Please be patient as the page loads
|
ORC1_MOUSE
|
||||||
| SwissProt Accessions | Q9Z1N2 | Gene names | Orc1l, Orc1 | |||
|
Domain Architecture |

|
|||||
| Description | Origin recognition complex subunit 1. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ORC1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.949520 (rank : 2) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q13415, Q13471, Q5T0F5 | Gene names | ORC1L, ORC1, PARC1 | |||
|
Domain Architecture |

|
|||||
| Description | Origin recognition complex subunit 1 (Replication control protein 1). | |||||
|
ORC1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9Z1N2 | Gene names | Orc1l, Orc1 | |||
|
Domain Architecture |

|
|||||
| Description | Origin recognition complex subunit 1. | |||||
|
CDC6_HUMAN
|
||||||
| θ value | 1.98146e-39 (rank : 3) | NC score | 0.756228 (rank : 4) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99741, Q8TB30 | Gene names | CDC6, CDC18L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division control protein 6 homolog (CDC6-related protein) (p62(cdc6)) (HsCDC6) (HsCDC18). | |||||
|
CDC6_MOUSE
|
||||||
| θ value | 4.41421e-39 (rank : 4) | NC score | 0.760107 (rank : 3) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O89033, Q3TMD0, Q8C3S5, Q9CZT0 | Gene names | Cdc6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division control protein 6 homolog (CDC6-related protein) (p62(cdc6)). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 0.000602161 (rank : 5) | NC score | 0.050732 (rank : 17) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
RAD17_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 6) | NC score | 0.123830 (rank : 5) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75943, O75714, Q7Z3S4, Q9UNK7, Q9UNR7, Q9UNR8, Q9UPF5 | Gene names | RAD17, R24L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell cycle checkpoint protein RAD17 (hRad17) (RF-C/activator 1 homolog). | |||||
|
NFH_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 7) | NC score | 0.054678 (rank : 16) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
SRRM1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 8) | NC score | 0.086182 (rank : 8) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
MAP1B_MOUSE
|
||||||
| θ value | 0.279714 (rank : 9) | NC score | 0.059696 (rank : 11) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
RHG12_MOUSE
|
||||||
| θ value | 0.279714 (rank : 10) | NC score | 0.026898 (rank : 36) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C0D4, Q8BVP8 | Gene names | Arhgap12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 12. | |||||
|
STX18_HUMAN
|
||||||
| θ value | 0.279714 (rank : 11) | NC score | 0.056539 (rank : 14) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9P2W9, Q596L3, Q5TZP5 | Gene names | STX18, GIG9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Syntaxin-18 (Growth-inhibiting gene 9 protein). | |||||
|
GOGA4_HUMAN
|
||||||
| θ value | 0.365318 (rank : 12) | NC score | 0.024292 (rank : 42) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
RAD17_MOUSE
|
||||||
| θ value | 0.365318 (rank : 13) | NC score | 0.092662 (rank : 7) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6NXW6, O88934, O89024 | Gene names | Rad17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell cycle checkpoint protein RAD17. | |||||
|
APC_MOUSE
|
||||||
| θ value | 0.47712 (rank : 14) | NC score | 0.056552 (rank : 13) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q61315, Q62044 | Gene names | Apc | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC) (mAPC). | |||||
|
MAP4_HUMAN
|
||||||
| θ value | 0.62314 (rank : 15) | NC score | 0.064864 (rank : 10) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P27816, Q13082, Q96A76 | Gene names | MAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
POLH_HUMAN
|
||||||
| θ value | 0.62314 (rank : 16) | NC score | 0.039454 (rank : 22) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y253, Q7L8E3, Q96BC4, Q9BX13 | Gene names | POLH, RAD30, RAD30A, XPV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase eta (EC 2.7.7.7) (RAD30 homolog A) (Xeroderma pigmentosum variant type protein). | |||||
|
PRG4_MOUSE
|
||||||
| θ value | 0.62314 (rank : 17) | NC score | 0.048342 (rank : 20) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
ANKR6_HUMAN
|
||||||
