Please be patient as the page loads
|
CRIS2_HUMAN
|
||||||
| SwissProt Accessions | P16562 | Gene names | CRISP2, TPX1 | |||
|
Domain Architecture |
|
|||||
| Description | Cysteine-rich secretory protein 2 precursor (CRISP-2) (Testis-specific protein TPX-1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CRIS2_HUMAN
|
||||||
| θ value | 1.86219e-138 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P16562 | Gene names | CRISP2, TPX1 | |||
|
Domain Architecture |

|
|||||
| Description | Cysteine-rich secretory protein 2 precursor (CRISP-2) (Testis-specific protein TPX-1). | |||||
|
CRIS2_MOUSE
|
||||||
| θ value | 9.32813e-106 (rank : 2) | NC score | 0.994617 (rank : 3) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P16563 | Gene names | Crisp2, Tpx-1, Tpx1 | |||
|
Domain Architecture |
|
|||||
| Description | Cysteine-rich secretory protein 2 precursor (CRISP-2) (Testis-specific protein TPX-1). | |||||
|
CRIS3_HUMAN
|
||||||
| θ value | 9.035e-101 (rank : 3) | NC score | 0.995661 (rank : 2) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P54108, Q15512, Q5JW83, Q9H108 | Gene names | CRISP3 | |||
|
Domain Architecture |
|
|||||
| Description | Cysteine-rich secretory protein 3 precursor (CRISP-3) (SGP28 protein). | |||||
|
CRIS1_MOUSE
|
||||||
| θ value | 5.69867e-79 (rank : 4) | NC score | 0.987120 (rank : 4) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q03401 | Gene names | Crisp1, Aeg-1, Aeg1 | |||
|
Domain Architecture |
|
|||||
| Description | Cysteine-rich secretory protein 1 precursor (CRISP-1) (Acidic epididymal glycoprotein 1) (Sperm-coating glycoprotein 1) (SCP 1). | |||||
|
CRIS3_MOUSE
|
||||||
| θ value | 8.55514e-59 (rank : 5) | NC score | 0.976755 (rank : 5) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q03402 | Gene names | Crisp3, Aeg-2, Aeg2 | |||
|
Domain Architecture |
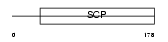
|
|||||
| Description | Cysteine-rich secretory protein 3 precursor (CRISP-3) (Acidic epididymal glycoprotein 2) (Sperm-coating glycoprotein 2) (SCP 2). | |||||
|
CRIS1_HUMAN
|
||||||
| θ value | 6.77864e-56 (rank : 6) | NC score | 0.971471 (rank : 6) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P54107, O00698, Q13248, Q14082, Q96SF6 | Gene names | CRISP1, AEGL1 | |||
|
Domain Architecture |
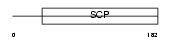
|
|||||
| Description | Cysteine-rich secretory protein 1 precursor (CRISP-1) (Acidic epididymal glycoprotein homolog) (AEG-like protein) (ARP). | |||||
|
GLIP1_MOUSE
|
||||||
| θ value | 1.33217e-27 (rank : 7) | NC score | 0.884150 (rank : 7) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9CWG1 | Gene names | Glipr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma pathogenesis-related protein 1 precursor (GliPR 1). | |||||
|
GLIP1_HUMAN
|
||||||
| θ value | 1.24688e-25 (rank : 8) | NC score | 0.866028 (rank : 8) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P48060, Q15409, Q969K2 | Gene names | GLIPR1, GLIPR, RTVP1 | |||
|
Domain Architecture |
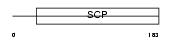
|
|||||
| Description | Glioma pathogenesis-related protein 1 precursor (GliPR 1) (RTVP-1 protein). | |||||
|
CRLD2_HUMAN
|
||||||
| θ value | 3.17815e-21 (rank : 9) | NC score | 0.739787 (rank : 9) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9H0B8, Q6P590, Q6UWH0, Q6UXC6, Q96IB1, Q96K61 | Gene names | CRISPLD2, CRISP11, LCRISP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich secretory protein LCCL domain-containing 2 precursor (LCCL domain-containing cysteine-rich secretory protein 2) (Cysteine- rich secretory protein 11) (CRISP-11). | |||||
|
CRLD1_HUMAN
|
||||||
| θ value | 4.15078e-21 (rank : 10) | NC score | 0.726114 (rank : 10) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9H336 | Gene names | CRISPLD1, CRISP10, LCRISP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich secretory protein LCCL domain-containing 1 precursor (LCCL domain-containing cysteine-rich secretory protein 1) (Cysteine- rich secretory protein 10) (CRISP-10) (Trypsin inhibitor Hl) (CocoaCrisp). | |||||
|
CRLD1_MOUSE
|
||||||
| θ value | 2.06002e-20 (rank : 11) | NC score | 0.719509 (rank : 11) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8CGD2, Q8K018, Q99MM6 | Gene names | Crispld1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich secretory protein LCCL domain-containing 1 precursor (CocoaCrisp). | |||||
|
GAPR1_MOUSE
|
||||||
| θ value | 9.59137e-10 (rank : 12) | NC score | 0.625408 (rank : 12) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9CYL5, Q3TJ42, Q8BTK4 | Gene names | Glipr2, Gapr1 | |||
|
Domain Architecture |
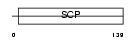
|
|||||
| Description | Golgi-associated plant pathogenesis-related protein 1 (Golgi- associated PR-1 protein) (GAPR-1) (Glioma pathogenesis-related protein 2) (GliPR 2). | |||||
|
GAPR1_HUMAN
|
||||||
| θ value | 1.25267e-09 (rank : 13) | NC score | 0.608405 (rank : 13) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9H4G4, Q5VZR1, Q8N2S6, Q8WWC9, Q8WX36 | Gene names | GLIPR2, C9orf19, GAPR1 | |||
|
Domain Architecture |
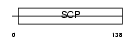
|
|||||
| Description | Golgi-associated plant pathogenesis-related protein 1 (Golgi- associated PR-1 protein) (GAPR-1) (Glioma pathogenesis-related protein 2) (GliPR 2). | |||||
|
MATN4_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 14) | NC score | 0.021364 (rank : 14) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O95460, Q5QPU2, Q5QPU3, Q5QPU4, Q8N2M5, Q8N2M7, Q9H1F8, Q9H1F9 | Gene names | MATN4 | |||
|
Domain Architecture |

