Domain Architecture |

|
|||||
| Description | TAR DNA-binding protein 43 (TDP-43). | |||||
|
IF2B2_HUMAN
|
||||||
| NC score | 0.061958 (rank : 154) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Y6M1 | Gene names | IGF2BP2, IMP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulin-like growth factor 2 mRNA-binding protein 2 (IGF-II mRNA- binding protein 2) (IMP-2) (Hepatocellular carcinoma autoantigen p62). | |||||
|
SART3_HUMAN
|
||||||
| NC score | 0.061917 (rank : 155) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 301 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q15020, Q58F06, Q8IUS1, Q96J95 | Gene names | SART3, KIAA0156, TIP110 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Squamous cell carcinoma antigen recognized by T-cells 3 (SART-3) (hSART-3) (Tat-interacting protein of 110 kDa) (Tip110). | |||||
|
SFRS5_HUMAN
|
||||||
| NC score | 0.061519 (rank : 156) | θ value | θ > 10 (rank : 181) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q13243, O14797, Q16662 | Gene names | SFRS5, HRS, SRP40 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 5 (Pre-mRNA-splicing factor SRP40) (Delayed-early protein HRS). | |||||
|
RB15B_MOUSE
|
||||||
| NC score | 0.060982 (rank : 157) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q6PHZ5, Q8C6G2 | Gene names | Rbm15b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative RNA-binding protein 15B (RNA-binding motif protein 15B). | |||||
|
THOC4_HUMAN
|
||||||
| NC score | 0.060875 (rank : 158) | θ value | θ > 10 (rank : 189) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q86V81, O43672 | Gene names | THOC4, ALY, BEF | |||
|
Domain Architecture |

|
|||||
| Description | THO complex subunit 4 (Tho4) (Ally of AML-1 and LEF-1) (Transcriptional coactivator Aly/REF) (bZIP-enhancing factor BEF). | |||||
|
ROA2_HUMAN
|
||||||
| NC score | 0.060257 (rank : 159) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P22626, P22627 | Gene names | HNRPA2B1 | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoproteins A2/B1 (hnRNP A2 / hnRNP B1). | |||||
|
ROA2_MOUSE
|
||||||
| NC score | 0.060188 (rank : 160) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O88569 | Gene names | Hnrpa2b1 | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoproteins A2/B1 (hnRNP A2 / hnRNP B1). | |||||
|
ROA1_MOUSE
|
||||||
| NC score | 0.060182 (rank : 161) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P49312, P97312, Q3V269 | Gene names | Hnrpa1, Fli-2, Tis | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein A1 (Helix-destabilizing protein) (Single-strand-binding protein) (hnRNP core protein A1) (HDP- 1) (Topoisomerase-inhibitor suppressed). | |||||
|
ROA1_HUMAN
|
||||||
| NC score | 0.059965 (rank : 162) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P09651, Q6PJZ7 | Gene names | HNRPA1 | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein A1 (Helix-destabilizing protein) (Single-strand RNA-binding protein) (hnRNP core protein A1). | |||||
|
HNRPM_MOUSE
|
||||||
| NC score | 0.059523 (rank : 163) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9D0E1, Q6P1B2, Q99JQ0 | Gene names | Hnrpm | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heterogeneous nuclear ribonucleoprotein M (hnRNP M). | |||||
|
SFRS4_MOUSE
|
||||||
| NC score | 0.059384 (rank : 164) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8VE97, Q9JJC3 | Gene names | Sfrs4 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4. | |||||
|
ROAA_MOUSE
|
||||||
| NC score | 0.059206 (rank : 165) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q99020, Q8BPE0 | Gene names | Hnrpab, Cbf-a, Cgbfa | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein A/B (hnRNP A/B) (CArG-binding factor-A) (CBF-A). | |||||
|
SFRS4_HUMAN
|
||||||
| NC score | 0.058233 (rank : 166) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q08170, Q5VXP1, Q9BUA4, Q9UEB5 | Gene names | SFRS4, SRP75 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4 (Pre-mRNA-splicing factor SRP75) (SRP001LB). | |||||
|
BOLL_MOUSE
|
||||||
| NC score | 0.058124 (rank : 167) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q924M5, Q9D4V2 | Gene names | Boll, Boule | |||
|
Domain Architecture |

