Please be patient as the page loads
|
COPD_HUMAN
|
||||||
| SwissProt Accessions | P48444, Q52M80 | Gene names | ARCN1, COPD | |||
|
Domain Architecture |

|
|||||
| Description | Coatomer subunit delta (Delta-coat protein) (Delta-COP) (Archain). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
COPD_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P48444, Q52M80 | Gene names | ARCN1, COPD | |||
|
Domain Architecture |

|
|||||
| Description | Coatomer subunit delta (Delta-coat protein) (Delta-COP) (Archain). | |||||
|
COPD_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999524 (rank : 2) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q5XJY5 | Gene names | Arcn1, Copd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coatomer subunit delta (Delta-coat protein) (Delta-COP) (Archain). | |||||
|
AP4M1_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 3) | NC score | 0.226202 (rank : 3) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O00189, Q9UHK9 | Gene names | AP4M1, MUARP2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-4 complex subunit mu-1 (Adapter-related protein complex 4 mu-1 subunit) (Mu subunit of AP-4) (AP-4 adapter complex mu subunit) (Mu- adaptin-related protein 2) (mu-ARP2) (mu4). | |||||
|
AP4M1_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 4) | NC score | 0.224768 (rank : 4) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9JKC7 | Gene names | Ap4m1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-4 complex subunit mu-1 (Adapter-related protein complex 4 mu-1 subunit) (Mu subunit of AP-4) (AP-4 adapter complex mu subunit) (Mu- adaptin-related protein 2) (mu-ARP2) (mu4). | |||||
|
AP1M1_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 5) | NC score | 0.189259 (rank : 7) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9BXS5 | Gene names | AP1M1, CLTNM | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit mu-1 (Adaptor-related protein complex 1 mu-1 subunit) (Mu-adaptin 1) (Adaptor protein complex AP-1 mu-1 subunit) (Golgi adaptor HA1/AP1 adaptin mu-1 subunit) (Clathrin assembly protein assembly protein complex 1 medium chain 1) (AP-mu chain family member mu1A) (Clathrin coat assembly protein AP47) (Clathrin coat- associated protein AP47). | |||||
|
AP1M1_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 6) | NC score | 0.189452 (rank : 6) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P35585 | Gene names | Ap1m1, Cltnm | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit mu-1 (Adaptor-related protein complex 1 mu-1 subunit) (Mu-adaptin 1) (Adaptor protein complex AP-1 mu-1 subunit) (Golgi adaptor HA1/AP1 adaptin mu-1 subunit) (Clathrin assembly protein assembly protein complex 1 medium chain 1) (AP-mu chain family member mu1A) (Clathrin coat assembly protein AP47) (Clathrin coat- associated protein AP47). | |||||
|
AP1M2_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 7) | NC score | 0.189471 (rank : 5) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y6Q5, Q9BSI8 | Gene names | AP1M2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit mu-2 (Adaptor-related protein complex 1 mu-2 subunit) (Mu-adaptin 2) (Adaptor protein complex AP-1 mu-2 subunit) (Golgi adaptor HA1/AP1 adaptin mu-2 subunit) (Clathrin assembly protein assembly protein complex 1 medium chain 2) (AP-mu chain family member mu1B). | |||||
|
PLEC1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 8) | NC score | 0.024672 (rank : 37) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
AP1M2_MOUSE
|
||||||
| θ value | 0.125558 (rank : 9) | NC score | 0.174651 (rank : 8) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9WVP1, Q99LA4, Q9CWP7 | Gene names | Ap1m2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit mu-2 (Adaptor-related protein complex 1 mu-2 subunit) (Mu-adaptin 2) (Adaptor protein complex AP-1 mu-2 subunit) (Golgi adaptor HA1/AP1 adaptin mu-2 subunit) (Clathrin assembly protein assembly protein complex 1 medium chain 2) (AP-mu chain family member mu1B). | |||||
|
AP2S1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 10) | NC score | 0.119491 (rank : 11) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P53680, O75977, Q6PK67 | Gene names | AP2S1, AP17, CLAPS2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-2 complex subunit sigma-1 (Adapter-related protein complex 2 sigma- 1 subunit) (Clathrin coat assembly protein AP17) (Clathrin coat- associated protein AP17) (Plasma membrane adaptor AP-2 17 kDa protein) (HA2 17 kDa subunit) (Clathrin assembly protein 2 small chain). | |||||
|
