Please be patient as the page loads
|
FANCA_HUMAN
|
||||||
| SwissProt Accessions | O15360, O75266, Q6PL10, Q92497, Q96H18, Q9UEA5, Q9UEL8, Q9UEL9, Q9UPK3, Q9Y6M2 | Gene names | FANCA, FAA | |||
|
Domain Architecture |

|
|||||
| Description | Fanconi anemia group A protein (Protein FACA). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
FANCA_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O15360, O75266, Q6PL10, Q92497, Q96H18, Q9UEA5, Q9UEL8, Q9UEL9, Q9UPK3, Q9Y6M2 | Gene names | FANCA, FAA | |||
|
Domain Architecture |

|
|||||
| Description | Fanconi anemia group A protein (Protein FACA). | |||||
|
FANCA_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.984257 (rank : 2) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JL70, Q9ESU9, Q9ESV3, Q9ET44, Q9JL64, Q9JL65, Q9JL66, Q9JL67, Q9JL68, Q9JL69 | Gene names | Fanca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group A protein homolog (Protein FACA). | |||||
|
NU160_HUMAN
|
||||||
| θ value | 1.81305 (rank : 3) | NC score | 0.030309 (rank : 3) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q12769, Q96GB3, Q9H660 | Gene names | NUP160, KIAA0197, NUP120 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear pore complex protein Nup160 (Nucleoporin Nup160) (160 kDa nucleoporin). | |||||
|
SPTB2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 4) | NC score | 0.013433 (rank : 8) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q01082, O60837, Q16057 | Gene names | SPTBN1, SPTB2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
SPTB2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 5) | NC score | 0.013372 (rank : 9) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q62261, Q5SQL8 | Gene names | Sptbn1, Spnb-2, Spnb2, Sptb2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
PTPRK_MOUSE
|
||||||
| θ value | 3.0926 (rank : 6) | NC score | 0.007386 (rank : 13) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P35822 | Gene names | Ptprk, Ptpk | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase kappa precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase kappa) (R-PTP-kappa). | |||||
|
SSFA2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 7) | NC score | 0.030243 (rank : 4) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q922B9, Q8VEF7 | Gene names | Ssfa2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-specific antigen 2. | |||||
|
MIB2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 8) | NC score | 0.006625 (rank : 15) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96AX9, Q7Z437, Q8IY62, Q8N897, Q8N8R2, Q8N911, Q8NB36, Q8NCY1, Q8NG59, Q8NG60, Q8NG61, Q8NI59, Q8WYN1 | Gene names | MIB2, SKD, ZZANK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein MIB2 (EC 6.3.2.-) (Mind bomb homolog 2) (Zinc finger ZZ type with ankyrin repeat domain protein 1) (Skeletrophin) (Novelzin) (Novel zinc finger protein) (Putative NF-kappa-B-activating protein 002N). | |||||
|
COG8_HUMAN
|
||||||
| θ value | 6.88961 (rank : 9) | NC score | 0.017859 (rank : 7) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96MW5, Q8WVV6, Q9H6F8 | Gene names | COG8 | |||
|
Domain Architecture |
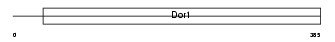
|
|||||
| Description | Conserved oligomeric Golgi complex component 8. | |||||
|
FGD3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 10) | NC score | 0.006959 (rank : 14) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O88842, Q8BQ72 | Gene names | Fgd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 3. | |||||
|
PDP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 11) | NC score | 0.020642 (rank : 5) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9P0J1, Q5U5K1 | Gene names | PDP1, PDP, PPM2C | |||
|
Domain Architecture |

