Please be patient as the page loads
|
LAT_MOUSE
|
||||||
| SwissProt Accessions | O54957 | Gene names | Lat | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Linker for activation of T-cells family member 1 (36 kDa phospho- tyrosine adapter protein) (pp36) (p36-38). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
LAT_MOUSE
|
||||||
| θ value | 2.30293e-96 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O54957 | Gene names | Lat | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Linker for activation of T-cells family member 1 (36 kDa phospho- tyrosine adapter protein) (pp36) (p36-38). | |||||
|
LAT_HUMAN
|
||||||
| θ value | 9.78833e-55 (rank : 2) | NC score | 0.959750 (rank : 2) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43561, O43919 | Gene names | LAT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Linker for activation of T-cells family member 1 (36 kDa phospho- tyrosine adapter protein) (pp36) (p36-38). | |||||
|
TP53B_MOUSE
|
||||||
| θ value | 0.279714 (rank : 3) | NC score | 0.059040 (rank : 7) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 0.365318 (rank : 4) | NC score | 0.065520 (rank : 6) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
FMN2_MOUSE
|
||||||
| θ value | 0.62314 (rank : 5) | NC score | 0.046323 (rank : 8) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JL04 | Gene names | Fmn2 | |||
|
Domain Architecture |

|
|||||
| Description | Formin-2. | |||||
|
CO039_HUMAN
|
||||||
| θ value | 1.06291 (rank : 6) | NC score | 0.076197 (rank : 3) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6ZRI6, Q71JB1, Q7L3S0, Q8N3F2, Q96FB6, Q9NTU5 | Gene names | C15orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf39. | |||||
|
NCOA6_MOUSE
|
||||||
| θ value | 1.38821 (rank : 7) | NC score | 0.068586 (rank : 5) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
CO039_MOUSE
|
||||||
| θ value | 1.81305 (rank : 8) | NC score | 0.070834 (rank : 4) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q3TEI4, Q3TMF2, Q3U253, Q3UM75, Q9DA93 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf39 homolog. | |||||
|
ELK4_MOUSE
|
||||||
| θ value | 2.36792 (rank : 9) | NC score | 0.030979 (rank : 14) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P41158 | Gene names | Elk4, Sap1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-4 (Serum response factor accessory protein 1) (SAP-1). | |||||
|
ELK4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 10) | NC score | 0.035784 (rank : 11) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P28324, P28323 | Gene names | ELK4, SAP1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-4 (Serum response factor accessory protein 1) (SAP-1). | |||||
|
NFAC2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 11) | NC score | 0.038448 (rank : 9) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13469, Q13468, Q5TFW7, Q5TFW8, Q9NPX6, Q9NQH3, Q9UJR2 | Gene names | NFATC2, NFAT1, NFATP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 2 (T cell transcription factor NFAT1) (NFAT pre-existing subunit) (NF-ATp). | |||||
|
PGFRA_MOUSE
|
||||||
| θ value | 3.0926 (rank : 12) | NC score | 0.010219 (rank : 27) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P26618, Q62046 | Gene names | Pdgfra | |||
|
Domain Architecture |

|
|||||
| Description | Alpha platelet-derived growth factor receptor precursor (EC 2.7.10.1) (PDGF-R-alpha) (CD140a antigen). | |||||
|
B4GN4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 13) | NC score | 0.024676 (rank : 18) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q76KP1, Q96LV2 | Gene names | B4GALNT4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 1 (EC 2.4.1.244) (NGalNAc-T1) (Beta- 1,4-N-acetylgalactosaminyltransferase IV) (Beta4GalNAc-T4) (Beta4GalNAcT4). | |||||
|
NFAC2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 14) | NC score | 0.035443 (rank : 12) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q60591, Q60984, Q60985 | Gene names | Nfatc2, Nfat1, Nfatp | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 2 (T cell transcription factor NFAT1) (NFAT pre-existing subunit) (NF-ATp). | |||||
|
PGFRA_HUMAN
|
||||||
| θ value | 4.03905 (rank : 15) | NC score | 0.009820 (rank : 28) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 980 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P16234, Q96KZ7 | Gene names | PDGFRA | |||
|
Domain Architecture |