AP2S1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 11) | NC score | 0.119491 (rank : 12) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P62743, P70626, P97626, Q00380 | Gene names | Ap2s1, Ap17, Claps2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-2 complex subunit sigma-1 (Adapter-related protein complex 2 sigma- 1 subunit) (Sigma-adaptin 3b) (Clathrin coat assembly protein AP17) (Clathrin coat-associated protein AP17) (Plasma membrane adaptor AP-2 17 kDa protein) (Clathrin assembly protein 2 small chain). | |||||
|
AP4S1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 12) | NC score | 0.118537 (rank : 13) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Y587 | Gene names | AP4S1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-4 complex subunit sigma-1 (Adapter-related protein complex 4 sigma- 1 subunit) (Sigma subunit of AP-4) (AP-4 adapter complex sigma subunit). | |||||
|
ICAL_MOUSE
|
||||||
| θ value | 0.813845 (rank : 13) | NC score | 0.064108 (rank : 25) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P51125, Q9EQV4, Q9EQV5, Q9QXQ3, Q9QXQ4, Q9R0N1 | Gene names | Cast | |||
|
Domain Architecture |

|
|||||
| Description | Calpastatin (Calpain inhibitor). | |||||
|
INCE_MOUSE
|
||||||
| θ value | 1.06291 (rank : 14) | NC score | 0.028664 (rank : 34) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9WU62, Q7TN28, Q8BGN4, Q8CGI4 | Gene names | Incenp | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
AP1S2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 15) | NC score | 0.098129 (rank : 14) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P56377, O95326, Q9H2N6 | Gene names | AP1S2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit sigma-2 (Adapter-related protein complex 1 sigma- 1B subunit) (Sigma-adaptin 1B) (Adaptor protein complex AP-1 sigma-1B subunit) (Golgi adaptor HA1/AP1 adaptin sigma-1B subunit) (Clathrin assembly protein complex 1 sigma-1B small chain) (Sigma 1B subunit of AP-1 clathrin). | |||||
|
PLVAP_MOUSE
|
||||||
| θ value | 1.38821 (rank : 16) | NC score | 0.054588 (rank : 26) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91VC4, Q99JB1 | Gene names | Plvap, Pv1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plasmalemma vesicle-associated protein (Plasmalemma vesicle protein 1) (PV-1) (MECA-32 antigen). | |||||
|
CROP_HUMAN
|
||||||
| θ value | 1.81305 (rank : 17) | NC score | 0.046882 (rank : 28) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O95232, Q6PHR9, Q9NUY0, Q9P2S7 | Gene names | CROP, CREAP1, O48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cisplatin resistance-associated overexpressed protein (cAMP regulatory element-associated protein 1) (CRE-associated protein 1) (CREAP-1) (Luc7A) (Okadaic acid-inducible phosphoprotein OA48-18). | |||||
|
CROP_MOUSE
|
||||||
| θ value | 1.81305 (rank : 18) | NC score | 0.046887 (rank : 27) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5SUF2, Q3U9D5, Q8BUJ5, Q921Z3, Q9CRS7, Q9CTY4 | Gene names | Crop | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cisplatin resistance-associated overexpressed protein. | |||||
|
ECHM_HUMAN
|
||||||
| θ value | 1.81305 (rank : 19) | NC score | 0.040627 (rank : 29) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P30084, O00739, Q5VWY1, Q96H54 | Gene names | ECHS1 | |||
|
Domain Architecture |

|
|||||
| Description | Enoyl-CoA hydratase, mitochondrial precursor (EC 4.2.1.17) (Short chain enoyl-CoA hydratase) (SCEH) (Enoyl-CoA hydratase 1). | |||||
|
IF16_HUMAN
|
||||||
| θ value | 1.81305 (rank : 20) | NC score | 0.033951 (rank : 32) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q16666, Q9UH78 | Gene names | IFI16, IFNGIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-interferon-inducible protein Ifi-16 (Interferon-inducible myeloid differentiation transcriptional activator) (IFI 16). | |||||
|
AP4S1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 21) | NC score | 0.096360 (rank : 15) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9WVL1 | Gene names | Ap4s1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-4 complex subunit sigma-1 (Adapter-related protein complex 4 sigma- 1 subunit) (Sigma subunit of AP-4) (AP-4 adapter complex sigma subunit). | |||||
|
AP1S2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 22) | NC score | 0.091629 (rank : 18) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9DB50, Q543R7 | Gene names | Ap1s2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit sigma-2 (Adapter-related protein complex 1 sigma- 1B subunit) (Sigma-adaptin 1B) (Adaptor protein complex AP-1 sigma-1B subunit) (Golgi adaptor HA1/AP1 adaptin sigma-1B subunit) (Clathrin assembly protein complex 1 sigma-1B small chain) (Sigma 1B subunit of AP-1 clathrin). | |||||
|
SGIP1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 23) | NC score | 0.038400 (rank : 30) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BQI5, Q4LE32, Q5VYE2, Q5VYE3, Q5VYE4, Q68D76, Q6MZY6, Q8IWC2 | Gene names | SGIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH3-containing GRB2-like protein 3-interacting protein 1 (Endophilin- 3-interacting protein). | |||||
