Please be patient as the page loads
|
PCY1A_HUMAN
|
||||||
| SwissProt Accessions | P49585 | Gene names | PCYT1A, CTPCT, PCYT1 | |||
|
Domain Architecture |
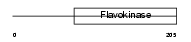
|
|||||
| Description | Choline-phosphate cytidylyltransferase A (EC 2.7.7.15) (Phosphorylcholine transferase A) (CTP:phosphocholine cytidylyltransferase A) (CT A) (CCT A) (CCT-alpha). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PCY1A_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P49585 | Gene names | PCYT1A, CTPCT, PCYT1 | |||
|
Domain Architecture |

|
|||||
| Description | Choline-phosphate cytidylyltransferase A (EC 2.7.7.15) (Phosphorylcholine transferase A) (CTP:phosphocholine cytidylyltransferase A) (CT A) (CCT A) (CCT-alpha). | |||||
|
PCY1A_MOUSE
|
||||||
| θ value | 4.38123e-180 (rank : 2) | NC score | 0.997115 (rank : 2) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P49586, Q542W4 | Gene names | Pcyt1a, Ctpct, Pcyt1 | |||
|
Domain Architecture |
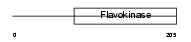
|
|||||
| Description | Choline-phosphate cytidylyltransferase A (EC 2.7.7.15) (Phosphorylcholine transferase A) (CTP:phosphocholine cytidylyltransferase A) (CT A) (CCT A) (CCT-alpha). | |||||
|
PCY1B_MOUSE
|
||||||
| θ value | 1.20984e-129 (rank : 3) | NC score | 0.985736 (rank : 4) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q811Q9, Q3UEW0, Q80Y63, Q811Q8, Q8BKD2, Q8C085 | Gene names | Pcyt1b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Choline-phosphate cytidylyltransferase B (EC 2.7.7.15) (Phosphorylcholine transferase B) (CTP:phosphocholine cytidylyltransferase B) (CT B) (CCT B) (CCT-beta). | |||||
|
PCY1B_HUMAN
|
||||||
| θ value | 1.5801e-129 (rank : 4) | NC score | 0.986578 (rank : 3) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y5K3, O60621, Q86XC9 | Gene names | PCYT1B, CCTB | |||
|
Domain Architecture |
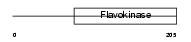
|
|||||
| Description | Choline-phosphate cytidylyltransferase B (EC 2.7.7.15) (Phosphorylcholine transferase B) (CTP:phosphocholine cytidylyltransferase B) (CT B) (CCT B) (CCT-beta). | |||||
|
PCY2_MOUSE
|
||||||
| θ value | 7.56453e-23 (rank : 5) | NC score | 0.723902 (rank : 6) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q922E4, Q99J50 | Gene names | Pcyt2 | |||
|
Domain Architecture |

|
|||||
| Description | Ethanolamine-phosphate cytidylyltransferase (EC 2.7.7.14) (Phosphorylethanolamine transferase) (CTP:phosphoethanolamine cytidylyltransferase). | |||||
|
PCY2_HUMAN
|
||||||
| θ value | 6.40375e-22 (rank : 6) | NC score | 0.724559 (rank : 5) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99447, Q96G08 | Gene names | PCYT2 | |||
|
Domain Architecture |
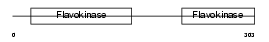
|
|||||
| Description | Ethanolamine-phosphate cytidylyltransferase (EC 2.7.7.14) (Phosphorylethanolamine transferase) (CTP:phosphoethanolamine cytidylyltransferase). | |||||
|
GOGA4_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 7) | NC score | 0.044490 (rank : 9) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
TAXB1_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 8) | NC score | 0.041492 (rank : 10) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q3UKC1, Q91YT6, Q9CVF0, Q9DC45 | Gene names | Tax1bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tax1-binding protein 1 homolog. | |||||
|
EEA1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 9) | NC score | 0.023854 (rank : 22) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
GOGA4_HUMAN
|
||||||
| θ value | 0.62314 (rank : 10) | NC score | 0.031566 (rank : 14) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
MRCKB_MOUSE
|
||||||
| θ value | 1.06291 (rank : 11) | NC score | 0.011056 (rank : 33) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 2180 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q7TT50, Q69ZR5, Q80W33 | Gene names | Cdc42bpb, Kiaa1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
CP135_MOUSE
|
||||||