|
|||||
| Description | Alpha platelet-derived growth factor receptor precursor (EC 2.7.10.1) (PDGF-R-alpha) (CD140a antigen). | |||||
|
RBM12_MOUSE
|
||||||
| θ value | 4.03905 (rank : 16) | NC score | 0.031005 (rank : 13) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8R4X3, Q8K373, Q8R302, Q9CS80 | Gene names | Rbm12 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 12 (RNA-binding motif protein 12) (SH3/WW domain anchor protein in the nucleus) (SWAN). | |||||
|
TMPSD_MOUSE
|
||||||
| θ value | 4.03905 (rank : 17) | NC score | 0.007317 (rank : 31) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5U405, Q8CFE0, Q91VQ8 | Gene names | Tmprss13, Msp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 13 (EC 3.4.21.-) (Mosaic serine protease) (Membrane-type mosaic serine protease). | |||||
|
ANX11_HUMAN
|
||||||
| θ value | 5.27518 (rank : 18) | NC score | 0.013685 (rank : 24) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P50995 | Gene names | ANXA11, ANX11 | |||
|
Domain Architecture |
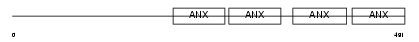
|
|||||
| Description | Annexin A11 (Annexin XI) (Calcyclin-associated annexin 50) (CAP-50) (56 kDa autoantigen). | |||||
|
BI2L1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 19) | NC score | 0.022259 (rank : 21) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9DBJ3 | Gene names | Baiap2l1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain-specific angiogenesis inhibitor 1-associated protein 2-like protein 1 (BAI1-associated protein 2-like protein 1). | |||||
|
PKHA6_MOUSE
|
||||||
| θ value | 5.27518 (rank : 20) | NC score | 0.026279 (rank : 16) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7TQG1, Q8K0J5 | Gene names | Plekha6, Pepp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
WNK2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 21) | NC score | 0.007963 (rank : 29) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
FMN2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 22) | NC score | 0.036263 (rank : 10) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 484 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NZ56, Q59GF6, Q5VU37, Q9NZ55 | Gene names | FMN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Formin-2. | |||||
|
HELZ_HUMAN
|
||||||
| θ value | 6.88961 (rank : 23) | NC score | 0.025161 (rank : 17) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P42694 | Gene names | HELZ, KIAA0054 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable helicase with zinc-finger domain (EC 3.6.1.-). | |||||
|
LIMD1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 24) | NC score | 0.011700 (rank : 26) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UGP4, Q9BQQ9, Q9NQ47 | Gene names | LIMD1 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain-containing protein 1. | |||||
|
MMP10_MOUSE
|
||||||
| θ value | 6.88961 (rank : 25) | NC score | 0.007575 (rank : 30) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O55123 | Gene names | Mmp10 | |||
|
Domain Architecture |
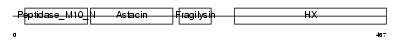
|
|||||
| Description | Stromelysin-2 precursor (EC 3.4.24.22) (Matrix metalloproteinase-10) (MMP-10) (Transin-2) (SL-2). | |||||
|
PELP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 26) | NC score | 0.030642 (rank : 15) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
BAZ2A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 27) | NC score | 0.016678 (rank : 23) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q91YE5 | Gene names | Baz2a, Tip5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5). | |||||
|
CBP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 28) | NC score | 0.023627 (rank : 19) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
MAP1B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 29) | NC score | 0.013344 (rank : 25) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
PKHA6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 30) | NC score | 0.023549 (rank : 20) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y2H5 | Gene names | PLEKHA6, KIAA0969, PEPP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
TAF4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 31) | NC score | 0.022150 (rank : 22) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
LAT_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 2.30293e-96 (rank : 1) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O54957 | Gene names | Lat | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Linker for activation of T-cells family member 1 (36 kDa phospho- tyrosine adapter protein) (pp36) (p36-38). | |||||
|
LAT_HUMAN
|
||||||
| NC score | 0.959750 (rank : 2) | θ value | 9.78833e-55 (rank : 2) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43561, O43919 | Gene names | LAT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Linker for activation of T-cells family member 1 (36 kDa phospho- tyrosine adapter protein) (pp36) (p36-38). | |||||
|
CO039_HUMAN
|
||||||
| NC score | 0.076197 (rank : 3) | θ value | 1.06291 (rank : 6) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6ZRI6, Q71JB1, Q7L3S0, Q8N3F2, Q96FB6, Q9NTU5 | Gene names | C15orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf39. | |||||
|
CO039_MOUSE
|
||||||
| NC score | 0.070834 (rank : 4) | θ value | 1.81305 (rank : 8) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q3TEI4, Q3TMF2, Q3U253, Q3UM75, Q9DA93 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf39 homolog. | |||||
|
NCOA6_MOUSE
|
||||||
| NC score | 0.068586 (rank : 5) | θ value | 1.38821 (rank : 7) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.065520 (rank : 6) | θ value | 0.365318 (rank : 4) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
TP53B_MOUSE
|
||||||
| NC score | 0.059040 (rank : 7) | θ value | 0.279714 (rank : 3) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
FMN2_MOUSE
|
||||||
| NC score | 0.046323 (rank : 8) | θ value | 0.62314 (rank : 5) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JL04 | Gene names | Fmn2 | |||
|
Domain Architecture |