| θ value | 0.813845 (rank : 18) | NC score | 0.013043 (rank : 59) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y2G4, Q9NU24, Q9UFQ9 | Gene names | ANKRD6, KIAA0957 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 6. | |||||
|
ABL1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 19) | NC score | 0.007165 (rank : 64) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1220 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P00519, Q13869, Q13870, Q16133, Q45F09 | Gene names | ABL1, ABL, JTK7 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase ABL1 (EC 2.7.10.2) (p150) (c- ABL) (Abelson murine leukemia viral oncogene homolog 1). | |||||
|
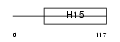
H11_HUMAN
|
||||||
| θ value | 1.38821 (rank : 20) | NC score | 0.036271 (rank : 24) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q02539 | Gene names | HIST1H1A, H1F1 | |||
|
Domain Architecture |
|
|||||
| Description | Histone H1.1. | |||||
|
NKTR_HUMAN
|
||||||
| θ value | 1.38821 (rank : 21) | NC score | 0.028790 (rank : 30) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P30414 | Gene names | NKTR | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
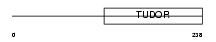
TDRD5_HUMAN
|
||||||
| θ value | 1.38821 (rank : 22) | NC score | 0.039446 (rank : 23) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NAT2, Q5VTV0 | Gene names | TDRD5 | |||
|
Domain Architecture |
|
|||||
| Description | Tudor domain-containing protein 5. | |||||
|
M3K6_HUMAN
|
||||||
| θ value | 1.81305 (rank : 23) | NC score | 0.004690 (rank : 66) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95382 | Gene names | MAP3K6, MAPKKK6 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 6 (EC 2.7.11.25). | |||||
|
PRG4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 24) | NC score | 0.048531 (rank : 19) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
BSN_HUMAN
|
||||||
| θ value | 2.36792 (rank : 25) | NC score | 0.029122 (rank : 28) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 26) | NC score | 0.020764 (rank : 51) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
MYH1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 27) | NC score | 0.025980 (rank : 39) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1478 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P12882, Q9Y622 | Gene names | MYH1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1) (Myosin heavy chain IIx/d) (MyHC-IIx/d). | |||||
|
UBF1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 28) | NC score | 0.028915 (rank : 29) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P17480 | Gene names | UBTF, UBF, UBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1) (Autoantigen NOR-90). | |||||
|
ABL1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 29) | NC score | 0.006371 (rank : 65) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P00520, P97896, Q61252, Q61253, Q61254, Q61255, Q61256, Q61257, Q61258, Q61259, Q61260, Q61261, Q6PCM5, Q8C1X4 | Gene names | Abl1, Abl | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase ABL1 (EC 2.7.10.2) (p150) (c- ABL) (Abelson murine leukemia viral oncogene homolog 1). | |||||
|
AFF4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 30) | NC score | 0.032833 (rank : 25) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
LRCH2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 31) | NC score | 0.009791 (rank : 60) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5VUJ6, Q9HA88, Q9P233 | Gene names | LRCH2, KIAA1495 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeats and calponin homology domain-containing protein 2. | |||||
|
MAP4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 32) | NC score | 0.056113 (rank : 15) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P27546 | Gene names | Map4, Mtap4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
MDC1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 33) | NC score | 0.043004 (rank : 21) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
MYH1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 34) | NC score | 0.026089 (rank : 38) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1461 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q5SX40 | Gene names | Myh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1). | |||||
|
MYH2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 35) | NC score | 0.022950 (rank : 46) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1491 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UKX2, Q14322, Q16229 | Gene names | MYH2, MYHSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, skeletal muscle, adult 2 (Myosin heavy chain IIa) (MyHC-IIa). | |||||