| θ value | 1.38821 (rank : 12) | NC score | 0.023897 (rank : 21) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 1278 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6P5D4, Q6A030 | Gene names | Cep135, Cep4, Kiaa0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
NUF2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 13) | NC score | 0.036750 (rank : 13) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 661 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9BZD4, Q8WU69, Q96HJ4, Q96Q78 | Gene names | CDCA1, NUF2, NUF2R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Nuf2 (hNuf2) (hNuf2R) (hsNuf2) (Cell division cycle-associated protein 1). | |||||
|
KIF23_HUMAN
|
||||||
| θ value | 3.0926 (rank : 14) | NC score | 0.015642 (rank : 29) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q02241 | Gene names | KIF23, KNSL5, MKLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF23 (Mitotic kinesin-like protein 1) (Kinesin- like protein 5). | |||||
|
MAF1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 15) | NC score | 0.051641 (rank : 7) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H063 | Gene names | MAF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Repressor of RNA polymerase III transcription MAF1 homolog. | |||||
|
TRI60_HUMAN
|
||||||
| θ value | 3.0926 (rank : 16) | NC score | 0.008596 (rank : 34) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q495X7, Q8NA35 | Gene names | TRIM60, RNF129 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 60 (RING finger protein 129). | |||||
|
IFT74_MOUSE
|
||||||
| θ value | 4.03905 (rank : 17) | NC score | 0.020849 (rank : 24) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 939 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BKE9, Q80ZI3, Q9CSY1, Q9CUS0, Q9D9T5 | Gene names | Ift74, Ccdc2, Cmg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 74 homolog (Coiled-coil domain-containing protein 2) (Capillary morphogenesis protein 1) (CMG-1). | |||||
|
K1C16_MOUSE
|
||||||
| θ value | 4.03905 (rank : 18) | NC score | 0.007541 (rank : 35) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Z2K1 | Gene names | Krt16, Krt1-16 | |||
|
Domain Architecture |
|
|||||
| Description | Keratin, type I cytoskeletal 16 (Cytokeratin-16) (CK-16) (Keratin-16) (K16). | |||||
|
MAF1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 19) | NC score | 0.050291 (rank : 8) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9D0U6, Q3U4U3, Q91W84 | Gene names | Maf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Repressor of RNA polymerase III transcription MAF1 homolog. | |||||
|
SPAT7_MOUSE
|
||||||
| θ value | 4.03905 (rank : 20) | NC score | 0.026152 (rank : 18) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80VP2, Q8BK23 | Gene names | Spata7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spermatogenesis-associated protein 7 homolog. | |||||
|
TAF11_HUMAN
|
||||||
| θ value | 4.03905 (rank : 21) | NC score | 0.024969 (rank : 19) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15544, Q9UHS0 | Gene names | TAF11, TAF2I | |||
|
Domain Architecture |
|
|||||
| Description | Transcription initiation factor TFIID subunit 11 (Transcription initiation factor TFIID 28 kDa subunit) (TAF(II)28) (TAFII-28) (TAFII28) (TFIID subunit p30-beta). | |||||
|
CTGE5_MOUSE
|
||||||
| θ value | 5.27518 (rank : 22) | NC score | 0.021749 (rank : 23) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 928 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8R311, Q8CIE3 | Gene names | Ctage5, Mea6, Mgea6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cutaneous T-cell lymphoma-associated antigen 5 homolog (cTAGE-5 protein) (Meningioma-expressed antigen 6). | |||||
|
IF37_HUMAN
|
||||||
| θ value | 5.27518 (rank : 23) | NC score | 0.038055 (rank : 11) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15371, Q5M9Q6 | Gene names | EIF3S7 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 7 (eIF-3 zeta) (eIF3 p66) (eIF3d). | |||||
|
IF37_MOUSE
|
||||||
| θ value | 5.27518 (rank : 24) | NC score | 0.037338 (rank : 12) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O70194 | Gene names | Eif3s7 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 7 (eIF-3 zeta) (eIF3 p66) (eIF3d). | |||||
