Please be patient as the page loads
|
CPLX3_MOUSE
|
||||||
| SwissProt Accessions | Q8R1B5, Q96AW7 | Gene names | Cplx3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complexin-3 (Complexin III) (CPX III). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CPLX3_MOUSE
|
||||||
| θ value | 3.13423e-69 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8R1B5, Q96AW7 | Gene names | Cplx3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complexin-3 (Complexin III) (CPX III). | |||||
|
CPLX3_HUMAN
|
||||||
| θ value | 6.53529e-67 (rank : 2) | NC score | 0.995430 (rank : 2) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8WVH0, Q8TEM6, Q9H818 | Gene names | CPLX3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complexin-3 (Complexin III) (CPX III). | |||||
|
CPLX4_MOUSE
|
||||||
| θ value | 4.2656e-42 (rank : 3) | NC score | 0.963498 (rank : 3) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q80WM3, Q8C8Y0, Q91WE5 | Gene names | Cplx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complexin-4 (Complexin IV) (CPX IV). | |||||
|
CPLX4_HUMAN
|
||||||
| θ value | 1.62093e-41 (rank : 4) | NC score | 0.958012 (rank : 4) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q7Z7G2 | Gene names | CPLX4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complexin-4 (Complexin IV) (CPX IV). | |||||
|
CPLX1_MOUSE
|
||||||
| θ value | 0.000461057 (rank : 5) | NC score | 0.422580 (rank : 5) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P63040, O09057, O09142, Q3UYB3, Q64276, Q91WJ3 | Gene names | Cplx1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complexin-1 (Complexin I) (CPX I) (Synaphin-2) (921-S). | |||||
|
CPLX1_HUMAN
|
||||||
| θ value | 0.000602161 (rank : 6) | NC score | 0.415642 (rank : 6) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O14810 | Gene names | CPLX1 | |||
|
Domain Architecture |

|
|||||
| Description | Complexin-1 (Complexin I) (CPX I) (Synaphin-2). | |||||
|
CPLX2_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 7) | NC score | 0.376713 (rank : 7) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6PUV4, O09056, Q13329, Q28184, Q52M15, Q64386 | Gene names | CPLX2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complexin-2 (Complexin II) (CPX II) (Synaphin-1). | |||||
|
CPLX2_MOUSE
|
||||||
| θ value | 0.000786445 (rank : 8) | NC score | 0.376713 (rank : 8) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P84086, O09056, Q13329, Q28184, Q32KK4, Q64386 | Gene names | Cplx2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complexin-2 (Complexin II) (CPX II) (Synaphin-1) (921-L). | |||||
|
RAD50_HUMAN
|
||||||
| θ value | 0.365318 (rank : 9) | NC score | 0.034995 (rank : 15) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q92878, O43254, Q6GMT7, Q6P5X3, Q9UP86 | Gene names | RAD50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (hRAD50). | |||||
|
CROCC_HUMAN
|
||||||
| θ value | 0.62314 (rank : 10) | NC score | 0.035864 (rank : 14) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 1409 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q5TZA2, Q2VHY3, Q66GT7, Q7Z2L4, Q7Z5D7 | Gene names | CROCC, KIAA0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
CTRO_HUMAN
|
||||||
| θ value | 0.813845 (rank : 11) | NC score | 0.013855 (rank : 26) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 2092 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O14578, Q6XUH8, Q86UQ9, Q9UPZ7 | Gene names | CIT, KIAA0949, STK21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
MYO10_HUMAN
|
||||||
| θ value | 0.813845 (rank : 12) | NC score | 0.020742 (rank : 22) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 920 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9HD67, Q9NYM7, Q9P110, Q9P111, Q9UHF6 | Gene names | MYO10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin X). | |||||
|
U2AF2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 13) | NC score | 0.046568 (rank : 9) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P26368 | Gene names | U2AF2, U2AF65 | |||
|
Domain Architecture |
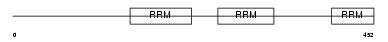
|
|||||
| Description | Splicing factor U2AF 65 kDa subunit (U2 auxiliary factor 65 kDa subunit) (U2 snRNP auxiliary factor large subunit) (hU2AF(65)). | |||||
|