|
|||||
| Description | [Pyruvate dehydrogenase [lipoamide]]-phosphatase 1, mitochondrial precursor (EC 3.1.3.43) (PDP 1) (Pyruvate dehydrogenase phosphatase, catalytic subunit 1) (PDPC 1) (Protein phosphatase 2C). | |||||
|
PDP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 12) | NC score | 0.020574 (rank : 6) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3UV70 | Gene names | Pdp1, Ppm2c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | [Pyruvate dehydrogenase [lipoamide]]-phosphatase 1, mitochondrial precursor (EC 3.1.3.43) (PDP 1) (Pyruvate dehydrogenase phosphatase, catalytic subunit 1) (PDPC 1) (Protein phosphatase 2C). | |||||
|
TCF8_HUMAN
|
||||||
| θ value | 6.88961 (rank : 13) | NC score | 0.001989 (rank : 17) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 1057 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P37275, Q12924, Q13800 | Gene names | TCF8, AREB6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 8 (NIL-2-A zinc finger protein) (Negative regulator of IL2). | |||||
|
AKA12_HUMAN
|
||||||
| θ value | 8.99809 (rank : 14) | NC score | 0.009458 (rank : 11) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
CE152_HUMAN
|
||||||
| θ value | 8.99809 (rank : 15) | NC score | 0.004408 (rank : 16) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 976 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O94986, Q6NTA0 | Gene names | CEP152, KIAA0912 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 152 kDa (Cep152 protein). | |||||
|
GLSL_HUMAN
|
||||||
| θ value | 8.99809 (rank : 16) | NC score | 0.009514 (rank : 10) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UI32, Q9NYY2, Q9UI31 | Gene names | GLS2, GA | |||
|
Domain Architecture |

|
|||||
| Description | Glutaminase liver isoform, mitochondrial precursor (EC 3.5.1.2) (GLS) (L-glutamine amidohydrolase) (L-glutaminase). | |||||
|
ITIH2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 17) | NC score | 0.008637 (rank : 12) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P19823, Q14659, Q15484 | Gene names | ITIH2, IGHEP2 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H2 precursor (ITI heavy chain H2) (Inter-alpha-inhibitor heavy chain 2) (Inter-alpha-trypsin inhibitor complex component II) (Serum-derived hyaluronan-associated protein) (SHAP). | |||||
|
O13C4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 18) | NC score | 0.000169 (rank : 18) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 721 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NGS5, Q96R41 | Gene names | OR13C4 | |||
|
Domain Architecture |

|
|||||
| Description | Olfactory receptor 13C4. | |||||
|
FANCA_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O15360, O75266, Q6PL10, Q92497, Q96H18, Q9UEA5, Q9UEL8, Q9UEL9, Q9UPK3, Q9Y6M2 | Gene names | FANCA, FAA | |||
|
Domain Architecture |

|
|||||
| Description | Fanconi anemia group A protein (Protein FACA). | |||||
|
FANCA_MOUSE
|
||||||
| NC score | 0.984257 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JL70, Q9ESU9, Q9ESV3, Q9ET44, Q9JL64, Q9JL65, Q9JL66, Q9JL67, Q9JL68, Q9JL69 | Gene names | Fanca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group A protein homolog (Protein FACA). | |||||
|
NU160_HUMAN
|
||||||
| NC score | 0.030309 (rank : 3) | θ value | 1.81305 (rank : 3) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q12769, Q96GB3, Q9H660 | Gene names | NUP160, KIAA0197, NUP120 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear pore complex protein Nup160 (Nucleoporin Nup160) (160 kDa nucleoporin). | |||||
|
SSFA2_MOUSE
|
||||||
| NC score | 0.030243 (rank : 4) | θ value | 4.03905 (rank : 7) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q922B9, Q8VEF7 | Gene names | Ssfa2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-specific antigen 2. | |||||
|
PDP1_HUMAN
|
||||||
| NC score | 0.020642 (rank : 5) | θ value | 6.88961 (rank : 11) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9P0J1, Q5U5K1 | Gene names | PDP1, PDP, PPM2C | |||
|
Domain Architecture |

|
|||||
| Description | [Pyruvate dehydrogenase [lipoamide]]-phosphatase 1, mitochondrial precursor (EC 3.1.3.43) (PDP 1) (Pyruvate dehydrogenase phosphatase, catalytic subunit 1) (PDPC 1) (Protein phosphatase 2C). | |||||
|
PDP1_MOUSE
|
||||||
| NC score | 0.020574 (rank : 6) | θ value | 6.88961 (rank : 12) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3UV70 | Gene names | Pdp1, Ppm2c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | [Pyruvate dehydrogenase [lipoamide]]-phosphatase 1, mitochondrial precursor (EC 3.1.3.43) (PDP 1) (Pyruvate dehydrogenase phosphatase, catalytic subunit 1) (PDPC 1) (Protein phosphatase 2C). | |||||
|
COG8_HUMAN
|
||||||
| NC score | 0.017859 (rank : 7) | θ value | 6.88961 (rank : 9) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96MW5, Q8WVV6, Q9H6F8 | Gene names | COG8 | |||
|
Domain Architecture |