|
MYH4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 36) | NC score | 0.025693 (rank : 41) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1377 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Y623 | Gene names | MYH4 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal) (Myosin heavy chain IIb) (MyHC-IIb). | |||||
|
MYH4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 37) | NC score | 0.025871 (rank : 40) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
SRGP1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 38) | NC score | 0.016429 (rank : 55) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q7Z6B7, Q9H8A3, Q9P2P2 | Gene names | SRGAP1, ARHGAP13, KIAA1304 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 1 (srGAP1) (Rho-GTPase- activating protein 13). | |||||
|
ZAN_HUMAN
|
||||||
| θ value | 3.0926 (rank : 39) | NC score | 0.020646 (rank : 52) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y493, O00218, Q96L85, Q96L86, Q96L87, Q96L88, Q96L89, Q96L90, Q9BXN9, Q9BZ83, Q9BZ84, Q9BZ85, Q9BZ86, Q9BZ87, Q9BZ88 | Gene names | ZAN | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
C8AP2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 40) | NC score | 0.028120 (rank : 33) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 548 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UKL3, Q6PH76, Q7LCQ7, Q86YD9, Q9NUQ4, Q9NZV9, Q9P2N1, Q9Y563 | Gene names | CASP8AP2, FLASH, KIAA1315, RIP25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CASP8-associated protein 2 (FLICE-associated huge protein). | |||||
|
MAP1B_HUMAN
|
||||||
| θ value | 4.03905 (rank : 41) | NC score | 0.059436 (rank : 12) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
MYO15_MOUSE
|
||||||
| θ value | 4.03905 (rank : 42) | NC score | 0.028651 (rank : 32) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9QZZ4, O70395, Q9QWL6 | Gene names | Myo15a, Myo15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
NDE1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 43) | NC score | 0.022002 (rank : 48) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 602 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9CZA6, Q3UBS6, Q3UIC1, Q9ERR0 | Gene names | Nde1, Nude | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear distribution protein nudE homolog 1 (NudE) (mNudE). | |||||
|
NKTR_MOUSE
|
||||||
| θ value | 4.03905 (rank : 44) | NC score | 0.026817 (rank : 37) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P30415 | Gene names | Nktr | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
PPOX_MOUSE
|
||||||
| θ value | 4.03905 (rank : 45) | NC score | 0.023785 (rank : 44) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P51175, P97344, Q99M34 | Gene names | Ppox | |||
|
Domain Architecture |
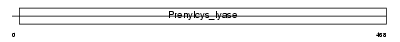
|
|||||
| Description | Protoporphyrinogen oxidase (EC 1.3.3.4) (PPO). | |||||
|
PRP4B_HUMAN
|
||||||
| θ value | 4.03905 (rank : 46) | NC score | 0.015194 (rank : 57) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1030 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q13523, Q8TDP2, Q96QT7, Q9UEE6 | Gene names | PRPF4B, KIAA0536, PRP4, PRP4H, PRP4K | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PRP4 homolog (EC 2.7.11.1) (PRP4 pre- mRNA-processing factor 4 homolog) (PRP4 kinase). | |||||
|
SNIP_MOUSE
|
||||||
| θ value | 4.03905 (rank : 47) | NC score | 0.032810 (rank : 26) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9QWI6, O70298 | Gene names | P140, Kiaa1684 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | p130Cas-associated protein (p140Cap) (SNAP-25-interacting protein) (SNIP). | |||||
|
TARA_HUMAN
|
||||||
| θ value | 4.03905 (rank : 48) | NC score | 0.048603 (rank : 18) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
XRCC4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 49) | NC score | 0.028740 (rank : 31) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q13426, Q9BS72, Q9UP94 | Gene names | XRCC4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein XRCC4 (X-ray repair cross-complementing protein 4). | |||||
|
NP1L3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 50) | NC score | 0.023830 (rank : 43) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q794H2, O54802 | Gene names | Nap1l3, Mb20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome assembly protein 1-like 3 (Brain-specific protein MB20). | |||||