|
LRRF2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 25) | NC score | 0.026724 (rank : 17) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 791 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y608, Q68CV3, Q9NXH5 | Gene names | LRRFIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 2 (LRR FLII- interacting protein 2). | |||||
|
LRRF2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 26) | NC score | 0.031039 (rank : 15) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q91WK0, Q8BVD1, Q9CU89 | Gene names | Lrrfip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 2 (LRR FLII- interacting protein 2). | |||||
|
MSRE_MOUSE
|
||||||
| θ value | 5.27518 (rank : 27) | NC score | 0.024345 (rank : 20) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P30204, Q923G0, Q9QZ56 | Gene names | Msr1, Scvr | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage scavenger receptor types I and II (Macrophage acetylated LDL receptor I and II) (Scavenger receptor type A) (SR-A). | |||||
|
TRAF5_MOUSE
|
||||||
| θ value | 5.27518 (rank : 28) | NC score | 0.012185 (rank : 32) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 355 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P70191, Q61480 | Gene names | Traf5 | |||
|
Domain Architecture |

|
|||||
| Description | TNF receptor-associated factor 5. | |||||
|
CL025_HUMAN
|
||||||
| θ value | 6.88961 (rank : 29) | NC score | 0.027546 (rank : 16) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IYM0, Q8TCP7, Q9H0L3 | Gene names | C12orf25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C12orf25. | |||||
|
PLCB1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 30) | NC score | 0.018601 (rank : 26) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Z1B3, Q62075, Q8K5A5, Q8K5A6, Q9Z0E5, Q9Z2T5 | Gene names | Plcb1, Plcb | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 1 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-1) (PLC-beta-1) (PLC-I) (PLC-154). | |||||
|
UBF1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 31) | NC score | 0.020289 (rank : 25) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P25976 | Gene names | Ubtf, Tcfubf, Ubf-1, Ubf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1). | |||||
|
CP250_HUMAN
|
||||||
| θ value | 8.99809 (rank : 32) | NC score | 0.014814 (rank : 30) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
K0831_MOUSE
|
||||||
| θ value | 8.99809 (rank : 33) | NC score | 0.012525 (rank : 31) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8CDJ3, Q69ZY1, Q6PFY6, Q8C6N0, Q8R3M3 | Gene names | Kiaa0831, D14Ertd436e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0831. | |||||
|
KINH_HUMAN
|
||||||
| θ value | 8.99809 (rank : 34) | NC score | 0.017705 (rank : 28) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 1231 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P33176, Q5VZ85 | Gene names | KIF5B, KNS, KNS1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain (Ubiquitous kinesin heavy chain) (UKHC). | |||||
|
KINH_MOUSE
|
||||||
| θ value | 8.99809 (rank : 35) | NC score | 0.018260 (rank : 27) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q61768, O08711, Q61580 | Gene names | Kif5b, Khcs, Kns1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain (Ubiquitous kinesin heavy chain) (UKHC). | |||||
|
NOTC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 36) | NC score | 0.003376 (rank : 38) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P46531 | Gene names | NOTCH1, TAN1 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 1 precursor (Notch 1) (hN1) (Translocation-associated notch protein TAN-1) [Contains: Notch 1 extracellular truncation; Notch 1 intracellular domain]. | |||||
|
NOTC1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 37) | NC score | 0.003577 (rank : 37) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q01705, Q06007, Q61905, Q99JC2, Q9QW58, Q9R0X7 | Gene names | Notch1, Motch | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 1 precursor (Notch 1) (Motch A) (mT14) (p300) [Contains: Notch 1 extracellular truncation; Notch 1 intracellular domain]. | |||||