|
AP2M1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 24) | NC score | 0.140744 (rank : 9) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96CW1, P20172, P53679 | Gene names | AP2M1, CLAPM1, KIAA0109 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-2 complex subunit mu-1 (Adaptin mu-1) (AP-2 mu-2 chain) (Clathrin coat assembly protein AP50) (Clathrin coat-associated protein AP50) (Plasma membrane adaptor AP-2 50 kDa protein) (HA2 50 kDa subunit) (Clathrin assembly protein complex 2 medium chain). | |||||
|
AP2M1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 25) | NC score | 0.140744 (rank : 10) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P84091, P20172, P53679 | Gene names | Ap2m1, Clapm1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-2 complex subunit mu-1 (Adaptin mu-1) (AP-2 mu-2 chain) (Clathrin coat assembly protein AP50) (Clathrin coat-associated protein AP50) (Plasma membrane adaptor AP-2 50 kDa protein) (Clathrin assembly protein complex 2 medium chain). | |||||
|
KI21A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 26) | NC score | 0.010321 (rank : 41) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1543 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q7Z4S6, Q6UKL9, Q7Z668, Q86WZ5, Q8IVZ8, Q9C0F5, Q9NXU4, Q9Y590 | Gene names | KIF21A, KIAA1708, KIF2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A (Kinesin-like protein KIF2) (NY-REN-62 antigen). | |||||
|
GLE1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 27) | NC score | 0.027336 (rank : 36) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q53GS7, O75458, Q53GT9, Q5VVU1, Q8NCP6, Q9UFL6 | Gene names | GLE1L, GLE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin GLE1 (GLE1-like protein) (hGLE1). | |||||
|
IDLC_MOUSE
|
||||||
| θ value | 6.88961 (rank : 28) | NC score | 0.038132 (rank : 31) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BVN8 | Gene names | Dnali1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Axonemal dynein light intermediate polypeptide 1 (Inner dynein arm light chain, axonemal). | |||||
|
PP16A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 29) | NC score | 0.009013 (rank : 42) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 340 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96I34 | Gene names | PPP1R16A, MYPT3 | |||
|
Domain Architecture |
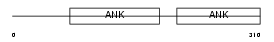
|
|||||
| Description | Protein phosphatase 1 regulatory subunit 16A (Myosin phosphatase- targeting subunit 3). | |||||
|
SGIP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 30) | NC score | 0.027639 (rank : 35) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VD37, Q3UFU3, Q3UGA0, Q8BXX4, Q8C034, Q9CXT2 | Gene names | Sgip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH3-containing GRB2-like protein 3-interacting protein 1 (Endophilin- 3-interacting protein). | |||||
|
SOX5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 31) | NC score | 0.012537 (rank : 39) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P35711, Q86UK8, Q8J017, Q8J018, Q8J019, Q8J020, Q8N1D9, Q8N7E0, Q8TEA4 | Gene names | SOX5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-5. | |||||
|
TRHY_HUMAN
|
||||||
| θ value | 6.88961 (rank : 32) | NC score | 0.007566 (rank : 43) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
AP1S1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 33) | NC score | 0.080802 (rank : 21) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P61966, P82267, Q00382, Q9BTN4, Q9UDW9 | Gene names | AP1S1, AP19, CLAPS1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit sigma-1A (Adapter-related protein complex 1 sigma-1A subunit) (Sigma-adaptin 1A) (Adaptor protein complex AP-1 sigma-1A subunit) (Golgi adaptor HA1/AP1 adaptin sigma-1A subunit) (Clathrin assembly protein complex 1 sigma-1A small chain) (Clathrin coat assembly protein AP19) (HA1 19 kDa subunit) (Sigma 1a subunit of AP-1 clathrin). | |||||
|
AP1S1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 34) | NC score | 0.080802 (rank : 22) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P61967, P82267, Q00382, Q9BTN4, Q9UDW9 | Gene names | Ap1s1, Ap19 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit sigma-1A (Adapter-related protein complex 1 sigma-1A subunit) (Sigma-adaptin 1A) (Adaptor protein complex AP-1 sigma-1A subunit) (Golgi adaptor HA1/AP1 adaptin sigma-1A subunit) (Clathrin assembly protein complex 1 sigma-1A small chain) (Clathrin coat assembly protein AP19) (HA1 19 kDa subunit) (Sigma 1a subunit of AP-1 clathrin). | |||||
|
