Please be patient as the page loads
|
GCST_HUMAN
|
||||||
| SwissProt Accessions | P48728 | Gene names | AMT, GCST | |||
|
Domain Architecture |
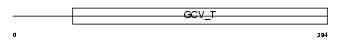
|
|||||
| Description | Aminomethyltransferase, mitochondrial precursor (EC 2.1.2.10) (Glycine cleavage system T protein) (GCVT). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
GCST_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P48728 | Gene names | AMT, GCST | |||
|
Domain Architecture |

|
|||||
| Description | Aminomethyltransferase, mitochondrial precursor (EC 2.1.2.10) (Glycine cleavage system T protein) (GCVT). | |||||
|
GCST_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.998151 (rank : 2) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8CFA2 | Gene names | Amt | |||
|
Domain Architecture |
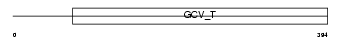
|
|||||
| Description | Aminomethyltransferase, mitochondrial precursor (EC 2.1.2.10) (Glycine cleavage system T protein) (GCVT). | |||||
|
M2GD_HUMAN
|
||||||
| θ value | 9.56915e-18 (rank : 3) | NC score | 0.586714 (rank : 3) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UI17 | Gene names | DMGDH | |||
|
Domain Architecture |

|
|||||
| Description | Dimethylglycine dehydrogenase, mitochondrial precursor (EC 1.5.99.2) (ME2GLYDH). | |||||
|
M2GD_MOUSE
|
||||||
| θ value | 2.60593e-15 (rank : 4) | NC score | 0.576322 (rank : 4) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9DBT9, Q8R1S7 | Gene names | Dmgdh | |||
|
Domain Architecture |

|
|||||
| Description | Dimethylglycine dehydrogenase, mitochondrial precursor (EC 1.5.99.2) (ME2GLYDH). | |||||
|
SARDH_HUMAN
|
||||||
| θ value | 4.59992e-12 (rank : 5) | NC score | 0.525442 (rank : 6) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UL12, Q9Y280, Q9Y2Y3 | Gene names | SARDH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcosine dehydrogenase, mitochondrial precursor (EC 1.5.99.1) (SarDH) (BPR-2). | |||||
|
SARDH_MOUSE
|
||||||
| θ value | 1.02475e-11 (rank : 6) | NC score | 0.527112 (rank : 5) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99LB7 | Gene names | Sardh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcosine dehydrogenase, mitochondrial precursor (EC 1.5.99.1) (SarDH). | |||||
|
CP250_MOUSE
|
||||||
| θ value | 2.36792 (rank : 7) | NC score | 0.006684 (rank : 10) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
GPDM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 8) | NC score | 0.058638 (rank : 9) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q64521, Q61507 | Gene names | Gpd2, Gdm1 | |||
|
Domain Architecture |

|
|||||
| Description | Glycerol-3-phosphate dehydrogenase, mitochondrial precursor (EC 1.1.99.5) (GPD-M) (GPDH-M). | |||||
|
SOX_HUMAN
|
||||||
| θ value | θ > 10 (rank : 9) | NC score | 0.099246 (rank : 7) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9P0Z9, Q96H28, Q9C070 | Gene names | PIPOX, LPIPOX, PSO | |||
|
Domain Architecture |
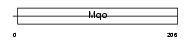
|
|||||
| Description | Peroxisomal sarcosine oxidase (EC 1.5.3.1) (PSO) (L-pipecolate oxidase) (EC 1.5.3.7) (L-pipecolic acid oxidase). | |||||
|
SOX_MOUSE
|
||||||
| θ value | θ > 10 (rank : 10) | NC score | 0.065533 (rank : 8) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D826, O55223 | Gene names | Pipox, Pso | |||
|
Domain Architecture |
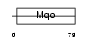
|
|||||
| Description | Peroxisomal sarcosine oxidase (EC 1.5.3.1) (PSO) (L-pipecolate oxidase) (EC 1.5.3.7) (L-pipecolic acid oxidase). | |||||
|
GCST_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P48728 | Gene names | AMT, GCST | |||
|
Domain Architecture |

|
|||||
| Description | Aminomethyltransferase, mitochondrial precursor (EC 2.1.2.10) (Glycine cleavage system T protein) (GCVT). | |||||
|
GCST_MOUSE
|
||||||
| NC score | 0.998151 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8CFA2 | Gene names | Amt | |||
|
Domain Architecture |

|
|||||
| Description | Aminomethyltransferase, mitochondrial precursor (EC 2.1.2.10) (Glycine cleavage system T protein) (GCVT). | |||||
|
M2GD_HUMAN
|
||||||
| NC score | 0.586714 (rank : 3) | θ value | 9.56915e-18 (rank : 3) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UI17 | Gene names | DMGDH | |||
|
Domain Architecture |

|
|||||
| Description | Dimethylglycine dehydrogenase, mitochondrial precursor (EC 1.5.99.2) (ME2GLYDH). | |||||
|
M2GD_MOUSE
|
||||||
| NC score | 0.576322 (rank : 4) | θ value | 2.60593e-15 (rank : 4) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9DBT9, Q8R1S7 | Gene names | Dmgdh | |||
|
Domain Architecture |

|
|||||
| Description | Dimethylglycine dehydrogenase, mitochondrial precursor (EC 1.5.99.2) (ME2GLYDH). | |||||
|
SARDH_MOUSE
|
||||||
| NC score | 0.527112 (rank : 5) | θ value | 1.02475e-11 (rank : 6) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99LB7 | Gene names | Sardh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcosine dehydrogenase, mitochondrial precursor (EC 1.5.99.1) (SarDH). | |||||
|
SARDH_HUMAN
|
||||||
| NC score | 0.525442 (rank : 6) | θ value | 4.59992e-12 (rank : 5) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UL12, Q9Y280, Q9Y2Y3 | Gene names | SARDH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcosine dehydrogenase, mitochondrial precursor (EC 1.5.99.1) (SarDH) (BPR-2). | |||||
|
SOX_HUMAN
|
||||||
| NC score | 0.099246 (rank : 7) | θ value | θ > 10 (rank : 9) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9P0Z9, Q96H28, Q9C070 | Gene names | PIPOX, LPIPOX, PSO | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal sarcosine oxidase (EC 1.5.3.1) (PSO) (L-pipecolate oxidase) (EC 1.5.3.7) (L-pipecolic acid oxidase). | |||||
|
SOX_MOUSE
|
||||||
| NC score | 0.065533 (rank : 8) | θ value | θ > 10 (rank : 10) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D826, O55223 | Gene names | Pipox, Pso | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal sarcosine oxidase (EC 1.5.3.1) (PSO) (L-pipecolate oxidase) (EC 1.5.3.7) (L-pipecolic acid oxidase). | |||||
|
GPDM_MOUSE
|
||||||
| NC score | 0.058638 (rank : 9) | θ value | θ > 10 (rank : 8) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q64521, Q61507 | Gene names | Gpd2, Gdm1 | |||
|
Domain Architecture |

|
|||||
| Description | Glycerol-3-phosphate dehydrogenase, mitochondrial precursor (EC 1.1.99.5) (GPD-M) (GPDH-M). | |||||
|
CP250_MOUSE
|
||||||
| NC score | 0.006684 (rank : 10) | θ value | 2.36792 (rank : 7) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||