Please be patient as the page loads
|
CO5_HUMAN
|
||||||
| SwissProt Accessions | P01031 | Gene names | C5 | |||
|
Domain Architecture |

|
|||||
| Description | Complement C5 precursor [Contains: Complement C5 beta chain; Complement C5 alpha chain; C5a anaphylatoxin; Complement C5 alpha' chain]. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CO3_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.954731 (rank : 3) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P01024 | Gene names | C3 | |||
|
Domain Architecture |

|
|||||
| Description | Complement C3 precursor [Contains: Complement C3 beta chain; Complement C3 alpha chain; C3a anaphylatoxin; Complement C3b alpha' chain; Complement C3c fragment; Complement C3dg fragment; Complement C3g fragment; Complement C3d fragment; Complement C3f fragment]. | |||||
|
CO3_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.954435 (rank : 4) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P01027, Q61370 | Gene names | C3 | |||
|
Domain Architecture |

|
|||||
| Description | Complement C3 precursor (HSE-MSF) [Contains: Complement C3 beta chain; Complement C3 alpha chain; C3a anaphylatoxin; Complement C3b alpha' chain; Complement C3c fragment; Complement C3dg fragment; Complement C3g fragment; Complement C3d fragment; Complement C3, isoform Short; Complement C3f fragment]. | |||||
|
CO5_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P01031 | Gene names | C5 | |||
|
Domain Architecture |

|
|||||
| Description | Complement C5 precursor [Contains: Complement C5 beta chain; Complement C5 alpha chain; C5a anaphylatoxin; Complement C5 alpha' chain]. | |||||
|
CO5_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.997130 (rank : 2) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P06684 | Gene names | C5, Hc | |||
|
Domain Architecture |

|
|||||
| Description | Complement C5 precursor (Hemolytic complement) [Contains: Complement C5 beta chain; Complement C5 alpha chain; C5a anaphylatoxin; Complement C5 alpha' chain]. | |||||
|
CO4B_MOUSE
|
||||||
| θ value | 6.19918e-134 (rank : 5) | NC score | 0.923614 (rank : 5) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P01029, Q31201, Q61372, Q61859, Q62353 | Gene names | C4b, C4 | |||
|
Domain Architecture |

|
|||||
| Description | Complement C4-B precursor [Contains: Complement C4 beta chain; Complement C4 alpha chain; C4a anaphylatoxin; Complement C4 gamma chain]. | |||||
|
CO4B_HUMAN
|
||||||
| θ value | 1.21547e-113 (rank : 6) | NC score | 0.917772 (rank : 6) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P0C0L5, P01028, P78445, Q13160, Q13906, Q14033, Q14835, Q9NPK5, Q9UIP5 | Gene names | C4B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complement C4-B precursor (Basic complement C4) [Contains: Complement C4 beta chain; Complement C4-B alpha chain; C4a anaphylatoxin; C4b-B; C4d-B; Complement C4 gamma chain]. | |||||
|
CO4A_HUMAN
|
||||||
| θ value | 2.07327e-113 (rank : 7) | NC score | 0.917608 (rank : 7) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P0C0L4, P01028, P78445, Q13160, Q13906, Q14033, Q14835, Q5JQM8, Q9NPK5, Q9UIP5 | Gene names | C4A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complement C4-A precursor (Acidic complement C4) [Contains: Complement C4 beta chain; Complement C4-A alpha chain; C4a anaphylatoxin; C4b-A; C4d-A; Complement C4 gamma chain]. | |||||
|
A2MG_HUMAN
|
||||||
| θ value | 4.83544e-70 (rank : 8) | NC score | 0.831344 (rank : 8) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P01023, Q13677, Q59F47, Q5QTS0, Q68DN2, Q6PIY3, Q6PN97 | Gene names | A2M | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-2-macroglobulin precursor (Alpha-2-M). | |||||
|
MUG1_MOUSE
|
||||||
| θ value | 2.24614e-67 (rank : 9) | NC score | 0.816871 (rank : 9) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P28665 | Gene names | Mug1, Mug-1 | |||
|
Domain Architecture |

|
|||||
| Description | Murinoglobulin-1 precursor (MuG1). | |||||
|
MUG2_MOUSE
|
||||||
| θ value | 2.85459e-46 (rank : 10) | NC score | 0.793166 (rank : 12) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P28666 | Gene names | Mug2, Mug-2 | |||
|
Domain Architecture |

|
|||||
| Description | Murinoglobulin-2 precursor (MuG2). | |||||
|
PZP_HUMAN
|
||||||
| θ value | 2.41655e-45 (rank : 11) | NC score | 0.812638 (rank : 10) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P20742, Q15273 | Gene names | PZP | |||
|
Domain Architecture |