CD2L1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 35) | NC score | 0.002841 (rank : 46) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1467 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P21127, O95265, Q12817, Q12818, Q12819, Q12820, Q12822, Q8N530, Q9NZS5, Q9UBJ0, Q9UBQ1, Q9UBR0, Q9UNY2, Q9UP57, Q9UP58, Q9UP59 | Gene names | CDC2L1, PITSLREA, PK58 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1) (CLK-1) (CDK11) (p58 CLK-1). | |||||
|
CD2L2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 36) | NC score | 0.002871 (rank : 45) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1439 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UQ88, O95227, O95228, O96012, Q12821, Q12853, Q12854, Q5QPR0, Q5QPR1, Q5QPR2, Q9UBC4, Q9UBI3, Q9UEI1, Q9UEI2, Q9UP53, Q9UP54, Q9UP55, Q9UP56, Q9UQ86, Q9UQ87, Q9UQ89 | Gene names | CDC2L2, PITSLREB | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L2 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 2) (CDK11). | |||||
|
DESM_HUMAN
|
||||||
| θ value | 8.99809 (rank : 37) | NC score | 0.005766 (rank : 44) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 495 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P17661, Q15787, Q8IZR1, Q8IZR6, Q8NES2, Q8NEU6, Q8TAC4, Q8TCX2, Q8TD99, Q9UHN5, Q9UJ80 | Gene names | DES | |||
|
Domain Architecture |
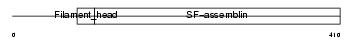
|
|||||
| Description | Desmin. | |||||
|
FREM1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 38) | NC score | 0.010386 (rank : 40) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5H8C1, Q5VV00, Q5VV01, Q6MZI4, Q8NEG9, Q96LI3 | Gene names | FREM1, C9orf154 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FRAS1-related extracellular matrix protein 1 precursor (Protein QBRICK). | |||||
|
IDLC_HUMAN
|
||||||
| θ value | 8.99809 (rank : 39) | NC score | 0.033712 (rank : 33) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O14645, Q5TGH0, Q7L0I5 | Gene names | DNALI1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Axonemal dynein light intermediate polypeptide 1 (Inner dynein arm light chain, axonemal) (hp28). | |||||
|
LRC27_HUMAN
|
||||||
| θ value | 8.99809 (rank : 40) | NC score | 0.018269 (rank : 38) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 710 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9C0I9, Q5SZH6, Q5SZH8, Q5SZH9, Q86XT5, Q8N7C8, Q8NA21 | Gene names | LRRC27, KIAA1674 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 27. | |||||
|
AP1S3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 41) | NC score | 0.067253 (rank : 23) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96PC3, Q8WTY1, Q96DD1 | Gene names | AP1S3 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit sigma-3 (Adapter-related protein complex 1 sigma- 1C subunit) (Sigma-adaptin 1C) (Adaptor protein complex AP-1 sigma-1C subunit) (Golgi adaptor HA1/AP1 adaptin sigma-1C subunit) (Clathrin assembly protein complex 1 sigma-1C small chain) (Sigma 1C subunit of AP-1 clathrin). | |||||
|
AP1S3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 42) | NC score | 0.066435 (rank : 24) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q7TN05 | Gene names | Ap1s3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-1 complex subunit sigma-3 (Adapter-related protein complex 1 sigma- 1C subunit) (Sigma-adaptin 1C) (Adaptor protein complex AP-1 sigma-1C subunit) (Golgi adaptor HA1/AP1 adaptin sigma-1C subunit) (Clathrin assembly protein complex 1 sigma-1C small chain) (Sigma 1C subunit of AP-1 clathrin). | |||||
|
AP3M1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 43) | NC score | 0.094890 (rank : 16) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Y2T2 | Gene names | AP3M1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit mu-1 (Adapter-related protein complex 3 mu-1 subunit) (Mu-adaptin 3A) (AP-3 adapter complex mu3A subunit). | |||||
|
AP3M1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 44) | NC score | 0.094755 (rank : 17) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9JKC8, Q5BKQ6 | Gene names | Ap3m1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit mu-1 (Adapter-related protein complex 3 mu-1 subunit) (Mu-adaptin 3A) (AP-3 adapter complex mu3A subunit). | |||||
|
AP3M2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 45) | NC score | 0.090675 (rank : 20) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P53677 | Gene names | AP3M2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit mu-2 (Adapter-related protein complex 3 mu-2 subunit) (Clathrin coat assembly protein AP47 homolog 2) (Clathrin coat-associated protein AP47 homolog 2) (Golgi adaptor AP-1 47 kDa protein homolog 2) (HA1 47 kDa subunit homolog 2) (Clathrin assembly protein assembly protein complex 1 medium chain homolog 2) (P47B). | |||||
|