|
|||||
| Description | Matrilin-4 precursor. | |||||
|
FBN1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 15) | NC score | 0.016835 (rank : 15) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P35555, Q15972, Q75N87 | Gene names | FBN1, FBN | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-1 precursor. | |||||
|
FBN1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 16) | NC score | 0.015996 (rank : 16) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 563 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61554, Q60826 | Gene names | Fbn1, Fbn-1 | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-1 precursor. | |||||
|
PROZ_HUMAN
|
||||||
| θ value | 1.81305 (rank : 17) | NC score | 0.015347 (rank : 17) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P22891, Q15213 | Gene names | PROZ | |||
|
Domain Architecture |
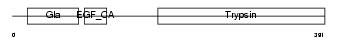
|
|||||
| Description | Vitamin K-dependent protein Z precursor. | |||||
|
ATS20_HUMAN
|
||||||
| θ value | 3.0926 (rank : 18) | NC score | 0.005700 (rank : 22) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P59510 | Gene names | ADAMTS20 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-20 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 20) (ADAM-TS 20) (ADAM-TS20). | |||||
|
LTBP4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 19) | NC score | 0.011956 (rank : 18) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8K4G1, Q8K4G0 | Gene names | Ltbp4 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 4 precursor (LTBP-4). | |||||
|
PROC_MOUSE
|
||||||
| θ value | 4.03905 (rank : 20) | NC score | 0.006834 (rank : 20) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P33587, O35498, Q91WN8, Q99PC6 | Gene names | Proc | |||
|
Domain Architecture |
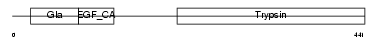
|
|||||
| Description | Vitamin K-dependent protein C precursor (EC 3.4.21.69) (Autoprothrombin IIA) (Anticoagulant protein C) (Blood coagulation factor XIV) [Contains: Vitamin K-dependent protein C light chain; Vitamin K-dependent protein C heavy chain; Activation peptide]. | |||||
|
GALT7_MOUSE
|
||||||
| θ value | 6.88961 (rank : 21) | NC score | 0.003768 (rank : 23) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80VA0, Q8BY62, Q8BZ70, Q8BZW0, Q91VW6, Q99MD7 | Gene names | Galnt7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetylgalactosaminyltransferase 7 (EC 2.4.1.-) (Protein-UDP acetylgalactosaminyltransferase 7) (UDP-GalNAc:polypeptide N- acetylgalactosaminyltransferase 7) (Polypeptide GalNAc transferase 7) (GalNAc-T7) (pp-GaNTase 7). | |||||
|
CORIN_MOUSE
|
||||||
| θ value | 8.99809 (rank : 22) | NC score | 0.003077 (rank : 24) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Z319 | Gene names | Corin, Crn, Lrp4 | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuteric peptide-converting enzyme (EC 3.4.21.-) (pro-ANP- converting enzyme) (Corin) (Low density lipoprotein receptor-related protein 4). | |||||
|
SSPO_MOUSE
|
||||||
| θ value | 8.99809 (rank : 23) | NC score | 0.006153 (rank : 21) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8CG65 | Gene names | Sspo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCO-spondin precursor. | |||||
|
TNR25_HUMAN
|
||||||
| θ value | 8.99809 (rank : 24) | NC score | 0.010511 (rank : 19) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q93038, O00275, O00276, O00277, O00278, O00279, O00280, O14865, O14866, P78507, P78515, Q92983, Q93036, Q93037, Q99722, Q99830, Q99831, Q9BY86, Q9UME0, Q9UME1, Q9UME5 | Gene names | TNFRSF25, APO3, DDR3, DR3, TNFRSF12, WSL, WSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 25 precursor (WSL-1 protein) (Apoptosis-mediating receptor DR3) (Apoptosis-mediating receptor TRAMP) (Death domain receptor 3) (WSL protein) (Apoptosis- inducing receptor AIR) (Apo-3) (Lymphocyte-associated receptor of death) (LARD). | |||||
|
CRIS2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 1.86219e-138 (rank : 1) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P16562 | Gene names | CRISP2, TPX1 | |||
|
Domain Architecture |

