Please be patient as the page loads
|
NFYA_MOUSE
|
||||||
| SwissProt Accessions | P23708 | Gene names | Nfya | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear transcription factor Y subunit alpha (Nuclear transcription factor Y subunit A) (NF-YA) (CAAT-box DNA-binding protein subunit A). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
NFYA_HUMAN
|
||||||
| θ value | 8.51556e-75 (rank : 1) | NC score | 0.996782 (rank : 2) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P23511, Q8IXU0 | Gene names | NFYA | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear transcription factor Y subunit alpha (Nuclear transcription factor Y subunit A) (NF-YA) (CAAT-box DNA-binding protein subunit A). | |||||
|
NFYA_MOUSE
|
||||||
| θ value | 8.51556e-75 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P23708 | Gene names | Nfya | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear transcription factor Y subunit alpha (Nuclear transcription factor Y subunit A) (NF-YA) (CAAT-box DNA-binding protein subunit A). | |||||
|
EP400_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 3) | NC score | 0.104934 (rank : 7) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
ATF1_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 4) | NC score | 0.158635 (rank : 3) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P18846, P25168 | Gene names | ATF1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-1 (Activating transcription factor 1) (TREB36 protein). | |||||
|
SP4_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 5) | NC score | 0.055603 (rank : 17) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q02446, O60402 | Gene names | SP4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Sp4 (SPR-1). | |||||
|
SP4_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 6) | NC score | 0.054430 (rank : 18) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q62445 | Gene names | Sp4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Sp4. | |||||
|
ZNF76_HUMAN
|
||||||
| θ value | 0.365318 (rank : 7) | NC score | 0.027157 (rank : 35) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 728 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P36508, Q9BQB2 | Gene names | ZNF76 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 76. | |||||
|
ACINU_MOUSE
|
||||||
| θ value | 0.47712 (rank : 8) | NC score | 0.073845 (rank : 14) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9JIX8, Q9CSN7, Q9CSR9, Q9CSX7, Q9R046, Q9R047 | Gene names | Acin1, Acinus | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
ZN335_HUMAN
|
||||||
| θ value | 0.47712 (rank : 9) | NC score | 0.028642 (rank : 32) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 973 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9H4Z2, Q9H684 | Gene names | ZNF335 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 335. | |||||
|
ZN628_MOUSE
|
||||||
| θ value | 0.47712 (rank : 10) | NC score | 0.025284 (rank : 37) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 815 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8CJ78, Q3U2L5, Q6P5B3 | Gene names | Znf628, Zec, Zfp628 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 628 (Zinc finger protein expressed in embryonal cells and certain adult organs). | |||||
|
ATF1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 11) | NC score | 0.139231 (rank : 4) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P81269 | Gene names | Atf1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-1 (Activating transcription factor 1) (TCR-ATF1). | |||||
|
PO6F2_MOUSE
|
||||||
| θ value | 0.62314 (rank : 12) | NC score | 0.049518 (rank : 20) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BJI4 | Gene names | Pou6f2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | POU domain, class 6, transcription factor 2. | |||||
|
ZN236_HUMAN
|
||||||
| θ value | 0.62314 (rank : 13) | NC score | 0.026140 (rank : 36) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 844 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UL36, Q9UL37 | Gene names | ZNF236 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 236. | |||||
|
SP2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 14) | NC score | 0.043493 (rank : 24) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q02086 | Gene names | SP2, KIAA0048 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Sp2. | |||||
|
SP3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 15) | NC score | 0.044040 (rank : 23) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 737 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q02447, Q8TD56, Q9BQR1 | Gene names | SP3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Sp3 (SPR-2). | |||||
|
YETS2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 16) | NC score | 0.096679 (rank : 9) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q3TUF7, Q6PGF8, Q80TI2, Q8CG86 | Gene names | Yeats2, Kiaa1197 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YEATS domain-containing protein 2. | |||||
|
COA1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 17) | NC score | 0.045323 (rank : 22) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13085 | Gene names | ACACA, ACAC, ACC1, ACCA | |||
|
Domain Architecture |

|
|||||
| Description | Acetyl-CoA carboxylase 1 (EC 6.4.1.2) (ACC-alpha) [Includes: Biotin carboxylase (EC 6.3.4.14)]. | |||||
|
CREB1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 18) | NC score | 0.115992 (rank : 6) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P16220, P21934, Q9UMA7 | Gene names | CREB1 | |||
|
Domain Architecture |