|
TRPC6_MOUSE
|
||||||
| θ value | 8.99809 (rank : 38) | NC score | 0.004668 (rank : 36) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61143, Q9Z2J1 | Gene names | Trpc6, Trp6, Trrp6 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 6 (TrpC6) (Calcium entry channel). | |||||
|
PCY1A_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P49585 | Gene names | PCYT1A, CTPCT, PCYT1 | |||
|
Domain Architecture |

|
|||||
| Description | Choline-phosphate cytidylyltransferase A (EC 2.7.7.15) (Phosphorylcholine transferase A) (CTP:phosphocholine cytidylyltransferase A) (CT A) (CCT A) (CCT-alpha). | |||||
|
PCY1A_MOUSE
|
||||||
| NC score | 0.997115 (rank : 2) | θ value | 4.38123e-180 (rank : 2) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P49586, Q542W4 | Gene names | Pcyt1a, Ctpct, Pcyt1 | |||
|
Domain Architecture |

|
|||||
| Description | Choline-phosphate cytidylyltransferase A (EC 2.7.7.15) (Phosphorylcholine transferase A) (CTP:phosphocholine cytidylyltransferase A) (CT A) (CCT A) (CCT-alpha). | |||||
|
PCY1B_HUMAN
|
||||||
| NC score | 0.986578 (rank : 3) | θ value | 1.5801e-129 (rank : 4) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y5K3, O60621, Q86XC9 | Gene names | PCYT1B, CCTB | |||
|
Domain Architecture |

|
|||||
| Description | Choline-phosphate cytidylyltransferase B (EC 2.7.7.15) (Phosphorylcholine transferase B) (CTP:phosphocholine cytidylyltransferase B) (CT B) (CCT B) (CCT-beta). | |||||
|
PCY1B_MOUSE
|
||||||
| NC score | 0.985736 (rank : 4) | θ value | 1.20984e-129 (rank : 3) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q811Q9, Q3UEW0, Q80Y63, Q811Q8, Q8BKD2, Q8C085 | Gene names | Pcyt1b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Choline-phosphate cytidylyltransferase B (EC 2.7.7.15) (Phosphorylcholine transferase B) (CTP:phosphocholine cytidylyltransferase B) (CT B) (CCT B) (CCT-beta). | |||||
|
PCY2_HUMAN
|
||||||
| NC score | 0.724559 (rank : 5) | θ value | 6.40375e-22 (rank : 6) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99447, Q96G08 | Gene names | PCYT2 | |||
|
Domain Architecture |

|
|||||
| Description | Ethanolamine-phosphate cytidylyltransferase (EC 2.7.7.14) (Phosphorylethanolamine transferase) (CTP:phosphoethanolamine cytidylyltransferase). | |||||
|
PCY2_MOUSE
|
||||||
| NC score | 0.723902 (rank : 6) | θ value | 7.56453e-23 (rank : 5) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q922E4, Q99J50 | Gene names | Pcyt2 | |||
|
Domain Architecture |

|
|||||
| Description | Ethanolamine-phosphate cytidylyltransferase (EC 2.7.7.14) (Phosphorylethanolamine transferase) (CTP:phosphoethanolamine cytidylyltransferase). | |||||
|
MAF1_HUMAN
|
||||||
| NC score | 0.051641 (rank : 7) | θ value | 3.0926 (rank : 15) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H063 | Gene names | MAF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Repressor of RNA polymerase III transcription MAF1 homolog. | |||||
|
MAF1_MOUSE
|
||||||
| NC score | 0.050291 (rank : 8) | θ value | 4.03905 (rank : 19) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9D0U6, Q3U4U3, Q91W84 | Gene names | Maf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Repressor of RNA polymerase III transcription MAF1 homolog. | |||||
|
GOGA4_MOUSE
|
||||||
| NC score | 0.044490 (rank : 9) | θ value | 0.0563607 (rank : 7) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
TAXB1_MOUSE
|
||||||
| NC score | 0.041492 (rank : 10) | θ value | 0.0563607 (rank : 8) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q3UKC1, Q91YT6, Q9CVF0, Q9DC45 | Gene names | Tax1bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tax1-binding protein 1 homolog. | |||||
|
IF37_HUMAN
|
||||||
| NC score | 0.038055 (rank : 11) | θ value | 5.27518 (rank : 23) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15371, Q5M9Q6 | Gene names | EIF3S7 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 7 (eIF-3 zeta) (eIF3 p66) (eIF3d). | |||||
|
IF37_MOUSE
|
||||||