AP3M2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 46) | NC score | 0.090900 (rank : 19) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8R2R9, Q3UYJ3, Q923G7 | Gene names | Ap3m2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit mu-2 (Adapter-related protein complex 3 mu-2 subunit) (Clathrin coat assembly protein AP47 homolog 2) (Clathrin coat-associated protein AP47 homolog 2) (Golgi adaptor AP-1 47 kDa protein homolog 2) (HA1 47 kDa subunit homolog 2) (Clathrin assembly protein assembly protein complex 1 medium chain homolog 2) (P47B) (m3B). | |||||
|
COPD_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P48444, Q52M80 | Gene names | ARCN1, COPD | |||
|
Domain Architecture |

|
|||||
| Description | Coatomer subunit delta (Delta-coat protein) (Delta-COP) (Archain). | |||||
|
COPD_MOUSE
|
||||||
| NC score | 0.999524 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q5XJY5 | Gene names | Arcn1, Copd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coatomer subunit delta (Delta-coat protein) (Delta-COP) (Archain). | |||||
|
AP4M1_HUMAN
|
||||||
| NC score | 0.226202 (rank : 3) | θ value | 0.00228821 (rank : 3) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O00189, Q9UHK9 | Gene names | AP4M1, MUARP2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-4 complex subunit mu-1 (Adapter-related protein complex 4 mu-1 subunit) (Mu subunit of AP-4) (AP-4 adapter complex mu subunit) (Mu- adaptin-related protein 2) (mu-ARP2) (mu4). | |||||
|
AP4M1_MOUSE
|
||||||
| NC score | 0.224768 (rank : 4) | θ value | 0.00228821 (rank : 4) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9JKC7 | Gene names | Ap4m1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-4 complex subunit mu-1 (Adapter-related protein complex 4 mu-1 subunit) (Mu subunit of AP-4) (AP-4 adapter complex mu subunit) (Mu- adaptin-related protein 2) (mu-ARP2) (mu4). | |||||
|
AP1M2_HUMAN
|
||||||
| NC score | 0.189471 (rank : 5) | θ value | 0.0113563 (rank : 7) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y6Q5, Q9BSI8 | Gene names | AP1M2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit mu-2 (Adaptor-related protein complex 1 mu-2 subunit) (Mu-adaptin 2) (Adaptor protein complex AP-1 mu-2 subunit) (Golgi adaptor HA1/AP1 adaptin mu-2 subunit) (Clathrin assembly protein assembly protein complex 1 medium chain 2) (AP-mu chain family member mu1B). | |||||
|
AP1M1_MOUSE
|
||||||
| NC score | 0.189452 (rank : 6) | θ value | 0.00869519 (rank : 6) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P35585 | Gene names | Ap1m1, Cltnm | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit mu-1 (Adaptor-related protein complex 1 mu-1 subunit) (Mu-adaptin 1) (Adaptor protein complex AP-1 mu-1 subunit) (Golgi adaptor HA1/AP1 adaptin mu-1 subunit) (Clathrin assembly protein assembly protein complex 1 medium chain 1) (AP-mu chain family member mu1A) (Clathrin coat assembly protein AP47) (Clathrin coat- associated protein AP47). | |||||
|
AP1M1_HUMAN
|
||||||
| NC score | 0.189259 (rank : 7) | θ value | 0.00869519 (rank : 5) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9BXS5 | Gene names | AP1M1, CLTNM | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit mu-1 (Adaptor-related protein complex 1 mu-1 subunit) (Mu-adaptin 1) (Adaptor protein complex AP-1 mu-1 subunit) (Golgi adaptor HA1/AP1 adaptin mu-1 subunit) (Clathrin assembly protein assembly protein complex 1 medium chain 1) (AP-mu chain family member mu1A) (Clathrin coat assembly protein AP47) (Clathrin coat- associated protein AP47). | |||||
|
AP1M2_MOUSE
|
||||||
| NC score | 0.174651 (rank : 8) | θ value | 0.125558 (rank : 9) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9WVP1, Q99LA4, Q9CWP7 | Gene names | Ap1m2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit mu-2 (Adaptor-related protein complex 1 mu-2 subunit) (Mu-adaptin 2) (Adaptor protein complex AP-1 mu-2 subunit) (Golgi adaptor HA1/AP1 adaptin mu-2 subunit) (Clathrin assembly protein assembly protein complex 1 medium chain 2) (AP-mu chain family member mu1B). | |||||
|
AP2M1_HUMAN
|
||||||
| NC score | 0.140744 (rank : 9) | θ value | 5.27518 (rank : 24) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96CW1, P20172, P53679 | Gene names | AP2M1, CLAPM1, KIAA0109 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-2 complex subunit mu-1 (Adaptin mu-1) (AP-2 mu-2 chain) (Clathrin coat assembly protein AP50) (Clathrin coat-associated protein AP50) (Plasma membrane adaptor AP-2 50 kDa protein) (HA2 50 kDa subunit) (Clathrin assembly protein complex 2 medium chain). | |||||
|
AP2M1_MOUSE
|
||||||
| NC score | 0.140744 (rank : 10) | θ value | 5.27518 (rank : 25) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P84091, P20172, P53679 | Gene names | Ap2m1, Clapm1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-2 complex subunit mu-1 (Adaptin mu-1) (AP-2 mu-2 chain) (Clathrin coat assembly protein AP50) (Clathrin coat-associated protein AP50) (Plasma membrane adaptor AP-2 50 kDa protein) (Clathrin assembly protein complex 2 medium chain). | |||||