|
|||||
| Description | cAMP response element-binding protein (CREB). | |||||
|
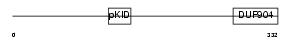
CREB1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 19) | NC score | 0.116036 (rank : 5) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q01147 | Gene names | Creb1, Creb-1 | |||
|
Domain Architecture |
|
|||||
| Description | cAMP response element-binding protein (CREB). | |||||
|
ZN687_MOUSE
|
||||||
| θ value | 1.38821 (rank : 20) | NC score | 0.033550 (rank : 30) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 913 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9D2D7, Q6PAP3, Q6ZPQ9 | Gene names | Znf687, Kiaa1441, Zfp687 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
EMSY_MOUSE
|
||||||
| θ value | 1.81305 (rank : 21) | NC score | 0.098875 (rank : 8) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BMB0, Q5FWK5, Q80XU1, Q8VDW9 | Gene names | Emsy | |||
|
Domain Architecture |

|
|||||
| Description | Protein EMSY. | |||||
|
YETS2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 22) | NC score | 0.086924 (rank : 11) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9ULM3, Q641P6, Q9NW96 | Gene names | YEATS2, KIAA1197 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YEATS domain-containing protein 2. | |||||
|
HCFC1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 23) | NC score | 0.052292 (rank : 19) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q61191, Q684R1, Q7TSB0, Q8C2D0, Q9QWH2 | Gene names | Hcfc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Host cell factor (HCF) (HCF-1) (C1 factor) [Contains: HCF N-terminal chain 1; HCF N-terminal chain 2; HCF N-terminal chain 3; HCF N- terminal chain 4; HCF N-terminal chain 5; HCF N-terminal chain 6; HCF C-terminal chain 1; HCF C-terminal chain 2; HCF C-terminal chain 3; HCF C-terminal chain 4; HCF C-terminal chain 5; HCF C-terminal chain 6]. | |||||
|
NCOA6_MOUSE
|
||||||
| θ value | 2.36792 (rank : 24) | NC score | 0.041618 (rank : 27) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
SP1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 25) | NC score | 0.042572 (rank : 26) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P08047, Q86TN8, Q9H3Q5, Q9NR51, Q9NY21, Q9NYE7 | Gene names | SP1, TSFP1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Sp1. | |||||
|
ZN628_HUMAN
|
||||||
| θ value | 2.36792 (rank : 26) | NC score | 0.022778 (rank : 41) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 814 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5EBL2, Q86X34 | Gene names | ZNF628 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 628. | |||||
|
EMSY_HUMAN
|
||||||
| θ value | 3.0926 (rank : 27) | NC score | 0.090272 (rank : 10) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q7Z589, Q4G109, Q8NBU6, Q8TE50, Q9H8I9, Q9NRH0 | Gene names | EMSY, C11orf30 | |||
|
Domain Architecture |

|
|||||
| Description | Protein EMSY. | |||||
|
ACINU_HUMAN
|
||||||
| θ value | 4.03905 (rank : 28) | NC score | 0.058522 (rank : 16) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UKV3, O75158, Q9UG91, Q9UKV1, Q9UKV2 | Gene names | ACIN1, ACINUS, KIAA0670 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
KIRR1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 29) | NC score | 0.023458 (rank : 40) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96J84, Q5XKC6, Q7Z696, Q7Z7N8, Q8TB15, Q9H9N1, Q9NVA5 | Gene names | KIRREL, NEPH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kin of IRRE-like protein 1 precursor (Kin of irregular chiasm-like protein 1) (Nephrin-like protein 1). | |||||
|
PAR10_HUMAN
|
||||||
| θ value | 4.03905 (rank : 30) | NC score | 0.042717 (rank : 25) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q53GL7, Q8N2I0, Q8WV05, Q96CH7 | Gene names | PARP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly [ADP-ribose] polymerase 10 (EC 2.4.2.30) (PARP-10). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 4.03905 (rank : 31) | NC score | 0.024841 (rank : 39) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
BPTF_HUMAN
|
||||||
| θ value | 5.27518 (rank : 32) | NC score | 0.027365 (rank : 34) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
POGZ_MOUSE
|
||||||
| θ value | 5.27518 (rank : 33) | NC score | 0.024920 (rank : 38) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BZH4, Q4VA94, Q80TZ8, Q8C0K1, Q8K294 | Gene names | Pogz, Kiaa0461 | |||
|
Domain Architecture |