|
|||||
| Description | Pregnancy zone protein precursor. | |||||
|
A2MG_MOUSE
|
||||||
| θ value | 3.61106e-41 (rank : 12) | NC score | 0.799315 (rank : 11) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q61838, Q60628 | Gene names | A2m | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-2-macroglobulin precursor (Alpha-2-M) [Contains: Alpha-2- macroglobulin 165 kDa subunit; Alpha-2-macroglobulin 35 kDa subunit]. | |||||
|
ITAX_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 13) | NC score | 0.013888 (rank : 15) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P20702, Q8IVA6 | Gene names | ITGAX, CD11C | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-X precursor (Leukocyte adhesion glycoprotein p150,95 alpha chain) (Leukocyte adhesion receptor p150,95) (Leu M5) (CD11c antigen). | |||||
|
DHX57_MOUSE
|
||||||
| θ value | 0.62314 (rank : 14) | NC score | 0.012142 (rank : 16) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6P5D3, Q3TS93, Q6NZK4, Q6P1B4, Q8BI63, Q8BIA2, Q8R360 | Gene names | Dhx57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX57 (EC 3.6.1.-) (DEAH box protein 57). | |||||
|
IL6RB_HUMAN
|
||||||
| θ value | 1.06291 (rank : 15) | NC score | 0.019642 (rank : 13) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P40189, Q9UQ41 | Gene names | IL6ST | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-6 receptor beta chain precursor (IL-6R-beta) (Interleukin 6 signal transducer) (Membrane glycoprotein 130) (gp130) (Oncostatin M receptor) (CD130 antigen) (CDw130). | |||||
|
CTR3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 16) | NC score | 0.009110 (rank : 17) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P70423 | Gene names | Slc7a3, Atrc3, Cat3 | |||
|
Domain Architecture |

|
|||||
| Description | Cationic amino acid transporter 3 (CAT-3) (Solute carrier family 7 member 3) (Cationic amino acid transporter y+). | |||||
|
DLDH_HUMAN
|
||||||
| θ value | 3.0926 (rank : 17) | NC score | 0.016750 (rank : 14) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P09622, Q14131, Q14167 | Gene names | DLD, GCSL, LAD, PHE3 | |||
|
Domain Architecture |

|
|||||
| Description | Dihydrolipoyl dehydrogenase, mitochondrial precursor (EC 1.8.1.4) (Dihydrolipoamide dehydrogenase) (Glycine cleavage system L protein). | |||||
|
KREM1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 18) | NC score | 0.008343 (rank : 18) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99N43 | Gene names | Kremen1, Kremen | |||
|
Domain Architecture |
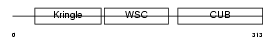
|
|||||
| Description | Kremen protein 1 precursor (Kringle-containing protein marking the eye and the nose) (Kringle domain-containing transmembrane protein 1) (Dickkopf receptor). | |||||
|
SSDH_HUMAN
|
||||||
| θ value | 4.03905 (rank : 19) | NC score | 0.006378 (rank : 24) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P51649 | Gene names | ALDH5A1, SSADH | |||
|
Domain Architecture |

|
|||||
| Description | Succinate semialdehyde dehydrogenase, mitochondrial precursor (EC 1.2.1.24) (NAD(+)-dependent succinic semialdehyde dehydrogenase). | |||||
|
PCTK2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 20) | NC score | -0.001471 (rank : 25) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 837 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q00537, Q8NEB8 | Gene names | PCTK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PCTAIRE-2 (EC 2.7.11.22) (PCTAIRE- motif protein kinase 2). | |||||
|
MGR1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 21) | NC score | 0.006962 (rank : 20) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13255, Q13256, Q14757, Q14758, Q5VTF7, Q5VTF8, Q9NU10, Q9UGS9, Q9UGT0 | Gene names | GRM1, GPRC1A, MGLUR1 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 1 precursor (mGluR1). | |||||
|
MGR1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 22) | NC score | 0.006952 (rank : 21) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P97772, Q6AXG4, Q9EPV6 | Gene names | Grm1, Gprc1a, Mglur1 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 1 precursor (mGluR1). | |||||
|
TMC1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 23) | NC score | 0.006591 (rank : 22) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TDI8 | Gene names | TMC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane cochlear-expressed protein 1. | |||||
|
K0586_HUMAN
|
||||||
| θ value | 8.99809 (rank : 24) | NC score | 0.007402 (rank : 19) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BVV6, O60328 | Gene names | KIAA0586 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0586. | |||||
|
MGR5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 25) | NC score | 0.006381 (rank : 23) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P41594 | Gene names | GRM5, GPRC1E, MGLUR5 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 5 precursor (mGluR5). | |||||
|
CO5_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P01031 | Gene names | C5 | |||
|
Domain Architecture |