|
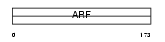
ARF6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 51) | NC score | 0.008479 (rank : 62) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P62330, P26438 | Gene names | ARF6 | |||
|
Domain Architecture |
|
|||||
| Description | ADP-ribosylation factor 6. | |||||
|
ARF6_MOUSE
|
||||||
| θ value | 6.88961 (rank : 52) | NC score | 0.008479 (rank : 63) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P62331, P26438 | Gene names | Arf6 | |||
|
Domain Architecture |
|
|||||
| Description | ADP-ribosylation factor 6. | |||||
|
MY18A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 53) | NC score | 0.021023 (rank : 50) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
MYH8_HUMAN
|
||||||
| θ value | 6.88961 (rank : 54) | NC score | 0.022187 (rank : 47) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1610 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P13535, Q14910 | Gene names | MYH8 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
P52K_MOUSE
|
||||||
| θ value | 6.88961 (rank : 55) | NC score | 0.014669 (rank : 58) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CUX1, Q80Y58 | Gene names | Prkrir, Thap0 | |||
|
Domain Architecture |

|
|||||
| Description | 52 kDa repressor of the inhibitor of the protein kinase (p58IPK- interacting protein) (58 kDa interferon-induced protein kinase- interacting protein) (P52rIPK) (THAP domain-containing protein 0). | |||||
|
REL1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 56) | NC score | 0.018617 (rank : 54) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P04808, Q99936, Q9UQJ1 | Gene names | RLN1 | |||
|
Domain Architecture |
|
|||||
| Description | Prorelaxin H1 precursor [Contains: Relaxin B chain; Relaxin A chain]. | |||||
|
SNX19_HUMAN
|
||||||
| θ value | 6.88961 (rank : 57) | NC score | 0.021652 (rank : 49) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q92543 | Gene names | SNX19, KIAA0254 | |||
|
Domain Architecture |

|
|||||
| Description | Sorting nexin-19. | |||||
|
UBF1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 58) | NC score | 0.027201 (rank : 35) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P25976 | Gene names | Ubtf, Tcfubf, Ubf-1, Ubf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1). | |||||
|
UBP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 59) | NC score | 0.023343 (rank : 45) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O94782, Q9UFR0, Q9UNJ3 | Gene names | USP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 1 (EC 3.1.2.15) (Ubiquitin thioesterase 1) (Ubiquitin-specific-processing protease 1) (Deubiquitinating enzyme 1) (hUBP). | |||||
|
ALEX_HUMAN
|
||||||
| θ value | 8.99809 (rank : 60) | NC score | 0.031039 (rank : 27) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P84996 | Gene names | GNAS, GNAS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALEX (Alternative gene product encoded by XL-exon). | |||||
|
BPTF_HUMAN
|
||||||
| θ value | 8.99809 (rank : 61) | NC score | 0.019980 (rank : 53) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
CNKR1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 62) | NC score | 0.016015 (rank : 56) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q969H4, O95381 | Gene names | CNKSR1, CNK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Connector enhancer of kinase suppressor of ras 1 (Connector enhancer of KSR1) (hCNK1) (Connector enhancer of KSR-like) (CNK homolog protein 1). | |||||
|
GP112_HUMAN
|
||||||
| θ value | 8.99809 (rank : 63) | NC score | 0.009643 (rank : 61) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8IZF6, Q86SM6 | Gene names | GPR112 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 112. | |||||
|
STK36_MOUSE
|
||||||
| θ value | 8.99809 (rank : 64) | NC score | 0.001543 (rank : 67) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 877 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q69ZM6, Q6PDI0, Q80XQ6 | Gene names | Stk36, Kiaa1278 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase 36 (EC 2.7.11.1) (Fused homolog). | |||||
|
TP53B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 65) | NC score | 0.027912 (rank : 34) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