|
|||||
| Description | Formin-2. | |||||
|
NFAC2_HUMAN
|
||||||
| NC score | 0.038448 (rank : 9) | θ value | 3.0926 (rank : 11) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13469, Q13468, Q5TFW7, Q5TFW8, Q9NPX6, Q9NQH3, Q9UJR2 | Gene names | NFATC2, NFAT1, NFATP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 2 (T cell transcription factor NFAT1) (NFAT pre-existing subunit) (NF-ATp). | |||||
|
FMN2_HUMAN
|
||||||
| NC score | 0.036263 (rank : 10) | θ value | 6.88961 (rank : 22) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 484 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NZ56, Q59GF6, Q5VU37, Q9NZ55 | Gene names | FMN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Formin-2. | |||||
|
ELK4_HUMAN
|
||||||
| NC score | 0.035784 (rank : 11) | θ value | 3.0926 (rank : 10) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P28324, P28323 | Gene names | ELK4, SAP1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-4 (Serum response factor accessory protein 1) (SAP-1). | |||||
|
NFAC2_MOUSE
|
||||||
| NC score | 0.035443 (rank : 12) | θ value | 4.03905 (rank : 14) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q60591, Q60984, Q60985 | Gene names | Nfatc2, Nfat1, Nfatp | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 2 (T cell transcription factor NFAT1) (NFAT pre-existing subunit) (NF-ATp). | |||||
|
RBM12_MOUSE
|
||||||
| NC score | 0.031005 (rank : 13) | θ value | 4.03905 (rank : 16) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8R4X3, Q8K373, Q8R302, Q9CS80 | Gene names | Rbm12 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 12 (RNA-binding motif protein 12) (SH3/WW domain anchor protein in the nucleus) (SWAN). | |||||
|
ELK4_MOUSE
|
||||||
| NC score | 0.030979 (rank : 14) | θ value | 2.36792 (rank : 9) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P41158 | Gene names | Elk4, Sap1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-4 (Serum response factor accessory protein 1) (SAP-1). | |||||
|
PELP1_MOUSE
|
||||||
| NC score | 0.030642 (rank : 15) | θ value | 6.88961 (rank : 26) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
PKHA6_MOUSE
|
||||||
| NC score | 0.026279 (rank : 16) | θ value | 5.27518 (rank : 20) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7TQG1, Q8K0J5 | Gene names | Plekha6, Pepp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
HELZ_HUMAN
|
||||||
| NC score | 0.025161 (rank : 17) | θ value | 6.88961 (rank : 23) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P42694 | Gene names | HELZ, KIAA0054 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable helicase with zinc-finger domain (EC 3.6.1.-). | |||||
|
B4GN4_HUMAN
|
||||||
| NC score | 0.024676 (rank : 18) | θ value | 4.03905 (rank : 13) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q76KP1, Q96LV2 | Gene names | B4GALNT4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 1 (EC 2.4.1.244) (NGalNAc-T1) (Beta- 1,4-N-acetylgalactosaminyltransferase IV) (Beta4GalNAc-T4) (Beta4GalNAcT4). | |||||
|
CBP_MOUSE
|
||||||
| NC score | 0.023627 (rank : 19) | θ value | 8.99809 (rank : 28) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
PKHA6_HUMAN
|
||||||
| NC score | 0.023549 (rank : 20) | θ value | 8.99809 (rank : 30) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y2H5 | Gene names | PLEKHA6, KIAA0969, PEPP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
BI2L1_MOUSE
|
||||||
| NC score | 0.022259 (rank : 21) | θ value | 5.27518 (rank : 19) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9DBJ3 | Gene names | Baiap2l1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain-specific angiogenesis inhibitor 1-associated protein 2-like protein 1 (BAI1-associated protein 2-like protein 1). | |||||
|
TAF4_HUMAN
|
||||||
| NC score | 0.022150 (rank : 22) | θ value | 8.99809 (rank : 31) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
BAZ2A_MOUSE
|
||||||
| NC score | 0.016678 (rank : 23) | θ value | 8.99809 (rank : 27) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q91YE5 | Gene names | Baz2a, Tip5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5). | |||||
|
ANX11_HUMAN
|
||||||
| NC score | 0.013685 (rank : 24) | θ value | 5.27518 (rank : 18) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P50995 | Gene names | ANXA11, ANX11 | |||
|
Domain Architecture |