| NC score | 0.037338 (rank : 12) | θ value | 5.27518 (rank : 24) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O70194 | Gene names | Eif3s7 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 7 (eIF-3 zeta) (eIF3 p66) (eIF3d). | |||||
|
NUF2_HUMAN
|
||||||
| NC score | 0.036750 (rank : 13) | θ value | 1.38821 (rank : 13) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 661 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9BZD4, Q8WU69, Q96HJ4, Q96Q78 | Gene names | CDCA1, NUF2, NUF2R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Nuf2 (hNuf2) (hNuf2R) (hsNuf2) (Cell division cycle-associated protein 1). | |||||
|
GOGA4_HUMAN
|
||||||
| NC score | 0.031566 (rank : 14) | θ value | 0.62314 (rank : 10) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
LRRF2_MOUSE
|
||||||
| NC score | 0.031039 (rank : 15) | θ value | 5.27518 (rank : 26) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q91WK0, Q8BVD1, Q9CU89 | Gene names | Lrrfip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 2 (LRR FLII- interacting protein 2). | |||||
|
CL025_HUMAN
|
||||||
| NC score | 0.027546 (rank : 16) | θ value | 6.88961 (rank : 29) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IYM0, Q8TCP7, Q9H0L3 | Gene names | C12orf25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C12orf25. | |||||
|
LRRF2_HUMAN
|
||||||
| NC score | 0.026724 (rank : 17) | θ value | 5.27518 (rank : 25) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 791 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y608, Q68CV3, Q9NXH5 | Gene names | LRRFIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 2 (LRR FLII- interacting protein 2). | |||||
|
SPAT7_MOUSE
|
||||||
| NC score | 0.026152 (rank : 18) | θ value | 4.03905 (rank : 20) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80VP2, Q8BK23 | Gene names | Spata7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spermatogenesis-associated protein 7 homolog. | |||||
|
TAF11_HUMAN
|
||||||
| NC score | 0.024969 (rank : 19) | θ value | 4.03905 (rank : 21) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15544, Q9UHS0 | Gene names | TAF11, TAF2I | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 11 (Transcription initiation factor TFIID 28 kDa subunit) (TAF(II)28) (TAFII-28) (TAFII28) (TFIID subunit p30-beta). | |||||
|
MSRE_MOUSE
|
||||||
| NC score | 0.024345 (rank : 20) | θ value | 5.27518 (rank : 27) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P30204, Q923G0, Q9QZ56 | Gene names | Msr1, Scvr | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage scavenger receptor types I and II (Macrophage acetylated LDL receptor I and II) (Scavenger receptor type A) (SR-A). | |||||
|
CP135_MOUSE
|
||||||
| NC score | 0.023897 (rank : 21) | θ value | 1.38821 (rank : 12) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 1278 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6P5D4, Q6A030 | Gene names | Cep135, Cep4, Kiaa0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
EEA1_HUMAN
|
||||||
| NC score | 0.023854 (rank : 22) | θ value | 0.47712 (rank : 9) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
CTGE5_MOUSE
|
||||||
| NC score | 0.021749 (rank : 23) | θ value | 5.27518 (rank : 22) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 928 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8R311, Q8CIE3 | Gene names | Ctage5, Mea6, Mgea6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cutaneous T-cell lymphoma-associated antigen 5 homolog (cTAGE-5 protein) (Meningioma-expressed antigen 6). | |||||
|
IFT74_MOUSE
|
||||||
| NC score | 0.020849 (rank : 24) | θ value | 4.03905 (rank : 17) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 939 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BKE9, Q80ZI3, Q9CSY1, Q9CUS0, Q9D9T5 | Gene names | Ift74, Ccdc2, Cmg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 74 homolog (Coiled-coil domain-containing protein 2) (Capillary morphogenesis protein 1) (CMG-1). | |||||
|
UBF1_MOUSE
|
||||||
| NC score | 0.020289 (rank : 25) | θ value | 6.88961 (rank : 31) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P25976 | Gene names | Ubtf, Tcfubf, Ubf-1, Ubf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1). | |||||