|
AP2S1_HUMAN
|
||||||
| NC score | 0.119491 (rank : 11) | θ value | 0.279714 (rank : 10) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P53680, O75977, Q6PK67 | Gene names | AP2S1, AP17, CLAPS2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-2 complex subunit sigma-1 (Adapter-related protein complex 2 sigma- 1 subunit) (Clathrin coat assembly protein AP17) (Clathrin coat- associated protein AP17) (Plasma membrane adaptor AP-2 17 kDa protein) (HA2 17 kDa subunit) (Clathrin assembly protein 2 small chain). | |||||
|
AP2S1_MOUSE
|
||||||
| NC score | 0.119491 (rank : 12) | θ value | 0.279714 (rank : 11) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P62743, P70626, P97626, Q00380 | Gene names | Ap2s1, Ap17, Claps2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-2 complex subunit sigma-1 (Adapter-related protein complex 2 sigma- 1 subunit) (Sigma-adaptin 3b) (Clathrin coat assembly protein AP17) (Clathrin coat-associated protein AP17) (Plasma membrane adaptor AP-2 17 kDa protein) (Clathrin assembly protein 2 small chain). | |||||
|
AP4S1_HUMAN
|
||||||
| NC score | 0.118537 (rank : 13) | θ value | 0.365318 (rank : 12) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Y587 | Gene names | AP4S1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-4 complex subunit sigma-1 (Adapter-related protein complex 4 sigma- 1 subunit) (Sigma subunit of AP-4) (AP-4 adapter complex sigma subunit). | |||||
|
AP1S2_HUMAN
|
||||||
| NC score | 0.098129 (rank : 14) | θ value | 1.38821 (rank : 15) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P56377, O95326, Q9H2N6 | Gene names | AP1S2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit sigma-2 (Adapter-related protein complex 1 sigma- 1B subunit) (Sigma-adaptin 1B) (Adaptor protein complex AP-1 sigma-1B subunit) (Golgi adaptor HA1/AP1 adaptin sigma-1B subunit) (Clathrin assembly protein complex 1 sigma-1B small chain) (Sigma 1B subunit of AP-1 clathrin). | |||||
|
AP4S1_MOUSE
|
||||||
| NC score | 0.096360 (rank : 15) | θ value | 2.36792 (rank : 21) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9WVL1 | Gene names | Ap4s1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-4 complex subunit sigma-1 (Adapter-related protein complex 4 sigma- 1 subunit) (Sigma subunit of AP-4) (AP-4 adapter complex sigma subunit). | |||||
|
AP3M1_HUMAN
|
||||||
| NC score | 0.094890 (rank : 16) | θ value | θ > 10 (rank : 43) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Y2T2 | Gene names | AP3M1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit mu-1 (Adapter-related protein complex 3 mu-1 subunit) (Mu-adaptin 3A) (AP-3 adapter complex mu3A subunit). | |||||
|
AP3M1_MOUSE
|
||||||
| NC score | 0.094755 (rank : 17) | θ value | θ > 10 (rank : 44) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9JKC8, Q5BKQ6 | Gene names | Ap3m1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit mu-1 (Adapter-related protein complex 3 mu-1 subunit) (Mu-adaptin 3A) (AP-3 adapter complex mu3A subunit). | |||||
|
AP1S2_MOUSE
|
||||||
| NC score | 0.091629 (rank : 18) | θ value | 3.0926 (rank : 22) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9DB50, Q543R7 | Gene names | Ap1s2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit sigma-2 (Adapter-related protein complex 1 sigma- 1B subunit) (Sigma-adaptin 1B) (Adaptor protein complex AP-1 sigma-1B subunit) (Golgi adaptor HA1/AP1 adaptin sigma-1B subunit) (Clathrin assembly protein complex 1 sigma-1B small chain) (Sigma 1B subunit of AP-1 clathrin). | |||||
|
AP3M2_MOUSE
|
||||||
| NC score | 0.090900 (rank : 19) | θ value | θ > 10 (rank : 46) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8R2R9, Q3UYJ3, Q923G7 | Gene names | Ap3m2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit mu-2 (Adapter-related protein complex 3 mu-2 subunit) (Clathrin coat assembly protein AP47 homolog 2) (Clathrin coat-associated protein AP47 homolog 2) (Golgi adaptor AP-1 47 kDa protein homolog 2) (HA1 47 kDa subunit homolog 2) (Clathrin assembly protein assembly protein complex 1 medium chain homolog 2) (P47B) (m3B). | |||||
|
AP3M2_HUMAN
|
||||||
| NC score | 0.090675 (rank : 20) | θ value | θ > 10 (rank : 45) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P53677 | Gene names | AP3M2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit mu-2 (Adapter-related protein complex 3 mu-2 subunit) (Clathrin coat assembly protein AP47 homolog 2) (Clathrin coat-associated protein AP47 homolog 2) (Golgi adaptor AP-1 47 kDa protein homolog 2) (HA1 47 kDa subunit homolog 2) (Clathrin assembly protein assembly protein complex 1 medium chain homolog 2) (P47B). | |||||
|
AP1S1_HUMAN
|
||||||