ORC4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.093355 (rank : 6) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43929, Q96C42 | Gene names | ORC4L | |||
|
Domain Architecture |
|
|||||
| Description | Origin recognition complex subunit 4. | |||||
|
ORC4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.074467 (rank : 9) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O88708, Q9QYX1 | Gene names | Orc4l, Orc4 | |||
|
Domain Architecture |
|
|||||
| Description | Origin recognition complex subunit 4. | |||||
|
ORC1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9Z1N2 | Gene names | Orc1l, Orc1 | |||
|
Domain Architecture |

|
|||||
| Description | Origin recognition complex subunit 1. | |||||
|
ORC1_HUMAN
|
||||||
| NC score | 0.949520 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q13415, Q13471, Q5T0F5 | Gene names | ORC1L, ORC1, PARC1 | |||
|
Domain Architecture |

|
|||||
| Description | Origin recognition complex subunit 1 (Replication control protein 1). | |||||
|
CDC6_MOUSE
|
||||||
| NC score | 0.760107 (rank : 3) | θ value | 4.41421e-39 (rank : 4) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O89033, Q3TMD0, Q8C3S5, Q9CZT0 | Gene names | Cdc6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division control protein 6 homolog (CDC6-related protein) (p62(cdc6)). | |||||
|
CDC6_HUMAN
|
||||||
| NC score | 0.756228 (rank : 4) | θ value | 1.98146e-39 (rank : 3) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99741, Q8TB30 | Gene names | CDC6, CDC18L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division control protein 6 homolog (CDC6-related protein) (p62(cdc6)) (HsCDC6) (HsCDC18). | |||||
|
RAD17_HUMAN
|
||||||
| NC score | 0.123830 (rank : 5) | θ value | 0.0330416 (rank : 6) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75943, O75714, Q7Z3S4, Q9UNK7, Q9UNR7, Q9UNR8, Q9UPF5 | Gene names | RAD17, R24L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell cycle checkpoint protein RAD17 (hRad17) (RF-C/activator 1 homolog). | |||||
|
ORC4_HUMAN
|
||||||
| NC score | 0.093355 (rank : 6) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43929, Q96C42 | Gene names | ORC4L | |||
|
Domain Architecture |

|
|||||
| Description | Origin recognition complex subunit 4. | |||||
|
RAD17_MOUSE
|
||||||
| NC score | 0.092662 (rank : 7) | θ value | 0.365318 (rank : 13) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6NXW6, O88934, O89024 | Gene names | Rad17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell cycle checkpoint protein RAD17. | |||||
|
SRRM1_HUMAN
|
||||||
| NC score | 0.086182 (rank : 8) | θ value | 0.21417 (rank : 8) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
ORC4_MOUSE
|
||||||
| NC score | 0.074467 (rank : 9) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O88708, Q9QYX1 | Gene names | Orc4l, Orc4 | |||
|
Domain Architecture |

|
|||||
| Description | Origin recognition complex subunit 4. | |||||
|
MAP4_HUMAN
|
||||||
| NC score | 0.064864 (rank : 10) | θ value | 0.62314 (rank : 15) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P27816, Q13082, Q96A76 | Gene names | MAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
MAP1B_MOUSE
|
||||||
| NC score | 0.059696 (rank : 11) | θ value | 0.279714 (rank : 9) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
MAP1B_HUMAN
|
||||||
| NC score | 0.059436 (rank : 12) | θ value | 4.03905 (rank : 41) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
APC_MOUSE
|
||||||
| NC score | 0.056552 (rank : 13) | θ value | 0.47712 (rank : 14) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q61315, Q62044 | Gene names | Apc | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC) (mAPC). | |||||
|
STX18_HUMAN
|
||||||
| NC score | 0.056539 (rank : 14) | θ value | 0.279714 (rank : 11) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9P2W9, Q596L3, Q5TZP5 | Gene names | STX18, GIG9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Syntaxin-18 (Growth-inhibiting gene 9 protein). | |||||
|
MAP4_MOUSE
|
||||||
| NC score | 0.056113 (rank : 15) | θ value | 3.0926 (rank : 32) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P27546 | Gene names | Map4, Mtap4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
NFH_HUMAN
|
||||||
| NC score | 0.054678 (rank : 16) | θ value | 0.0736092 (rank : 7) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.050732 (rank : 17) | θ value | 0.000602161 (rank : 5) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.048603 (rank : 18) | θ value | 4.03905 (rank : 48) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