|
|||||
| Description | Cysteine-rich secretory protein 2 precursor (CRISP-2) (Testis-specific protein TPX-1). | |||||
|
CRIS3_HUMAN
|
||||||
| NC score | 0.995661 (rank : 2) | θ value | 9.035e-101 (rank : 3) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P54108, Q15512, Q5JW83, Q9H108 | Gene names | CRISP3 | |||
|
Domain Architecture |

|
|||||
| Description | Cysteine-rich secretory protein 3 precursor (CRISP-3) (SGP28 protein). | |||||
|
CRIS2_MOUSE
|
||||||
| NC score | 0.994617 (rank : 3) | θ value | 9.32813e-106 (rank : 2) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P16563 | Gene names | Crisp2, Tpx-1, Tpx1 | |||
|
Domain Architecture |

|
|||||
| Description | Cysteine-rich secretory protein 2 precursor (CRISP-2) (Testis-specific protein TPX-1). | |||||
|
CRIS1_MOUSE
|
||||||
| NC score | 0.987120 (rank : 4) | θ value | 5.69867e-79 (rank : 4) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q03401 | Gene names | Crisp1, Aeg-1, Aeg1 | |||
|
Domain Architecture |

|
|||||
| Description | Cysteine-rich secretory protein 1 precursor (CRISP-1) (Acidic epididymal glycoprotein 1) (Sperm-coating glycoprotein 1) (SCP 1). | |||||
|
CRIS3_MOUSE
|
||||||
| NC score | 0.976755 (rank : 5) | θ value | 8.55514e-59 (rank : 5) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q03402 | Gene names | Crisp3, Aeg-2, Aeg2 | |||
|
Domain Architecture |