| NC score | 0.080802 (rank : 21) | θ value | 8.99809 (rank : 33) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P61966, P82267, Q00382, Q9BTN4, Q9UDW9 | Gene names | AP1S1, AP19, CLAPS1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit sigma-1A (Adapter-related protein complex 1 sigma-1A subunit) (Sigma-adaptin 1A) (Adaptor protein complex AP-1 sigma-1A subunit) (Golgi adaptor HA1/AP1 adaptin sigma-1A subunit) (Clathrin assembly protein complex 1 sigma-1A small chain) (Clathrin coat assembly protein AP19) (HA1 19 kDa subunit) (Sigma 1a subunit of AP-1 clathrin). | |||||
|
AP1S1_MOUSE
|
||||||
| NC score | 0.080802 (rank : 22) | θ value | 8.99809 (rank : 34) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P61967, P82267, Q00382, Q9BTN4, Q9UDW9 | Gene names | Ap1s1, Ap19 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit sigma-1A (Adapter-related protein complex 1 sigma-1A subunit) (Sigma-adaptin 1A) (Adaptor protein complex AP-1 sigma-1A subunit) (Golgi adaptor HA1/AP1 adaptin sigma-1A subunit) (Clathrin assembly protein complex 1 sigma-1A small chain) (Clathrin coat assembly protein AP19) (HA1 19 kDa subunit) (Sigma 1a subunit of AP-1 clathrin). | |||||
|
AP1S3_HUMAN
|
||||||
| NC score | 0.067253 (rank : 23) | θ value | θ > 10 (rank : 41) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96PC3, Q8WTY1, Q96DD1 | Gene names | AP1S3 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit sigma-3 (Adapter-related protein complex 1 sigma- 1C subunit) (Sigma-adaptin 1C) (Adaptor protein complex AP-1 sigma-1C subunit) (Golgi adaptor HA1/AP1 adaptin sigma-1C subunit) (Clathrin assembly protein complex 1 sigma-1C small chain) (Sigma 1C subunit of AP-1 clathrin). | |||||
|
AP1S3_MOUSE
|
||||||
| NC score | 0.066435 (rank : 24) | θ value | θ > 10 (rank : 42) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q7TN05 | Gene names | Ap1s3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-1 complex subunit sigma-3 (Adapter-related protein complex 1 sigma- 1C subunit) (Sigma-adaptin 1C) (Adaptor protein complex AP-1 sigma-1C subunit) (Golgi adaptor HA1/AP1 adaptin sigma-1C subunit) (Clathrin assembly protein complex 1 sigma-1C small chain) (Sigma 1C subunit of AP-1 clathrin). | |||||
|
ICAL_MOUSE
|
||||||
| NC score | 0.064108 (rank : 25) | θ value | 0.813845 (rank : 13) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P51125, Q9EQV4, Q9EQV5, Q9QXQ3, Q9QXQ4, Q9R0N1 | Gene names | Cast | |||
|
Domain Architecture |

|
|||||
| Description | Calpastatin (Calpain inhibitor). | |||||
|
PLVAP_MOUSE
|
||||||
| NC score | 0.054588 (rank : 26) | θ value | 1.38821 (rank : 16) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91VC4, Q99JB1 | Gene names | Plvap, Pv1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plasmalemma vesicle-associated protein (Plasmalemma vesicle protein 1) (PV-1) (MECA-32 antigen). | |||||
|
CROP_MOUSE
|
||||||
| NC score | 0.046887 (rank : 27) | θ value | 1.81305 (rank : 18) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5SUF2, Q3U9D5, Q8BUJ5, Q921Z3, Q9CRS7, Q9CTY4 | Gene names | Crop | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cisplatin resistance-associated overexpressed protein. | |||||
|
CROP_HUMAN
|
||||||
| NC score | 0.046882 (rank : 28) | θ value | 1.81305 (rank : 17) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O95232, Q6PHR9, Q9NUY0, Q9P2S7 | Gene names | CROP, CREAP1, O48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cisplatin resistance-associated overexpressed protein (cAMP regulatory element-associated protein 1) (CRE-associated protein 1) (CREAP-1) (Luc7A) (Okadaic acid-inducible phosphoprotein OA48-18). | |||||
|
ECHM_HUMAN
|
||||||
| NC score | 0.040627 (rank : 29) | θ value | 1.81305 (rank : 19) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P30084, O00739, Q5VWY1, Q96H54 | Gene names | ECHS1 | |||
|
Domain Architecture |

|
|||||
| Description | Enoyl-CoA hydratase, mitochondrial precursor (EC 4.2.1.17) (Short chain enoyl-CoA hydratase) (SCEH) (Enoyl-CoA hydratase 1). | |||||
|
SGIP1_HUMAN
|
||||||
| NC score | 0.038400 (rank : 30) | θ value | 4.03905 (rank : 23) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BQI5, Q4LE32, Q5VYE2, Q5VYE3, Q5VYE4, Q68D76, Q6MZY6, Q8IWC2 | Gene names | SGIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH3-containing GRB2-like protein 3-interacting protein 1 (Endophilin- 3-interacting protein). | |||||
|
IDLC_MOUSE
|
||||||
| NC score | 0.038132 (rank : 31) | θ value | 6.88961 (rank : 28) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BVN8 | Gene names | Dnali1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Axonemal dynein light intermediate polypeptide 1 (Inner dynein arm light chain, axonemal). | |||||
|
IF16_HUMAN
|
||||||
| NC score | 0.033951 (rank : 32) | θ value | 1.81305 (rank : 20) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q16666, Q9UH78 | Gene names | IFI16, IFNGIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-interferon-inducible protein Ifi-16 (Interferon-inducible myeloid differentiation transcriptional activator) (IFI 16). | |||||