|
|||||
| Description | Pogo transposable element with ZNF domain. | |||||
|
KIRR1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 34) | NC score | 0.020890 (rank : 42) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80W68, Q8CIJ4, Q8CJ59, Q923L4 | Gene names | Kirrel1, Kirrel, Neph1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kin of IRRE-like protein 1 precursor (Kin of irregular chiasm-like protein 1) (Nephrin-like protein 1). | |||||
|
NFYC_HUMAN
|
||||||
| θ value | 6.88961 (rank : 35) | NC score | 0.027365 (rank : 33) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13952, Q5T6K8, Q5T6L1, Q5TZR6, Q92869, Q9HBX1, Q9NXB5, Q9UM67, Q9UML0, Q9UMT7 | Gene names | NFYC | |||
|
Domain Architecture |
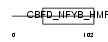
|
|||||
| Description | Nuclear transcription factor Y subunit gamma (Nuclear transcription factor Y subunit C) (NF-YC) (CAAT-box DNA-binding protein subunit C) (Transactivator HSM-1/2). | |||||
|
PO6F2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 36) | NC score | 0.033875 (rank : 29) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P78424, P78425, Q75ME8, Q86UM6, Q9UDS7 | Gene names | POU6F2, RPF1 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 6, transcription factor 2 (Retina-derived POU-domain factor 1) (RPF-1). | |||||
|
RCL1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 37) | NC score | 0.031512 (rank : 31) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y2P8, Q5VYW9, Q9H9D0, Q9NY00, Q9P044 | Gene names | RCL1, RNAC, RPC2, RTC2 | |||
|
Domain Architecture |
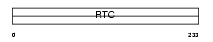
|
|||||
| Description | RNA 3'-terminal phosphate cyclase-like protein. | |||||
|
SP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 38) | NC score | 0.040570 (rank : 28) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 730 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O89090, O89087, Q62251, Q64167 | Gene names | Sp1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Sp1. | |||||
|
SRRM1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 39) | NC score | 0.020602 (rank : 43) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
EP400_HUMAN
|
||||||
| θ value | 8.99809 (rank : 40) | NC score | 0.066528 (rank : 15) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96L91, O15411, Q6P2F5, Q8N8Q7, Q8NE05, Q96JK7, Q9P230 | Gene names | EP400, CAGH32, KIAA1498, KIAA1818, TNRC12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (hDomino) (CAG repeat protein 32) (Trinucleotide repeat-containing gene 12 protein). | |||||
|
HCFC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 41) | NC score | 0.046412 (rank : 21) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P51610, Q6P4G5 | Gene names | HCFC1, HCF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Host cell factor (HCF) (HCF-1) (C1 factor) (VP16 accessory protein) (VCAF) (CFF) [Contains: HCF N-terminal chain 1; HCF N-terminal chain 2; HCF N-terminal chain 3; HCF N-terminal chain 4; HCF N-terminal chain 5; HCF N-terminal chain 6; HCF C-terminal chain 1; HCF C- terminal chain 2; HCF C-terminal chain 3; HCF C-terminal chain 4; HCF C-terminal chain 5; HCF C-terminal chain 6]. | |||||
|
TNIK_HUMAN
|
||||||
| θ value | 8.99809 (rank : 42) | NC score | 0.001024 (rank : 44) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 1570 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UKE5, O60298, Q8WUY7, Q9UKD8, Q9UKD9, Q9UKE0, Q9UKE1, Q9UKE2, Q9UKE3, Q9UKE4 | Gene names | TNIK, KIAA0551 | |||
|
Domain Architecture |

|
|||||
| Description | TRAF2 and NCK-interacting protein kinase (EC 2.7.11.1). | |||||
|
CREM_HUMAN
|
||||||
| θ value | θ > 10 (rank : 43) | NC score | 0.075826 (rank : 12) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q03060, Q16114, Q16116, Q9NZB9 | Gene names | CREM | |||
|
Domain Architecture |
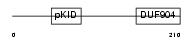
|
|||||
| Description | cAMP-responsive element modulator. | |||||
|
CREM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 44) | NC score | 0.074636 (rank : 13) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P27699, P27698 | Gene names | Crem | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-responsive element modulator. | |||||
|
NFYA_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 8.51556e-75 (rank : 2) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P23708 | Gene names | Nfya | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear transcription factor Y subunit alpha (Nuclear transcription factor Y subunit A) (NF-YA) (CAAT-box DNA-binding protein subunit A). | |||||
|
NFYA_HUMAN
|
||||||
| NC score | 0.996782 (rank : 2) | θ value | 8.51556e-75 (rank : 1) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P23511, Q8IXU0 | Gene names | NFYA | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear transcription factor Y subunit alpha (Nuclear transcription factor Y subunit A) (NF-YA) (CAAT-box DNA-binding protein subunit A). | |||||
|
ATF1_HUMAN
|
||||||
| NC score | 0.158635 (rank : 3) | θ value | 0.0431538 (rank : 4) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P18846, P25168 | Gene names | ATF1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-1 (Activating transcription factor 1) (TREB36 protein). | |||||
|
ATF1_MOUSE
|
||||||
| NC score | 0.139231 (rank : 4) | θ value | 0.62314 (rank : 11) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P81269 | Gene names | Atf1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-1 (Activating transcription factor 1) (TCR-ATF1). | |||||
|
CREB1_MOUSE
|
||||||
| NC score | 0.116036 (rank : 5) | θ value | 1.38821 (rank : 19) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q01147 | Gene names | Creb1, Creb-1 | |||
|
Domain Architecture |