PRG4_HUMAN
|
||||||
| NC score | 0.048531 (rank : 19) | θ value | 1.81305 (rank : 24) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PRG4_MOUSE
|
||||||
| NC score | 0.048342 (rank : 20) | θ value | 0.62314 (rank : 17) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
MDC1_MOUSE
|
||||||
| NC score | 0.043004 (rank : 21) | θ value | 3.0926 (rank : 33) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
POLH_HUMAN
|
||||||
| NC score | 0.039454 (rank : 22) | θ value | 0.62314 (rank : 16) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y253, Q7L8E3, Q96BC4, Q9BX13 | Gene names | POLH, RAD30, RAD30A, XPV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase eta (EC 2.7.7.7) (RAD30 homolog A) (Xeroderma pigmentosum variant type protein). | |||||
|
TDRD5_HUMAN
|
||||||
| NC score | 0.039446 (rank : 23) | θ value | 1.38821 (rank : 22) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NAT2, Q5VTV0 | Gene names | TDRD5 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 5. | |||||
|
H11_HUMAN
|
||||||
| NC score | 0.036271 (rank : 24) | θ value | 1.38821 (rank : 20) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q02539 | Gene names | HIST1H1A, H1F1 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.1. | |||||
|
AFF4_MOUSE
|
||||||
| NC score | 0.032833 (rank : 25) | θ value | 3.0926 (rank : 30) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
SNIP_MOUSE
|
||||||
| NC score | 0.032810 (rank : 26) | θ value | 4.03905 (rank : 47) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9QWI6, O70298 | Gene names | P140, Kiaa1684 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | p130Cas-associated protein (p140Cap) (SNAP-25-interacting protein) (SNIP). | |||||
|
ALEX_HUMAN
|
||||||
| NC score | 0.031039 (rank : 27) | θ value | 8.99809 (rank : 60) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P84996 | Gene names | GNAS, GNAS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALEX (Alternative gene product encoded by XL-exon). | |||||
|
BSN_HUMAN
|
||||||
| NC score | 0.029122 (rank : 28) | θ value | 2.36792 (rank : 25) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
UBF1_HUMAN
|
||||||
| NC score | 0.028915 (rank : 29) | θ value | 2.36792 (rank : 28) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P17480 | Gene names | UBTF, UBF, UBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1) (Autoantigen NOR-90). | |||||
|
NKTR_HUMAN
|
||||||
| NC score | 0.028790 (rank : 30) | θ value | 1.38821 (rank : 21) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P30414 | Gene names | NKTR | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
XRCC4_HUMAN
|
||||||
| NC score | 0.028740 (rank : 31) | θ value | 4.03905 (rank : 49) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q13426, Q9BS72, Q9UP94 | Gene names | XRCC4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein XRCC4 (X-ray repair cross-complementing protein 4). | |||||
|
MYO15_MOUSE
|
||||||
| NC score | 0.028651 (rank : 32) | θ value | 4.03905 (rank : 42) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9QZZ4, O70395, Q9QWL6 | Gene names | Myo15a, Myo15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
C8AP2_HUMAN
|
||||||
| NC score | 0.028120 (rank : 33) | θ value | 4.03905 (rank : 40) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 548 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UKL3, Q6PH76, Q7LCQ7, Q86YD9, Q9NUQ4, Q9NZV9, Q9P2N1, Q9Y563 | Gene names | CASP8AP2, FLASH, KIAA1315, RIP25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CASP8-associated protein 2 (FLICE-associated huge protein). | |||||
|
TP53B_MOUSE
|
||||||
| NC score | 0.027912 (rank : 34) | θ value | 8.99809 (rank : 65) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
UBF1_MOUSE
|
||||||
| NC score | 0.027201 (rank : 35) | θ value | 6.88961 (rank : 58) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P25976 | Gene names | Ubtf, Tcfubf, Ubf-1, Ubf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1). | |||||
|
RHG12_MOUSE
|
||||||
| NC score | 0.026898 (rank : 36) | θ value | 0.279714 (rank : 10) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C0D4, Q8BVP8 | Gene names | Arhgap12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 12. | |||||
|
NKTR_MOUSE
|
||||||
| NC score | 0.026817 (rank : 37) | θ value | 4.03905 (rank : 44) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P30415 | Gene names | Nktr | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