|
|||||
| Description | Cysteine-rich secretory protein 3 precursor (CRISP-3) (Acidic epididymal glycoprotein 2) (Sperm-coating glycoprotein 2) (SCP 2). | |||||
|
CRIS1_HUMAN
|
||||||
| NC score | 0.971471 (rank : 6) | θ value | 6.77864e-56 (rank : 6) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P54107, O00698, Q13248, Q14082, Q96SF6 | Gene names | CRISP1, AEGL1 | |||
|
Domain Architecture |

|
|||||
| Description | Cysteine-rich secretory protein 1 precursor (CRISP-1) (Acidic epididymal glycoprotein homolog) (AEG-like protein) (ARP). | |||||
|
GLIP1_MOUSE
|
||||||
| NC score | 0.884150 (rank : 7) | θ value | 1.33217e-27 (rank : 7) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9CWG1 | Gene names | Glipr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma pathogenesis-related protein 1 precursor (GliPR 1). | |||||
|
GLIP1_HUMAN
|
||||||
| NC score | 0.866028 (rank : 8) | θ value | 1.24688e-25 (rank : 8) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P48060, Q15409, Q969K2 | Gene names | GLIPR1, GLIPR, RTVP1 | |||
|
Domain Architecture |

|
|||||
| Description | Glioma pathogenesis-related protein 1 precursor (GliPR 1) (RTVP-1 protein). | |||||
|
CRLD2_HUMAN
|
||||||
| NC score | 0.739787 (rank : 9) | θ value | 3.17815e-21 (rank : 9) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9H0B8, Q6P590, Q6UWH0, Q6UXC6, Q96IB1, Q96K61 | Gene names | CRISPLD2, CRISP11, LCRISP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich secretory protein LCCL domain-containing 2 precursor (LCCL domain-containing cysteine-rich secretory protein 2) (Cysteine- rich secretory protein 11) (CRISP-11). | |||||
|
CRLD1_HUMAN
|
||||||
| NC score | 0.726114 (rank : 10) | θ value | 4.15078e-21 (rank : 10) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9H336 | Gene names | CRISPLD1, CRISP10, LCRISP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich secretory protein LCCL domain-containing 1 precursor (LCCL domain-containing cysteine-rich secretory protein 1) (Cysteine- rich secretory protein 10) (CRISP-10) (Trypsin inhibitor Hl) (CocoaCrisp). | |||||
|
CRLD1_MOUSE
|
||||||
| NC score | 0.719509 (rank : 11) | θ value | 2.06002e-20 (rank : 11) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8CGD2, Q8K018, Q99MM6 | Gene names | Crispld1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich secretory protein LCCL domain-containing 1 precursor (CocoaCrisp). | |||||
|
GAPR1_MOUSE
|
||||||
| NC score | 0.625408 (rank : 12) | θ value | 9.59137e-10 (rank : 12) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9CYL5, Q3TJ42, Q8BTK4 | Gene names | Glipr2, Gapr1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgi-associated plant pathogenesis-related protein 1 (Golgi- associated PR-1 protein) (GAPR-1) (Glioma pathogenesis-related protein 2) (GliPR 2). | |||||
|
GAPR1_HUMAN
|
||||||
| NC score | 0.608405 (rank : 13) | θ value | 1.25267e-09 (rank : 13) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9H4G4, Q5VZR1, Q8N2S6, Q8WWC9, Q8WX36 | Gene names | GLIPR2, C9orf19, GAPR1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgi-associated plant pathogenesis-related protein 1 (Golgi- associated PR-1 protein) (GAPR-1) (Glioma pathogenesis-related protein 2) (GliPR 2). | |||||
|
MATN4_HUMAN
|
||||||
| NC score | 0.021364 (rank : 14) | θ value | 0.0736092 (rank : 14) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O95460, Q5QPU2, Q5QPU3, Q5QPU4, Q8N2M5, Q8N2M7, Q9H1F8, Q9H1F9 | Gene names | MATN4 | |||
|
Domain Architecture |