|
|||||
| Description | cAMP response element-binding protein (CREB). | |||||
|
CREB1_HUMAN
|
||||||
| NC score | 0.115992 (rank : 6) | θ value | 1.38821 (rank : 18) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P16220, P21934, Q9UMA7 | Gene names | CREB1 | |||
|
Domain Architecture |

|
|||||
| Description | cAMP response element-binding protein (CREB). | |||||
|
EP400_MOUSE
|
||||||
| NC score | 0.104934 (rank : 7) | θ value | 0.00298849 (rank : 3) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
EMSY_MOUSE
|
||||||
| NC score | 0.098875 (rank : 8) | θ value | 1.81305 (rank : 21) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BMB0, Q5FWK5, Q80XU1, Q8VDW9 | Gene names | Emsy | |||
|
Domain Architecture |

|
|||||
| Description | Protein EMSY. | |||||
|
YETS2_MOUSE
|
||||||
| NC score | 0.096679 (rank : 9) | θ value | 1.06291 (rank : 16) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q3TUF7, Q6PGF8, Q80TI2, Q8CG86 | Gene names | Yeats2, Kiaa1197 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YEATS domain-containing protein 2. | |||||
|
EMSY_HUMAN
|
||||||
| NC score | 0.090272 (rank : 10) | θ value | 3.0926 (rank : 27) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q7Z589, Q4G109, Q8NBU6, Q8TE50, Q9H8I9, Q9NRH0 | Gene names | EMSY, C11orf30 | |||
|
Domain Architecture |