U2AF2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 14) | NC score | 0.046419 (rank : 10) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P26369 | Gene names | U2af2, U2af65 | |||
|
Domain Architecture |
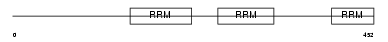
|
|||||
| Description | Splicing factor U2AF 65 kDa subunit (U2 auxiliary factor 65 kDa subunit) (U2 snRNP auxiliary factor large subunit). | |||||
|
CROCC_MOUSE
|
||||||
| θ value | 2.36792 (rank : 15) | NC score | 0.031672 (rank : 16) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 1460 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8CJ40, Q7TQL2, Q80U01, Q8R0B9 | Gene names | Crocc, Kiaa0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
GP73_MOUSE
|
||||||
| θ value | 3.0926 (rank : 16) | NC score | 0.041377 (rank : 12) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q91XA2 | Gene names | Golph2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi phosphoprotein 2 (Golgi membrane protein GP73). | |||||
|
ITSN2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 17) | NC score | 0.023003 (rank : 19) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 1173 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NZM3, O95062, Q15812, Q9HAK4, Q9NXE6, Q9NYG0, Q9NZM2, Q9ULG4 | Gene names | ITSN2, KIAA1256, SH3D1B, SWAP | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (SH3P18) (SH3P18-like WASP-associated protein). | |||||
|
ITSN2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 18) | NC score | 0.022835 (rank : 20) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 1371 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Z0R6, Q9Z0R5 | Gene names | Itsn2, Ese2, Sh3d1B | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (EH and SH3 domains protein 2) (EH domain and SH3 domain regulator of endocytosis 2). | |||||
|
CTRO_MOUSE
|
||||||
| θ value | 4.03905 (rank : 19) | NC score | 0.015687 (rank : 25) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 2344 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P49025, O88528, O88937, O88938, Q3UM99, Q8CIJ1 | Gene names | Cit | |||
|
Domain Architecture |

|
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
ALG2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 20) | NC score | 0.036976 (rank : 13) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9DBE8, Q7TN30, Q9CWI6 | Gene names | Alg2 | |||
|
Domain Architecture |
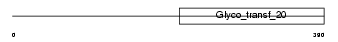
|
|||||
| Description | Alpha-1,3-mannosyltransferase ALG2 (EC 2.4.1.-) (GDP- Man:Man(1)GlcNAc(2)-PP-dolichol mannosyltransferase). | |||||
|
CCD46_MOUSE
|
||||||
| θ value | 5.27518 (rank : 21) | NC score | 0.022025 (rank : 21) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5PR68, Q80ZN6, Q99JS4, Q9D9T1 | Gene names | Ccdc46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled coil domain-containing protein 46. | |||||
|
INVO_HUMAN
|
||||||
| θ value | 5.27518 (rank : 22) | NC score | 0.028388 (rank : 18) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P07476 | Gene names | IVL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Involucrin. | |||||
|
AHSP_MOUSE
|
||||||
| θ value | 6.88961 (rank : 23) | NC score | 0.041897 (rank : 11) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9CY02, O70427 | Gene names | Ahsp, Edrf, Eraf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-hemoglobin-stabilizing protein (Erythroid-associated factor) (Erythroid differentiation-related factor). | |||||
|
LIPB2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 24) | NC score | 0.019253 (rank : 24) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 599 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8ND30, O75337, Q8WW26 | Gene names | PPFIBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-beta-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein-binding protein 2) (PTPRF-interacting protein-binding protein 2). | |||||
|
RBM25_HUMAN
|
||||||
| θ value | 6.88961 (rank : 25) | NC score | 0.028484 (rank : 17) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P49756, Q9UEQ5, Q9UIE9 | Gene names | RBM25, RNPC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable RNA-binding protein 25 (RNA-binding motif protein 25) (RNA- binding region-containing protein 7) (Protein S164). | |||||
|
CCD46_HUMAN
|
||||||