MYH1_MOUSE
|
||||||
| NC score | 0.026089 (rank : 38) | θ value | 3.0926 (rank : 34) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1461 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q5SX40 | Gene names | Myh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1). | |||||
|
MYH1_HUMAN
|
||||||
| NC score | 0.025980 (rank : 39) | θ value | 2.36792 (rank : 27) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1478 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P12882, Q9Y622 | Gene names | MYH1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1) (Myosin heavy chain IIx/d) (MyHC-IIx/d). | |||||
|
MYH4_MOUSE
|
||||||
| NC score | 0.025871 (rank : 40) | θ value | 3.0926 (rank : 37) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
MYH4_HUMAN
|
||||||
| NC score | 0.025693 (rank : 41) | θ value | 3.0926 (rank : 36) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1377 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Y623 | Gene names | MYH4 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal) (Myosin heavy chain IIb) (MyHC-IIb). | |||||
|
GOGA4_HUMAN
|
||||||
| NC score | 0.024292 (rank : 42) | θ value | 0.365318 (rank : 12) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
NP1L3_MOUSE
|
||||||
| NC score | 0.023830 (rank : 43) | θ value | 5.27518 (rank : 50) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q794H2, O54802 | Gene names | Nap1l3, Mb20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome assembly protein 1-like 3 (Brain-specific protein MB20). | |||||
|
PPOX_MOUSE
|
||||||
| NC score | 0.023785 (rank : 44) | θ value | 4.03905 (rank : 45) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P51175, P97344, Q99M34 | Gene names | Ppox | |||
|
Domain Architecture |
|
|||||
| Description | Protoporphyrinogen oxidase (EC 1.3.3.4) (PPO). | |||||
|
UBP1_HUMAN
|
||||||
| NC score | 0.023343 (rank : 45) | θ value | 6.88961 (rank : 59) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O94782, Q9UFR0, Q9UNJ3 | Gene names | USP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 1 (EC 3.1.2.15) (Ubiquitin thioesterase 1) (Ubiquitin-specific-processing protease 1) (Deubiquitinating enzyme 1) (hUBP). | |||||
|
MYH2_HUMAN
|
||||||
| NC score | 0.022950 (rank : 46) | θ value | 3.0926 (rank : 35) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1491 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UKX2, Q14322, Q16229 | Gene names | MYH2, MYHSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, skeletal muscle, adult 2 (Myosin heavy chain IIa) (MyHC-IIa). | |||||
|
MYH8_HUMAN
|
||||||
| NC score | 0.022187 (rank : 47) | θ value | 6.88961 (rank : 54) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1610 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P13535, Q14910 | Gene names | MYH8 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
NDE1_MOUSE
|
||||||
| NC score | 0.022002 (rank : 48) | θ value | 4.03905 (rank : 43) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 602 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9CZA6, Q3UBS6, Q3UIC1, Q9ERR0 | Gene names | Nde1, Nude | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear distribution protein nudE homolog 1 (NudE) (mNudE). | |||||
|
SNX19_HUMAN
|
||||||
| NC score | 0.021652 (rank : 49) | θ value | 6.88961 (rank : 57) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q92543 | Gene names | SNX19, KIAA0254 | |||
|
Domain Architecture |

|
|||||
| Description | Sorting nexin-19. | |||||
|
MY18A_MOUSE
|
||||||
| NC score | 0.021023 (rank : 50) | θ value | 6.88961 (rank : 53) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.020764 (rank : 51) | θ value | 2.36792 (rank : 26) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
ZAN_HUMAN
|
||||||
| NC score | 0.020646 (rank : 52) | θ value | 3.0926 (rank : 39) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y493, O00218, Q96L85, Q96L86, Q96L87, Q96L88, Q96L89, Q96L90, Q9BXN9, Q9BZ83, Q9BZ84, Q9BZ85, Q9BZ86, Q9BZ87, Q9BZ88 | Gene names | ZAN | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
BPTF_HUMAN
|
||||||
| NC score | 0.019980 (rank : 53) | θ value | 8.99809 (rank : 61) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
REL1_HUMAN
|
||||||
| NC score | 0.018617 (rank : 54) | θ value | 6.88961 (rank : 56) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P04808, Q99936, Q9UQJ1 | Gene names | RLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Prorelaxin H1 precursor [Contains: Relaxin B chain; Relaxin A chain]. | |||||