|
|||||
| Description | Complement C5 precursor [Contains: Complement C5 beta chain; Complement C5 alpha chain; C5a anaphylatoxin; Complement C5 alpha' chain]. | |||||
|
CO5_MOUSE
|
||||||
| NC score | 0.997130 (rank : 2) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P06684 | Gene names | C5, Hc | |||
|
Domain Architecture |

|
|||||
| Description | Complement C5 precursor (Hemolytic complement) [Contains: Complement C5 beta chain; Complement C5 alpha chain; C5a anaphylatoxin; Complement C5 alpha' chain]. | |||||
|
CO3_HUMAN
|
||||||
| NC score | 0.954731 (rank : 3) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P01024 | Gene names | C3 | |||
|
Domain Architecture |

|
|||||
| Description | Complement C3 precursor [Contains: Complement C3 beta chain; Complement C3 alpha chain; C3a anaphylatoxin; Complement C3b alpha' chain; Complement C3c fragment; Complement C3dg fragment; Complement C3g fragment; Complement C3d fragment; Complement C3f fragment]. | |||||
|
CO3_MOUSE
|
||||||
| NC score | 0.954435 (rank : 4) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P01027, Q61370 | Gene names | C3 | |||
|
Domain Architecture |

|
|||||
| Description | Complement C3 precursor (HSE-MSF) [Contains: Complement C3 beta chain; Complement C3 alpha chain; C3a anaphylatoxin; Complement C3b alpha' chain; Complement C3c fragment; Complement C3dg fragment; Complement C3g fragment; Complement C3d fragment; Complement C3, isoform Short; Complement C3f fragment]. | |||||
|
CO4B_MOUSE
|
||||||
| NC score | 0.923614 (rank : 5) | θ value | 6.19918e-134 (rank : 5) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P01029, Q31201, Q61372, Q61859, Q62353 | Gene names | C4b, C4 | |||
|
Domain Architecture |

|
|||||
| Description | Complement C4-B precursor [Contains: Complement C4 beta chain; Complement C4 alpha chain; C4a anaphylatoxin; Complement C4 gamma chain]. | |||||
|
CO4B_HUMAN
|
||||||
| NC score | 0.917772 (rank : 6) | θ value | 1.21547e-113 (rank : 6) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P0C0L5, P01028, P78445, Q13160, Q13906, Q14033, Q14835, Q9NPK5, Q9UIP5 | Gene names | C4B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complement C4-B precursor (Basic complement C4) [Contains: Complement C4 beta chain; Complement C4-B alpha chain; C4a anaphylatoxin; C4b-B; C4d-B; Complement C4 gamma chain]. | |||||
|
CO4A_HUMAN
|
||||||
| NC score | 0.917608 (rank : 7) | θ value | 2.07327e-113 (rank : 7) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P0C0L4, P01028, P78445, Q13160, Q13906, Q14033, Q14835, Q5JQM8, Q9NPK5, Q9UIP5 | Gene names | C4A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complement C4-A precursor (Acidic complement C4) [Contains: Complement C4 beta chain; Complement C4-A alpha chain; C4a anaphylatoxin; C4b-A; C4d-A; Complement C4 gamma chain]. | |||||
|
A2MG_HUMAN
|
||||||
| NC score | 0.831344 (rank : 8) | θ value | 4.83544e-70 (rank : 8) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P01023, Q13677, Q59F47, Q5QTS0, Q68DN2, Q6PIY3, Q6PN97 | Gene names | A2M | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-2-macroglobulin precursor (Alpha-2-M). | |||||
|
MUG1_MOUSE
|
||||||
| NC score | 0.816871 (rank : 9) | θ value | 2.24614e-67 (rank : 9) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P28665 | Gene names | Mug1, Mug-1 | |||
|
Domain Architecture |

|
|||||
| Description | Murinoglobulin-1 precursor (MuG1). | |||||
|
PZP_HUMAN
|
||||||
| NC score | 0.812638 (rank : 10) | θ value | 2.41655e-45 (rank : 11) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P20742, Q15273 | Gene names | PZP | |||
|
Domain Architecture |

|
|||||
| Description | Pregnancy zone protein precursor. | |||||
|
A2MG_MOUSE
|
||||||
| NC score | 0.799315 (rank : 11) | θ value | 3.61106e-41 (rank : 12) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q61838, Q60628 | Gene names | A2m | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-2-macroglobulin precursor (Alpha-2-M) [Contains: Alpha-2- macroglobulin 165 kDa subunit; Alpha-2-macroglobulin 35 kDa subunit]. | |||||
|
MUG2_MOUSE
|
||||||
| NC score | 0.793166 (rank : 12) | θ value | 2.85459e-46 (rank : 10) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P28666 | Gene names | Mug2, Mug-2 | |||
|
Domain Architecture |