| θ value | 8.99809 (rank : 26) | NC score | 0.020674 (rank : 23) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 1430 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8N8E3, Q6PIB5, Q8NCR4, Q8NFR4 | Gene names | CCDC46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 46. | |||||
|
CPLX3_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 3.13423e-69 (rank : 1) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8R1B5, Q96AW7 | Gene names | Cplx3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complexin-3 (Complexin III) (CPX III). | |||||
|
CPLX3_HUMAN
|
||||||
| NC score | 0.995430 (rank : 2) | θ value | 6.53529e-67 (rank : 2) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8WVH0, Q8TEM6, Q9H818 | Gene names | CPLX3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complexin-3 (Complexin III) (CPX III). | |||||
|
CPLX4_MOUSE
|
||||||
| NC score | 0.963498 (rank : 3) | θ value | 4.2656e-42 (rank : 3) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q80WM3, Q8C8Y0, Q91WE5 | Gene names | Cplx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complexin-4 (Complexin IV) (CPX IV). | |||||
|
CPLX4_HUMAN
|
||||||
| NC score | 0.958012 (rank : 4) | θ value | 1.62093e-41 (rank : 4) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q7Z7G2 | Gene names | CPLX4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complexin-4 (Complexin IV) (CPX IV). | |||||
|
CPLX1_MOUSE
|
||||||
| NC score | 0.422580 (rank : 5) | θ value | 0.000461057 (rank : 5) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P63040, O09057, O09142, Q3UYB3, Q64276, Q91WJ3 | Gene names | Cplx1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complexin-1 (Complexin I) (CPX I) (Synaphin-2) (921-S). | |||||
|
CPLX1_HUMAN
|
||||||
| NC score | 0.415642 (rank : 6) | θ value | 0.000602161 (rank : 6) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O14810 | Gene names | CPLX1 | |||
|
Domain Architecture |

|
|||||
| Description | Complexin-1 (Complexin I) (CPX I) (Synaphin-2). | |||||
|
CPLX2_HUMAN
|
||||||
| NC score | 0.376713 (rank : 7) | θ value | 0.000786445 (rank : 7) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6PUV4, O09056, Q13329, Q28184, Q52M15, Q64386 | Gene names | CPLX2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complexin-2 (Complexin II) (CPX II) (Synaphin-1). | |||||
|
CPLX2_MOUSE
|
||||||
| NC score | 0.376713 (rank : 8) | θ value | 0.000786445 (rank : 8) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P84086, O09056, Q13329, Q28184, Q32KK4, Q64386 | Gene names | Cplx2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complexin-2 (Complexin II) (CPX II) (Synaphin-1) (921-L). | |||||
|
U2AF2_HUMAN
|
||||||
| NC score | 0.046568 (rank : 9) | θ value | 1.81305 (rank : 13) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P26368 | Gene names | U2AF2, U2AF65 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor U2AF 65 kDa subunit (U2 auxiliary factor 65 kDa subunit) (U2 snRNP auxiliary factor large subunit) (hU2AF(65)). | |||||
|
U2AF2_MOUSE
|
||||||
| NC score | 0.046419 (rank : 10) | θ value | 1.81305 (rank : 14) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P26369 | Gene names | U2af2, U2af65 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor U2AF 65 kDa subunit (U2 auxiliary factor 65 kDa subunit) (U2 snRNP auxiliary factor large subunit). | |||||
|
AHSP_MOUSE
|
||||||
| NC score | 0.041897 (rank : 11) | θ value | 6.88961 (rank : 23) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9CY02, O70427 | Gene names | Ahsp, Edrf, Eraf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-hemoglobin-stabilizing protein (Erythroid-associated factor) (Erythroid differentiation-related factor). | |||||
|
GP73_MOUSE
|
||||||
| NC score | 0.041377 (rank : 12) | θ value | 3.0926 (rank : 16) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q91XA2 | Gene names | Golph2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi phosphoprotein 2 (Golgi membrane protein GP73). | |||||
|
ALG2_MOUSE
|
||||||
| NC score | 0.036976 (rank : 13) | θ value | 5.27518 (rank : 20) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9DBE8, Q7TN30, Q9CWI6 | Gene names | Alg2 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-1,3-mannosyltransferase ALG2 (EC 2.4.1.-) (GDP- Man:Man(1)GlcNAc(2)-PP-dolichol mannosyltransferase). | |||||
|
CROCC_HUMAN
|
||||||