|
|||||
| Description | Annexin A11 (Annexin XI) (Calcyclin-associated annexin 50) (CAP-50) (56 kDa autoantigen). | |||||
|
MAP1B_HUMAN
|
||||||
| NC score | 0.013344 (rank : 25) | θ value | 8.99809 (rank : 29) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
LIMD1_HUMAN
|
||||||
| NC score | 0.011700 (rank : 26) | θ value | 6.88961 (rank : 24) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UGP4, Q9BQQ9, Q9NQ47 | Gene names | LIMD1 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain-containing protein 1. | |||||
|
PGFRA_MOUSE
|
||||||
| NC score | 0.010219 (rank : 27) | θ value | 3.0926 (rank : 12) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P26618, Q62046 | Gene names | Pdgfra | |||
|
Domain Architecture |

|
|||||
| Description | Alpha platelet-derived growth factor receptor precursor (EC 2.7.10.1) (PDGF-R-alpha) (CD140a antigen). | |||||
|
PGFRA_HUMAN
|
||||||
| NC score | 0.009820 (rank : 28) | θ value | 4.03905 (rank : 15) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 980 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P16234, Q96KZ7 | Gene names | PDGFRA | |||
|
Domain Architecture |

|
|||||
| Description | Alpha platelet-derived growth factor receptor precursor (EC 2.7.10.1) (PDGF-R-alpha) (CD140a antigen). | |||||
|
WNK2_HUMAN
|
||||||
| NC score | 0.007963 (rank : 29) | θ value | 5.27518 (rank : 21) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
MMP10_MOUSE
|
||||||
| NC score | 0.007575 (rank : 30) | θ value | 6.88961 (rank : 25) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O55123 | Gene names | Mmp10 | |||
|
Domain Architecture |

|
|||||
| Description | Stromelysin-2 precursor (EC 3.4.24.22) (Matrix metalloproteinase-10) (MMP-10) (Transin-2) (SL-2). | |||||
|
TMPSD_MOUSE
|
||||||
| NC score | 0.007317 (rank : 31) | θ value | 4.03905 (rank : 17) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5U405, Q8CFE0, Q91VQ8 | Gene names | Tmprss13, Msp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 13 (EC 3.4.21.-) (Mosaic serine protease) (Membrane-type mosaic serine protease). | |||||