|
|||||
| Description | Conserved oligomeric Golgi complex component 8. | |||||
|
SPTB2_HUMAN
|
||||||
| NC score | 0.013433 (rank : 8) | θ value | 1.81305 (rank : 4) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q01082, O60837, Q16057 | Gene names | SPTBN1, SPTB2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
SPTB2_MOUSE
|
||||||
| NC score | 0.013372 (rank : 9) | θ value | 1.81305 (rank : 5) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q62261, Q5SQL8 | Gene names | Sptbn1, Spnb-2, Spnb2, Sptb2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
GLSL_HUMAN
|
||||||
| NC score | 0.009514 (rank : 10) | θ value | 8.99809 (rank : 16) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UI32, Q9NYY2, Q9UI31 | Gene names | GLS2, GA | |||
|
Domain Architecture |

|
|||||
| Description | Glutaminase liver isoform, mitochondrial precursor (EC 3.5.1.2) (GLS) (L-glutamine amidohydrolase) (L-glutaminase). | |||||
|
AKA12_HUMAN
|
||||||
| NC score | 0.009458 (rank : 11) | θ value | 8.99809 (rank : 14) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
ITIH2_HUMAN
|
||||||
| NC score | 0.008637 (rank : 12) | θ value | 8.99809 (rank : 17) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P19823, Q14659, Q15484 | Gene names | ITIH2, IGHEP2 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H2 precursor (ITI heavy chain H2) (Inter-alpha-inhibitor heavy chain 2) (Inter-alpha-trypsin inhibitor complex component II) (Serum-derived hyaluronan-associated protein) (SHAP). | |||||
|
PTPRK_MOUSE
|
||||||
| NC score | 0.007386 (rank : 13) | θ value | 3.0926 (rank : 6) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P35822 | Gene names | Ptprk, Ptpk | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase kappa precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase kappa) (R-PTP-kappa). | |||||
|
FGD3_MOUSE
|
||||||
| NC score | 0.006959 (rank : 14) | θ value | 6.88961 (rank : 10) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O88842, Q8BQ72 | Gene names | Fgd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 3. | |||||
|
MIB2_HUMAN
|
||||||
| NC score | 0.006625 (rank : 15) | θ value | 5.27518 (rank : 8) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96AX9, Q7Z437, Q8IY62, Q8N897, Q8N8R2, Q8N911, Q8NB36, Q8NCY1, Q8NG59, Q8NG60, Q8NG61, Q8NI59, Q8WYN1 | Gene names | MIB2, SKD, ZZANK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein MIB2 (EC 6.3.2.-) (Mind bomb homolog 2) (Zinc finger ZZ type with ankyrin repeat domain protein 1) (Skeletrophin) (Novelzin) (Novel zinc finger protein) (Putative NF-kappa-B-activating protein 002N). | |||||
|
CE152_HUMAN
|
||||||
| NC score | 0.004408 (rank : 16) | θ value | 8.99809 (rank : 15) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 976 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O94986, Q6NTA0 | Gene names | CEP152, KIAA0912 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 152 kDa (Cep152 protein). | |||||
|
TCF8_HUMAN
|
||||||
| NC score | 0.001989 (rank : 17) | θ value | 6.88961 (rank : 13) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 1057 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P37275, Q12924, Q13800 | Gene names | TCF8, AREB6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 8 (NIL-2-A zinc finger protein) (Negative regulator of IL2). | |||||
|
O13C4_HUMAN
|
||||||
| NC score | 0.000169 (rank : 18) | θ value | 8.99809 (rank : 18) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 721 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NGS5, Q96R41 | Gene names | OR13C4 | |||
|
Domain Architecture |

|
|||||
| Description | Olfactory receptor 13C4. | |||||