|
|||||
| Description | Protein EMSY. | |||||
|
YETS2_HUMAN
|
||||||
| NC score | 0.086924 (rank : 11) | θ value | 1.81305 (rank : 22) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9ULM3, Q641P6, Q9NW96 | Gene names | YEATS2, KIAA1197 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YEATS domain-containing protein 2. | |||||
|
CREM_HUMAN
|
||||||
| NC score | 0.075826 (rank : 12) | θ value | θ > 10 (rank : 43) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q03060, Q16114, Q16116, Q9NZB9 | Gene names | CREM | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-responsive element modulator. | |||||
|
CREM_MOUSE
|
||||||
| NC score | 0.074636 (rank : 13) | θ value | θ > 10 (rank : 44) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P27699, P27698 | Gene names | Crem | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-responsive element modulator. | |||||
|
ACINU_MOUSE
|
||||||
| NC score | 0.073845 (rank : 14) | θ value | 0.47712 (rank : 8) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9JIX8, Q9CSN7, Q9CSR9, Q9CSX7, Q9R046, Q9R047 | Gene names | Acin1, Acinus | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
EP400_HUMAN
|
||||||
| NC score | 0.066528 (rank : 15) | θ value | 8.99809 (rank : 40) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96L91, O15411, Q6P2F5, Q8N8Q7, Q8NE05, Q96JK7, Q9P230 | Gene names | EP400, CAGH32, KIAA1498, KIAA1818, TNRC12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (hDomino) (CAG repeat protein 32) (Trinucleotide repeat-containing gene 12 protein). | |||||
|
ACINU_HUMAN
|
||||||
| NC score | 0.058522 (rank : 16) | θ value | 4.03905 (rank : 28) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UKV3, O75158, Q9UG91, Q9UKV1, Q9UKV2 | Gene names | ACIN1, ACINUS, KIAA0670 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
SP4_HUMAN
|
||||||
| NC score | 0.055603 (rank : 17) | θ value | 0.0736092 (rank : 5) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q02446, O60402 | Gene names | SP4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Sp4 (SPR-1). | |||||
|
SP4_MOUSE
|
||||||
| NC score | 0.054430 (rank : 18) | θ value | 0.0961366 (rank : 6) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q62445 | Gene names | Sp4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Sp4. | |||||
|
HCFC1_MOUSE
|
||||||
| NC score | 0.052292 (rank : 19) | θ value | 2.36792 (rank : 23) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q61191, Q684R1, Q7TSB0, Q8C2D0, Q9QWH2 | Gene names | Hcfc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Host cell factor (HCF) (HCF-1) (C1 factor) [Contains: HCF N-terminal chain 1; HCF N-terminal chain 2; HCF N-terminal chain 3; HCF N- terminal chain 4; HCF N-terminal chain 5; HCF N-terminal chain 6; HCF C-terminal chain 1; HCF C-terminal chain 2; HCF C-terminal chain 3; HCF C-terminal chain 4; HCF C-terminal chain 5; HCF C-terminal chain 6]. | |||||
|
PO6F2_MOUSE
|
||||||
| NC score | 0.049518 (rank : 20) | θ value | 0.62314 (rank : 12) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BJI4 | Gene names | Pou6f2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | POU domain, class 6, transcription factor 2. | |||||
|
HCFC1_HUMAN
|
||||||
| NC score | 0.046412 (rank : 21) | θ value | 8.99809 (rank : 41) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P51610, Q6P4G5 | Gene names | HCFC1, HCF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Host cell factor (HCF) (HCF-1) (C1 factor) (VP16 accessory protein) (VCAF) (CFF) [Contains: HCF N-terminal chain 1; HCF N-terminal chain 2; HCF N-terminal chain 3; HCF N-terminal chain 4; HCF N-terminal chain 5; HCF N-terminal chain 6; HCF C-terminal chain 1; HCF C- terminal chain 2; HCF C-terminal chain 3; HCF C-terminal chain 4; HCF C-terminal chain 5; HCF C-terminal chain 6]. | |||||
|
COA1_HUMAN
|
||||||
| NC score | 0.045323 (rank : 22) | θ value | 1.38821 (rank : 17) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13085 | Gene names | ACACA, ACAC, ACC1, ACCA | |||
|
Domain Architecture |

|
|||||
| Description | Acetyl-CoA carboxylase 1 (EC 6.4.1.2) (ACC-alpha) [Includes: Biotin carboxylase (EC 6.3.4.14)]. | |||||
|
SP3_HUMAN
|
||||||
| NC score | 0.044040 (rank : 23) | θ value | 1.06291 (rank : 15) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 737 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q02447, Q8TD56, Q9BQR1 | Gene names | SP3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Sp3 (SPR-2). | |||||
|
SP2_HUMAN
|
||||||
| NC score | 0.043493 (rank : 24) | θ value | 0.813845 (rank : 14) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q02086 | Gene names | SP2, KIAA0048 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Sp2. | |||||
|
PAR10_HUMAN
|
||||||
| NC score | 0.042717 (rank : 25) | θ value | 4.03905 (rank : 30) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q53GL7, Q8N2I0, Q8WV05, Q96CH7 | Gene names | PARP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly [ADP-ribose] polymerase 10 (EC 2.4.2.30) (PARP-10). | |||||
|
SP1_HUMAN
|
||||||
| NC score | 0.042572 (rank : 26) | θ value | 2.36792 (rank : 25) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P08047, Q86TN8, Q9H3Q5, Q9NR51, Q9NY21, Q9NYE7 | Gene names | SP1, TSFP1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Sp1. | |||||
|
NCOA6_MOUSE
|
||||||
| NC score | 0.041618 (rank : 27) | θ value | 2.36792 (rank : 24) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
SP1_MOUSE
|
||||||
| NC score | 0.040570 (rank : 28) | θ value | 6.88961 (rank : 38) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 730 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O89090, O89087, Q62251, Q64167 | Gene names | Sp1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Sp1. | |||||
|
PO6F2_HUMAN
|
||||||
| NC score | 0.033875 (rank : 29) | θ value | 6.88961 (rank : 36) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P78424, P78425, Q75ME8, Q86UM6, Q9UDS7 | Gene names | POU6F2, RPF1 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 6, transcription factor 2 (Retina-derived POU-domain factor 1) (RPF-1). | |||||
|
ZN687_MOUSE
|
||||||
| NC score | 0.033550 (rank : 30) | θ value | 1.38821 (rank : 20) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 913 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9D2D7, Q6PAP3, Q6ZPQ9 | Gene names | Znf687, Kiaa1441, Zfp687 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
RCL1_HUMAN
|
||||||
| NC score | 0.031512 (rank : 31) | θ value | 6.88961 (rank : 37) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y2P8, Q5VYW9, Q9H9D0, Q9NY00, Q9P044 | Gene names | RCL1, RNAC, RPC2, RTC2 | |||
|
Domain Architecture |