|
|||||
| Description | Matrilin-4 precursor. | |||||
|
FBN1_HUMAN
|
||||||
| NC score | 0.016835 (rank : 15) | θ value | 1.81305 (rank : 15) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P35555, Q15972, Q75N87 | Gene names | FBN1, FBN | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-1 precursor. | |||||
|
FBN1_MOUSE
|
||||||
| NC score | 0.015996 (rank : 16) | θ value | 1.81305 (rank : 16) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 563 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61554, Q60826 | Gene names | Fbn1, Fbn-1 | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-1 precursor. | |||||
|
PROZ_HUMAN
|
||||||
| NC score | 0.015347 (rank : 17) | θ value | 1.81305 (rank : 17) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P22891, Q15213 | Gene names | PROZ | |||
|
Domain Architecture |

|
|||||
| Description | Vitamin K-dependent protein Z precursor. | |||||
|
LTBP4_MOUSE
|
||||||
| NC score | 0.011956 (rank : 18) | θ value | 3.0926 (rank : 19) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8K4G1, Q8K4G0 | Gene names | Ltbp4 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 4 precursor (LTBP-4). | |||||
|
TNR25_HUMAN
|
||||||
| NC score | 0.010511 (rank : 19) | θ value | 8.99809 (rank : 24) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q93038, O00275, O00276, O00277, O00278, O00279, O00280, O14865, O14866, P78507, P78515, Q92983, Q93036, Q93037, Q99722, Q99830, Q99831, Q9BY86, Q9UME0, Q9UME1, Q9UME5 | Gene names | TNFRSF25, APO3, DDR3, DR3, TNFRSF12, WSL, WSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 25 precursor (WSL-1 protein) (Apoptosis-mediating receptor DR3) (Apoptosis-mediating receptor TRAMP) (Death domain receptor 3) (WSL protein) (Apoptosis- inducing receptor AIR) (Apo-3) (Lymphocyte-associated receptor of death) (LARD). | |||||
|
PROC_MOUSE
|
||||||
| NC score | 0.006834 (rank : 20) | θ value | 4.03905 (rank : 20) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P33587, O35498, Q91WN8, Q99PC6 | Gene names | Proc | |||
|
Domain Architecture |

|
|||||
| Description | Vitamin K-dependent protein C precursor (EC 3.4.21.69) (Autoprothrombin IIA) (Anticoagulant protein C) (Blood coagulation factor XIV) [Contains: Vitamin K-dependent protein C light chain; Vitamin K-dependent protein C heavy chain; Activation peptide]. | |||||
|
SSPO_MOUSE
|
||||||
| NC score | 0.006153 (rank : 21) | θ value | 8.99809 (rank : 23) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8CG65 | Gene names | Sspo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCO-spondin precursor. | |||||
|
ATS20_HUMAN
|
||||||
| NC score | 0.005700 (rank : 22) | θ value | 3.0926 (rank : 18) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P59510 | Gene names | ADAMTS20 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-20 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 20) (ADAM-TS 20) (ADAM-TS20). | |||||
|
GALT7_MOUSE
|
||||||
| NC score | 0.003768 (rank : 23) | θ value | 6.88961 (rank : 21) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80VA0, Q8BY62, Q8BZ70, Q8BZW0, Q91VW6, Q99MD7 | Gene names | Galnt7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetylgalactosaminyltransferase 7 (EC 2.4.1.-) (Protein-UDP acetylgalactosaminyltransferase 7) (UDP-GalNAc:polypeptide N- acetylgalactosaminyltransferase 7) (Polypeptide GalNAc transferase 7) (GalNAc-T7) (pp-GaNTase 7). | |||||
|
CORIN_MOUSE
|
||||||
| NC score | 0.003077 (rank : 24) | θ value | 8.99809 (rank : 22) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Z319 | Gene names | Corin, Crn, Lrp4 | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuteric peptide-converting enzyme (EC 3.4.21.-) (pro-ANP- converting enzyme) (Corin) (Low density lipoprotein receptor-related protein 4). | |||||