|
|||||
| Description | Protein boule-like. | |||||
|
MYEF2_MOUSE
|
||||||
| NC score | 0.057325 (rank : 168) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8C854, Q60690, Q6P930, Q6ZPT3, Q8QZZ1 | Gene names | Myef2, Kiaa1341, Mef2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin expression factor 2 (MyEF-2) (MEF-2). | |||||
|
BOLL_HUMAN
|
||||||
| NC score | 0.057324 (rank : 169) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8N9W6, Q969U3 | Gene names | BOLL, BOULE | |||
|
Domain Architecture |

|
|||||
| Description | Protein boule-like. | |||||
|
PPIL4_MOUSE
|
||||||
| NC score | 0.056630 (rank : 170) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9CXG3, Q3TJX1, Q68FC7, Q9CT22 | Gene names | Ppil4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peptidyl-prolyl cis-trans isomerase-like 4 (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-like protein PPIL4). | |||||
|
ROAA_HUMAN
|
||||||
| NC score | 0.056551 (rank : 171) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q99729, Q04150, Q8N7U3, Q9BQ99 | Gene names | HNRPAB, ABBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein A/B (hnRNP A/B) (APOBEC-1- binding protein 1) (ABBP-1). | |||||
|
PPIL4_HUMAN
|
||||||
| NC score | 0.056527 (rank : 172) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8WUA2, Q7Z3Q5 | Gene names | PPIL4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peptidyl-prolyl cis-trans isomerase-like 4 (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-like protein PPIL4). | |||||
|
MINT_HUMAN
|
||||||
| NC score | 0.055656 (rank : 173) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
MINT_MOUSE
|
||||||
| NC score | 0.053973 (rank : 174) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
MYEF2_HUMAN
|
||||||
| NC score | 0.053585 (rank : 175) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9P2K5, Q6NUM5, Q7L388, Q7Z4B7, Q9H922, Q9NUQ1 | Gene names | MYEF2, KIAA1341 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin expression factor 2 (MyEF-2) (MST156). | |||||
|
ROA0_HUMAN
|
||||||
| NC score | 0.053354 (rank : 176) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q13151 | Gene names | HNRPA0 | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein A0 (hnRNP A0). | |||||
|
ROA3_HUMAN
|
||||||
| NC score | 0.053116 (rank : 177) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P51991, Q6URK5 | Gene names | HNRPA3 | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein A3 (hnRNP A3). | |||||
|
ROA3_MOUSE
|
||||||
| NC score | 0.053106 (rank : 178) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8BG05, Q8BHF8 | Gene names | Hnrpa3, Hnrnpa3 | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein A3 (hnRNP A3). | |||||
|
HNRPM_HUMAN
|
||||||
| NC score | 0.053094 (rank : 179) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P52272, Q15584, Q8WZ44, Q96H56, Q9BWL9, Q9Y492 | Gene names | HNRPM | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein M (hnRNP M). | |||||
|
IF2B2_MOUSE
|
||||||
| NC score | 0.052961 (rank : 180) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q5SF07, Q3TCU4, Q7TQF9 | Gene names | Igf2bp2, Imp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulin-like growth factor 2 mRNA-binding protein 2 (IGF-II mRNA- binding protein 2) (IMP-2). | |||||
|
REFP2_MOUSE
|
||||||
| NC score | 0.052958 (rank : 181) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9JJW6, Q8C869 | Gene names | Refbp2, Ref2 | |||
|
Domain Architecture |

|
|||||
| Description | RNA and export factor-binding protein 2. | |||||
|
RBM15_HUMAN
|
||||||
| NC score | 0.051820 (rank : 182) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 309 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q96T37, Q5D058, Q96PE4, Q96SC5, Q96SC6, Q96SC9, Q96SD0, Q96T38, Q9BRA5, Q9H6R8, Q9H9Y0 | Gene names | RBM15, OTT | |||
|
Domain Architecture |