|
IDLC_HUMAN
|
||||||
| NC score | 0.033712 (rank : 33) | θ value | 8.99809 (rank : 39) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O14645, Q5TGH0, Q7L0I5 | Gene names | DNALI1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Axonemal dynein light intermediate polypeptide 1 (Inner dynein arm light chain, axonemal) (hp28). | |||||
|
INCE_MOUSE
|
||||||
| NC score | 0.028664 (rank : 34) | θ value | 1.06291 (rank : 14) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9WU62, Q7TN28, Q8BGN4, Q8CGI4 | Gene names | Incenp | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
SGIP1_MOUSE
|
||||||
| NC score | 0.027639 (rank : 35) | θ value | 6.88961 (rank : 30) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VD37, Q3UFU3, Q3UGA0, Q8BXX4, Q8C034, Q9CXT2 | Gene names | Sgip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH3-containing GRB2-like protein 3-interacting protein 1 (Endophilin- 3-interacting protein). | |||||
|
GLE1_HUMAN
|
||||||
| NC score | 0.027336 (rank : 36) | θ value | 6.88961 (rank : 27) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q53GS7, O75458, Q53GT9, Q5VVU1, Q8NCP6, Q9UFL6 | Gene names | GLE1L, GLE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin GLE1 (GLE1-like protein) (hGLE1). | |||||
|
PLEC1_HUMAN
|
||||||
| NC score | 0.024672 (rank : 37) | θ value | 0.0961366 (rank : 8) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
LRC27_HUMAN
|
||||||
| NC score | 0.018269 (rank : 38) | θ value | 8.99809 (rank : 40) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 710 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9C0I9, Q5SZH6, Q5SZH8, Q5SZH9, Q86XT5, Q8N7C8, Q8NA21 | Gene names | LRRC27, KIAA1674 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 27. | |||||
|
SOX5_HUMAN
|
||||||
| NC score | 0.012537 (rank : 39) | θ value | 6.88961 (rank : 31) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P35711, Q86UK8, Q8J017, Q8J018, Q8J019, Q8J020, Q8N1D9, Q8N7E0, Q8TEA4 | Gene names | SOX5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-5. | |||||
|
FREM1_HUMAN
|
||||||
| NC score | 0.010386 (rank : 40) | θ value | 8.99809 (rank : 38) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5H8C1, Q5VV00, Q5VV01, Q6MZI4, Q8NEG9, Q96LI3 | Gene names | FREM1, C9orf154 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FRAS1-related extracellular matrix protein 1 precursor (Protein QBRICK). | |||||
|
KI21A_HUMAN
|
||||||
| NC score | 0.010321 (rank : 41) | θ value | 5.27518 (rank : 26) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1543 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q7Z4S6, Q6UKL9, Q7Z668, Q86WZ5, Q8IVZ8, Q9C0F5, Q9NXU4, Q9Y590 | Gene names | KIF21A, KIAA1708, KIF2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A (Kinesin-like protein KIF2) (NY-REN-62 antigen). | |||||
|
PP16A_HUMAN
|
||||||
| NC score | 0.009013 (rank : 42) | θ value | 6.88961 (rank : 29) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 340 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96I34 | Gene names | PPP1R16A, MYPT3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 16A (Myosin phosphatase- targeting subunit 3). | |||||
|
TRHY_HUMAN
|
||||||
| NC score | 0.007566 (rank : 43) | θ value | 6.88961 (rank : 32) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
DESM_HUMAN
|
||||||
| NC score | 0.005766 (rank : 44) | θ value | 8.99809 (rank : 37) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 495 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P17661, Q15787, Q8IZR1, Q8IZR6, Q8NES2, Q8NEU6, Q8TAC4, Q8TCX2, Q8TD99, Q9UHN5, Q9UJ80 | Gene names | DES | |||
|
Domain Architecture |

|
|||||
| Description | Desmin. | |||||
|
CD2L2_HUMAN
|
||||||
| NC score | 0.002871 (rank : 45) | θ value | 8.99809 (rank : 36) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1439 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UQ88, O95227, O95228, O96012, Q12821, Q12853, Q12854, Q5QPR0, Q5QPR1, Q5QPR2, Q9UBC4, Q9UBI3, Q9UEI1, Q9UEI2, Q9UP53, Q9UP54, Q9UP55, Q9UP56, Q9UQ86, Q9UQ87, Q9UQ89 | Gene names | CDC2L2, PITSLREB | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L2 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 2) (CDK11). | |||||
|
CD2L1_HUMAN
|
||||||
| NC score | 0.002841 (rank : 46) | θ value | 8.99809 (rank : 35) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1467 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P21127, O95265, Q12817, Q12818, Q12819, Q12820, Q12822, Q8N530, Q9NZS5, Q9UBJ0, Q9UBQ1, Q9UBR0, Q9UNY2, Q9UP57, Q9UP58, Q9UP59 | Gene names | CDC2L1, PITSLREA, PK58 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1) (CLK-1) (CDK11) (p58 CLK-1). | |||||