|
SRGP1_HUMAN
|
||||||
| NC score | 0.016429 (rank : 55) | θ value | 3.0926 (rank : 38) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q7Z6B7, Q9H8A3, Q9P2P2 | Gene names | SRGAP1, ARHGAP13, KIAA1304 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 1 (srGAP1) (Rho-GTPase- activating protein 13). | |||||
|
CNKR1_HUMAN
|
||||||
| NC score | 0.016015 (rank : 56) | θ value | 8.99809 (rank : 62) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q969H4, O95381 | Gene names | CNKSR1, CNK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Connector enhancer of kinase suppressor of ras 1 (Connector enhancer of KSR1) (hCNK1) (Connector enhancer of KSR-like) (CNK homolog protein 1). | |||||
|
PRP4B_HUMAN
|
||||||
| NC score | 0.015194 (rank : 57) | θ value | 4.03905 (rank : 46) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1030 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q13523, Q8TDP2, Q96QT7, Q9UEE6 | Gene names | PRPF4B, KIAA0536, PRP4, PRP4H, PRP4K | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PRP4 homolog (EC 2.7.11.1) (PRP4 pre- mRNA-processing factor 4 homolog) (PRP4 kinase). | |||||
|
P52K_MOUSE
|
||||||
| NC score | 0.014669 (rank : 58) | θ value | 6.88961 (rank : 55) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CUX1, Q80Y58 | Gene names | Prkrir, Thap0 | |||
|
Domain Architecture |

|
|||||
| Description | 52 kDa repressor of the inhibitor of the protein kinase (p58IPK- interacting protein) (58 kDa interferon-induced protein kinase- interacting protein) (P52rIPK) (THAP domain-containing protein 0). | |||||
|
ANKR6_HUMAN
|
||||||
| NC score | 0.013043 (rank : 59) | θ value | 0.813845 (rank : 18) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y2G4, Q9NU24, Q9UFQ9 | Gene names | ANKRD6, KIAA0957 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 6. | |||||
|
LRCH2_HUMAN
|
||||||
| NC score | 0.009791 (rank : 60) | θ value | 3.0926 (rank : 31) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5VUJ6, Q9HA88, Q9P233 | Gene names | LRCH2, KIAA1495 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeats and calponin homology domain-containing protein 2. | |||||
|
GP112_HUMAN
|
||||||
| NC score | 0.009643 (rank : 61) | θ value | 8.99809 (rank : 63) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8IZF6, Q86SM6 | Gene names | GPR112 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 112. | |||||
|
ARF6_HUMAN
|
||||||
| NC score | 0.008479 (rank : 62) | θ value | 6.88961 (rank : 51) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P62330, P26438 | Gene names | ARF6 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor 6. | |||||
|
ARF6_MOUSE
|
||||||
| NC score | 0.008479 (rank : 63) | θ value | 6.88961 (rank : 52) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P62331, P26438 | Gene names | Arf6 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor 6. | |||||
|
ABL1_HUMAN
|
||||||
| NC score | 0.007165 (rank : 64) | θ value | 1.38821 (rank : 19) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1220 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P00519, Q13869, Q13870, Q16133, Q45F09 | Gene names | ABL1, ABL, JTK7 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase ABL1 (EC 2.7.10.2) (p150) (c- ABL) (Abelson murine leukemia viral oncogene homolog 1). | |||||
|
ABL1_MOUSE
|
||||||
| NC score | 0.006371 (rank : 65) | θ value | 3.0926 (rank : 29) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P00520, P97896, Q61252, Q61253, Q61254, Q61255, Q61256, Q61257, Q61258, Q61259, Q61260, Q61261, Q6PCM5, Q8C1X4 | Gene names | Abl1, Abl | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase ABL1 (EC 2.7.10.2) (p150) (c- ABL) (Abelson murine leukemia viral oncogene homolog 1). | |||||
|
M3K6_HUMAN
|
||||||
| NC score | 0.004690 (rank : 66) | θ value | 1.81305 (rank : 23) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95382 | Gene names | MAP3K6, MAPKKK6 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 6 (EC 2.7.11.25). | |||||
|
STK36_MOUSE
|
||||||
| NC score | 0.001543 (rank : 67) | θ value | 8.99809 (rank : 64) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 877 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q69ZM6, Q6PDI0, Q80XQ6 | Gene names | Stk36, Kiaa1278 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase 36 (EC 2.7.11.1) (Fused homolog). | |||||