| NC score | 0.035864 (rank : 14) | θ value | 0.62314 (rank : 10) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 1409 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q5TZA2, Q2VHY3, Q66GT7, Q7Z2L4, Q7Z5D7 | Gene names | CROCC, KIAA0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
RAD50_HUMAN
|
||||||
| NC score | 0.034995 (rank : 15) | θ value | 0.365318 (rank : 9) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q92878, O43254, Q6GMT7, Q6P5X3, Q9UP86 | Gene names | RAD50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (hRAD50). | |||||
|
CROCC_MOUSE
|
||||||
| NC score | 0.031672 (rank : 16) | θ value | 2.36792 (rank : 15) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 1460 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8CJ40, Q7TQL2, Q80U01, Q8R0B9 | Gene names | Crocc, Kiaa0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
RBM25_HUMAN
|
||||||
| NC score | 0.028484 (rank : 17) | θ value | 6.88961 (rank : 25) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P49756, Q9UEQ5, Q9UIE9 | Gene names | RBM25, RNPC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable RNA-binding protein 25 (RNA-binding motif protein 25) (RNA- binding region-containing protein 7) (Protein S164). | |||||
|
INVO_HUMAN
|
||||||
| NC score | 0.028388 (rank : 18) | θ value | 5.27518 (rank : 22) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P07476 | Gene names | IVL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Involucrin. | |||||
|
ITSN2_HUMAN
|
||||||
| NC score | 0.023003 (rank : 19) | θ value | 3.0926 (rank : 17) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 1173 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NZM3, O95062, Q15812, Q9HAK4, Q9NXE6, Q9NYG0, Q9NZM2, Q9ULG4 | Gene names | ITSN2, KIAA1256, SH3D1B, SWAP | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (SH3P18) (SH3P18-like WASP-associated protein). | |||||
|
ITSN2_MOUSE
|
||||||
| NC score | 0.022835 (rank : 20) | θ value | 3.0926 (rank : 18) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 1371 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Z0R6, Q9Z0R5 | Gene names | Itsn2, Ese2, Sh3d1B | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (EH and SH3 domains protein 2) (EH domain and SH3 domain regulator of endocytosis 2). | |||||
|
CCD46_MOUSE
|
||||||
| NC score | 0.022025 (rank : 21) | θ value | 5.27518 (rank : 21) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5PR68, Q80ZN6, Q99JS4, Q9D9T1 | Gene names | Ccdc46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled coil domain-containing protein 46. | |||||
|
MYO10_HUMAN
|
||||||
| NC score | 0.020742 (rank : 22) | θ value | 0.813845 (rank : 12) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 920 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9HD67, Q9NYM7, Q9P110, Q9P111, Q9UHF6 | Gene names | MYO10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin X). | |||||
|
CCD46_HUMAN
|
||||||
| NC score | 0.020674 (rank : 23) | θ value | 8.99809 (rank : 26) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 1430 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8N8E3, Q6PIB5, Q8NCR4, Q8NFR4 | Gene names | CCDC46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 46. | |||||
|
LIPB2_HUMAN
|
||||||
| NC score | 0.019253 (rank : 24) | θ value | 6.88961 (rank : 24) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 599 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8ND30, O75337, Q8WW26 | Gene names | PPFIBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-beta-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein-binding protein 2) (PTPRF-interacting protein-binding protein 2). | |||||
|
CTRO_MOUSE
|
||||||
| NC score | 0.015687 (rank : 25) | θ value | 4.03905 (rank : 19) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 2344 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P49025, O88528, O88937, O88938, Q3UM99, Q8CIJ1 | Gene names | Cit | |||
|
Domain Architecture |

|
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
CTRO_HUMAN
|
||||||
| NC score | 0.013855 (rank : 26) | θ value | 0.813845 (rank : 11) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 2092 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O14578, Q6XUH8, Q86UQ9, Q9UPZ7 | Gene names | CIT, KIAA0949, STK21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||