|
|||||
| Description | RNA 3'-terminal phosphate cyclase-like protein. | |||||
|
ZN335_HUMAN
|
||||||
| NC score | 0.028642 (rank : 32) | θ value | 0.47712 (rank : 9) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 973 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9H4Z2, Q9H684 | Gene names | ZNF335 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 335. | |||||
|
NFYC_HUMAN
|
||||||
| NC score | 0.027365 (rank : 33) | θ value | 6.88961 (rank : 35) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13952, Q5T6K8, Q5T6L1, Q5TZR6, Q92869, Q9HBX1, Q9NXB5, Q9UM67, Q9UML0, Q9UMT7 | Gene names | NFYC | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear transcription factor Y subunit gamma (Nuclear transcription factor Y subunit C) (NF-YC) (CAAT-box DNA-binding protein subunit C) (Transactivator HSM-1/2). | |||||
|
BPTF_HUMAN
|
||||||
| NC score | 0.027365 (rank : 34) | θ value | 5.27518 (rank : 32) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
ZNF76_HUMAN
|
||||||
| NC score | 0.027157 (rank : 35) | θ value | 0.365318 (rank : 7) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 728 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P36508, Q9BQB2 | Gene names | ZNF76 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 76. | |||||
|
ZN236_HUMAN
|
||||||
| NC score | 0.026140 (rank : 36) | θ value | 0.62314 (rank : 13) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 844 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UL36, Q9UL37 | Gene names | ZNF236 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 236. | |||||
|
ZN628_MOUSE
|
||||||
| NC score | 0.025284 (rank : 37) | θ value | 0.47712 (rank : 10) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 815 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8CJ78, Q3U2L5, Q6P5B3 | Gene names | Znf628, Zec, Zfp628 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 628 (Zinc finger protein expressed in embryonal cells and certain adult organs). | |||||
|
POGZ_MOUSE
|
||||||
| NC score | 0.024920 (rank : 38) | θ value | 5.27518 (rank : 33) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BZH4, Q4VA94, Q80TZ8, Q8C0K1, Q8K294 | Gene names | Pogz, Kiaa0461 | |||
|
Domain Architecture |

|
|||||
| Description | Pogo transposable element with ZNF domain. | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.024841 (rank : 39) | θ value | 4.03905 (rank : 31) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
KIRR1_HUMAN
|
||||||
| NC score | 0.023458 (rank : 40) | θ value | 4.03905 (rank : 29) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96J84, Q5XKC6, Q7Z696, Q7Z7N8, Q8TB15, Q9H9N1, Q9NVA5 | Gene names | KIRREL, NEPH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kin of IRRE-like protein 1 precursor (Kin of irregular chiasm-like protein 1) (Nephrin-like protein 1). | |||||
|
ZN628_HUMAN
|
||||||
| NC score | 0.022778 (rank : 41) | θ value | 2.36792 (rank : 26) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 814 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5EBL2, Q86X34 | Gene names | ZNF628 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 628. | |||||
|
KIRR1_MOUSE
|
||||||
| NC score | 0.020890 (rank : 42) | θ value | 6.88961 (rank : 34) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80W68, Q8CIJ4, Q8CJ59, Q923L4 | Gene names | Kirrel1, Kirrel, Neph1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kin of IRRE-like protein 1 precursor (Kin of irregular chiasm-like protein 1) (Nephrin-like protein 1). | |||||
|
SRRM1_HUMAN
|
||||||
| NC score | 0.020602 (rank : 43) | θ value | 6.88961 (rank : 39) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
TNIK_HUMAN
|
||||||
| NC score | 0.001024 (rank : 44) | θ value | 8.99809 (rank : 42) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 1570 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UKE5, O60298, Q8WUY7, Q9UKD8, Q9UKD9, Q9UKE0, Q9UKE1, Q9UKE2, Q9UKE3, Q9UKE4 | Gene names | TNIK, KIAA0551 | |||
|
Domain Architecture |

|
|||||
| Description | TRAF2 and NCK-interacting protein kinase (EC 2.7.11.1). | |||||