|
PLCB1_MOUSE
|
||||||
| NC score | 0.018601 (rank : 26) | θ value | 6.88961 (rank : 30) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Z1B3, Q62075, Q8K5A5, Q8K5A6, Q9Z0E5, Q9Z2T5 | Gene names | Plcb1, Plcb | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 1 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-1) (PLC-beta-1) (PLC-I) (PLC-154). | |||||
|
KINH_MOUSE
|
||||||
| NC score | 0.018260 (rank : 27) | θ value | 8.99809 (rank : 35) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q61768, O08711, Q61580 | Gene names | Kif5b, Khcs, Kns1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain (Ubiquitous kinesin heavy chain) (UKHC). | |||||
|
KINH_HUMAN
|
||||||
| NC score | 0.017705 (rank : 28) | θ value | 8.99809 (rank : 34) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 1231 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P33176, Q5VZ85 | Gene names | KIF5B, KNS, KNS1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain (Ubiquitous kinesin heavy chain) (UKHC). | |||||
|
KIF23_HUMAN
|
||||||
| NC score | 0.015642 (rank : 29) | θ value | 3.0926 (rank : 14) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q02241 | Gene names | KIF23, KNSL5, MKLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF23 (Mitotic kinesin-like protein 1) (Kinesin- like protein 5). | |||||
|
CP250_HUMAN
|
||||||
| NC score | 0.014814 (rank : 30) | θ value | 8.99809 (rank : 32) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
K0831_MOUSE
|
||||||
| NC score | 0.012525 (rank : 31) | θ value | 8.99809 (rank : 33) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8CDJ3, Q69ZY1, Q6PFY6, Q8C6N0, Q8R3M3 | Gene names | Kiaa0831, D14Ertd436e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0831. | |||||
|
TRAF5_MOUSE
|
||||||
| NC score | 0.012185 (rank : 32) | θ value | 5.27518 (rank : 28) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 355 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P70191, Q61480 | Gene names | Traf5 | |||
|
Domain Architecture |

|
|||||
| Description | TNF receptor-associated factor 5. | |||||
|
MRCKB_MOUSE
|
||||||
| NC score | 0.011056 (rank : 33) | θ value | 1.06291 (rank : 11) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 2180 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q7TT50, Q69ZR5, Q80W33 | Gene names | Cdc42bpb, Kiaa1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
TRI60_HUMAN
|
||||||
| NC score | 0.008596 (rank : 34) | θ value | 3.0926 (rank : 16) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q495X7, Q8NA35 | Gene names | TRIM60, RNF129 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 60 (RING finger protein 129). | |||||
|
K1C16_MOUSE
|
||||||
| NC score | 0.007541 (rank : 35) | θ value | 4.03905 (rank : 18) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Z2K1 | Gene names | Krt16, Krt1-16 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 16 (Cytokeratin-16) (CK-16) (Keratin-16) (K16). | |||||
|
TRPC6_MOUSE
|
||||||
| NC score | 0.004668 (rank : 36) | θ value | 8.99809 (rank : 38) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61143, Q9Z2J1 | Gene names | Trpc6, Trp6, Trrp6 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 6 (TrpC6) (Calcium entry channel). | |||||
|
NOTC1_MOUSE
|
||||||
| NC score | 0.003577 (rank : 37) | θ value | 8.99809 (rank : 37) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q01705, Q06007, Q61905, Q99JC2, Q9QW58, Q9R0X7 | Gene names | Notch1, Motch | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 1 precursor (Notch 1) (Motch A) (mT14) (p300) [Contains: Notch 1 extracellular truncation; Notch 1 intracellular domain]. | |||||
|
NOTC1_HUMAN
|
||||||
| NC score | 0.003376 (rank : 38) | θ value | 8.99809 (rank : 36) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P46531 | Gene names | NOTCH1, TAN1 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 1 precursor (Notch 1) (hN1) (Translocation-associated notch protein TAN-1) [Contains: Notch 1 extracellular truncation; Notch 1 intracellular domain]. | |||||