|
|||||
| Description | Putative RNA-binding protein 15 (RNA-binding motif protein 15) (One- twenty two protein). | |||||
|
HNRCL_HUMAN
|
||||||
| NC score | 0.050583 (rank : 183) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O60812 | Gene names | HNRPCL1 | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein C-like 1 (hnRNP core protein C-like 1). | |||||
|
RBPMS_MOUSE
|
||||||
| NC score | 0.050432 (rank : 184) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9WVB0 | Gene names | Rbpms, Hermes | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein with multiple splicing (RBP-MS) (HEart, RRM Expressed Sequence) (Hermes). | |||||
|
IF39_MOUSE
|
||||||
| NC score | 0.043686 (rank : 185) | θ value | 8.99809 (rank : 83) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8JZQ9, Q922K2 | Gene names | Eif3s9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 9 (eIF-3 eta) (eIF3 p116). | |||||
|
SFR12_HUMAN
|
||||||
| NC score | 0.025740 (rank : 186) | θ value | 8.99809 (rank : 88) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8WXA9, Q86X37 | Gene names | SFRS12, SRRP86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86) (Splicing regulatory protein 508) (SRrp508). | |||||
|
SYTC_HUMAN
|
||||||
| NC score | 0.022281 (rank : 187) | θ value | 5.27518 (rank : 71) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P26639, Q96FP5, Q9BWA6 | Gene names | TARS | |||
|
Domain Architecture |

|
|||||
| Description | Threonyl-tRNA synthetase, cytoplasmic (EC 6.1.1.3) (Threonine--tRNA ligase) (ThrRS). | |||||
|
P3C2G_MOUSE
|
||||||
| NC score | 0.021797 (rank : 188) | θ value | 2.36792 (rank : 57) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O70167, O70168 | Gene names | Pik3c2g | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing gamma polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-gamma) (PtdIns-3-kinase C2 gamma) (PI3K-C2gamma). | |||||
|
SPT16_MOUSE
|
||||||
| NC score | 0.021600 (rank : 189) | θ value | 5.27518 (rank : 70) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q920B9, Q3TZ48, Q3UFH0, Q921H4 | Gene names | Supt16h, Fact140, Factp140 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FACT complex subunit SPT16 (Facilitates chromatin transcription complex subunit SPT16) (FACT 140 kDa subunit) (FACTp140) (Chromatin- specific transcription elongation factor 140 kDa subunit). | |||||
|
SYTC_MOUSE
|
||||||
| NC score | 0.020442 (rank : 190) | θ value | 6.88961 (rank : 79) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D0R2, Q3TL42, Q7TMQ2, Q8BMI6, Q9CX03 | Gene names | Tars | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Threonyl-tRNA synthetase, cytoplasmic (EC 6.1.1.3) (Threonine--tRNA ligase) (ThrRS). | |||||
|
SPT16_HUMAN
|
||||||
| NC score | 0.019830 (rank : 191) | θ value | 6.88961 (rank : 78) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y5B9, Q6GMT8, Q6P2F1, Q6PJM1, Q9NRX0 | Gene names | SUPT16H, FACT140, FACTP140 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FACT complex subunit SPT16 (Facilitates chromatin transcription complex subunit SPT16) (hSPT16) (FACT 140 kDa subunit) (FACTp140) (Chromatin-specific transcription elongation factor 140 kDa subunit). | |||||
|
CA103_HUMAN
|
||||||
| NC score | 0.019285 (rank : 192) | θ value | 5.27518 (rank : 68) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5T3J3, Q86XS4, Q8N3B6, Q96HT4, Q9NUM5, Q9NV32 | Gene names | C1orf103, RIF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf103 (Receptor-interacting factor 1). | |||||
|
EAN57_HUMAN
|
||||||
| NC score | 0.018688 (rank : 193) | θ value | 6.88961 (rank : 74) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43247, Q6ICF2, Q9Y4V8 | Gene names | EAN57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein EAN57. | |||||
|
PAR14_MOUSE
|
||||||
| NC score | 0.015035 (rank : 194) | θ value | 3.0926 (rank : 61) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q2EMV9, Q571C6, Q8BN35, Q8VDT6 | Gene names | Parp14, Kiaa1268 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly [ADP-ribose] polymerase 14 (EC 2.4.2.30) (PARP-14) (Collaborator of STAT6) (CoaSt6). | |||||
|
NEP_MOUSE
|
||||||
| NC score | 0.013005 (rank : 195) | θ value | 2.36792 (rank : 56) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61391, Q6NXX5, Q8K251 | Gene names | Mme | |||
|
Domain Architecture |

|
|||||
| Description | Neprilysin (EC 3.4.24.11) (Neutral endopeptidase) (NEP) (Enkephalinase) (Neutral endopeptidase 24.11) (Atriopeptidase) (CD10 antigen). | |||||
|
UBF1_HUMAN
|
||||||
| NC score | 0.011332 (rank : 196) | θ value | 8.99809 (rank : 89) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P17480 | Gene names | UBTF, UBF, UBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1) (Autoantigen NOR-90). | |||||