|
|||||
| Description | Murinoglobulin-2 precursor (MuG2). | |||||
|
IL6RB_HUMAN
|
||||||
| NC score | 0.019642 (rank : 13) | θ value | 1.06291 (rank : 15) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P40189, Q9UQ41 | Gene names | IL6ST | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-6 receptor beta chain precursor (IL-6R-beta) (Interleukin 6 signal transducer) (Membrane glycoprotein 130) (gp130) (Oncostatin M receptor) (CD130 antigen) (CDw130). | |||||
|
DLDH_HUMAN
|
||||||
| NC score | 0.016750 (rank : 14) | θ value | 3.0926 (rank : 17) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P09622, Q14131, Q14167 | Gene names | DLD, GCSL, LAD, PHE3 | |||
|
Domain Architecture |

|
|||||
| Description | Dihydrolipoyl dehydrogenase, mitochondrial precursor (EC 1.8.1.4) (Dihydrolipoamide dehydrogenase) (Glycine cleavage system L protein). | |||||
|
ITAX_HUMAN
|
||||||
| NC score | 0.013888 (rank : 15) | θ value | 0.0961366 (rank : 13) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P20702, Q8IVA6 | Gene names | ITGAX, CD11C | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-X precursor (Leukocyte adhesion glycoprotein p150,95 alpha chain) (Leukocyte adhesion receptor p150,95) (Leu M5) (CD11c antigen). | |||||
|
DHX57_MOUSE
|
||||||
| NC score | 0.012142 (rank : 16) | θ value | 0.62314 (rank : 14) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6P5D3, Q3TS93, Q6NZK4, Q6P1B4, Q8BI63, Q8BIA2, Q8R360 | Gene names | Dhx57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX57 (EC 3.6.1.-) (DEAH box protein 57). | |||||
|
CTR3_MOUSE
|
||||||
| NC score | 0.009110 (rank : 17) | θ value | 3.0926 (rank : 16) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P70423 | Gene names | Slc7a3, Atrc3, Cat3 | |||
|
Domain Architecture |

|
|||||
| Description | Cationic amino acid transporter 3 (CAT-3) (Solute carrier family 7 member 3) (Cationic amino acid transporter y+). | |||||
|
KREM1_MOUSE
|
||||||
| NC score | 0.008343 (rank : 18) | θ value | 4.03905 (rank : 18) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99N43 | Gene names | Kremen1, Kremen | |||
|
Domain Architecture |

|
|||||
| Description | Kremen protein 1 precursor (Kringle-containing protein marking the eye and the nose) (Kringle domain-containing transmembrane protein 1) (Dickkopf receptor). | |||||
|
K0586_HUMAN
|
||||||
| NC score | 0.007402 (rank : 19) | θ value | 8.99809 (rank : 24) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BVV6, O60328 | Gene names | KIAA0586 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0586. | |||||
|
MGR1_HUMAN
|
||||||
| NC score | 0.006962 (rank : 20) | θ value | 6.88961 (rank : 21) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13255, Q13256, Q14757, Q14758, Q5VTF7, Q5VTF8, Q9NU10, Q9UGS9, Q9UGT0 | Gene names | GRM1, GPRC1A, MGLUR1 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 1 precursor (mGluR1). | |||||
|
MGR1_MOUSE
|
||||||
| NC score | 0.006952 (rank : 21) | θ value | 6.88961 (rank : 22) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P97772, Q6AXG4, Q9EPV6 | Gene names | Grm1, Gprc1a, Mglur1 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 1 precursor (mGluR1). | |||||
|
TMC1_HUMAN
|
||||||
| NC score | 0.006591 (rank : 22) | θ value | 6.88961 (rank : 23) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TDI8 | Gene names | TMC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane cochlear-expressed protein 1. | |||||
|
MGR5_HUMAN
|
||||||
| NC score | 0.006381 (rank : 23) | θ value | 8.99809 (rank : 25) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P41594 | Gene names | GRM5, GPRC1E, MGLUR5 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 5 precursor (mGluR5). | |||||
|
SSDH_HUMAN
|
||||||
| NC score | 0.006378 (rank : 24) | θ value | 4.03905 (rank : 19) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P51649 | Gene names | ALDH5A1, SSADH | |||
|
Domain Architecture |

|
|||||
| Description | Succinate semialdehyde dehydrogenase, mitochondrial precursor (EC 1.2.1.24) (NAD(+)-dependent succinic semialdehyde dehydrogenase). | |||||
|
PCTK2_HUMAN
|
||||||
| NC score | -0.001471 (rank : 25) | θ value | 5.27518 (rank : 20) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 837 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q00537, Q8NEB8 | Gene names | PCTK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PCTAIRE-2 (EC 2.7.11.22) (PCTAIRE- motif protein kinase 2). | |||||