Please be patient as the page loads
|
CLD19_HUMAN
|
||||||
| SwissProt Accessions | Q8N6F1, Q8N8X0 | Gene names | CLDN19 | |||
|
Domain Architecture |

|
|||||
| Description | Claudin-19. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CLD19_MOUSE
|
||||||
| θ value | 2.06847e-121 (rank : 1) | NC score | 0.996180 (rank : 2) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9ET38, Q8R4B7 | Gene names | Cldn19 | |||
|
Domain Architecture |

|
|||||
| Description | Claudin-19. | |||||
|
CLD19_HUMAN
|
||||||
| θ value | 1.2549e-118 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q8N6F1, Q8N8X0 | Gene names | CLDN19 | |||
|
Domain Architecture |

|
|||||
| Description | Claudin-19. | |||||
|
CLD7_HUMAN
|
||||||
| θ value | 6.11685e-65 (rank : 3) | NC score | 0.985818 (rank : 6) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O95471, Q6IPN3, Q7Z4Y7, Q9BVN0 | Gene names | CLDN7 | |||
|
Domain Architecture |

|
|||||
| Description | Claudin-7 (CLDN-7). | |||||
|
CLD1_HUMAN
|
||||||
| θ value | 1.07972e-61 (rank : 4) | NC score | 0.991304 (rank : 3) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O95832 | Gene names | CLDN1, CLD1, SEMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Claudin-1 (Senescence-associated epithelial membrane protein). | |||||
|
CLD1_MOUSE
|
||||||
| θ value | 3.47335e-60 (rank : 5) | NC score | 0.990988 (rank : 4) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O88551 | Gene names | Cldn1 | |||
|
Domain Architecture |

|
|||||
| Description | Claudin-1. | |||||
|
CLD7_MOUSE
|
||||||
| θ value | 5.0155e-59 (rank : 6) | NC score | 0.988385 (rank : 5) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9Z261, Q3TJX4 | Gene names | Cldn7 | |||
|
Domain Architecture |

|
|||||
| Description | Claudin-7. | |||||
|
CLD4_MOUSE
|
||||||
| θ value | 8.87376e-48 (rank : 7) | NC score | 0.973589 (rank : 15) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O35054 | Gene names | Cldn4, Cper, Cpetr1 | |||
|
Domain Architecture |

|
|||||
| Description | Claudin-4 (Clostridium perfringens enterotoxin receptor) (CPE- receptor) (CPE-R). | |||||
|
CLD9_HUMAN
|
||||||
| θ value | 3.48949e-44 (rank : 8) | NC score | 0.977097 (rank : 9) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O95484 | Gene names | CLDN9 | |||
|
Domain Architecture |
|
|||||
| Description | Claudin-9. | |||||
|
CLD3_HUMAN
|
||||||
| θ value | 1.73182e-43 (rank : 9) | NC score | 0.975814 (rank : 13) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O15551 | Gene names | CLDN3, C7orf1, CPETR2 | |||
|
Domain Architecture |

|
|||||
| Description | Claudin-3 (Clostridium perfringens enterotoxin receptor 2) (CPE- receptor 2) (CPE-R 2) (Ventral prostate.1 protein homolog) (HRVP1). | |||||
|
CLD9_MOUSE
|
||||||
| θ value | 2.50074e-42 (rank : 10) | NC score | 0.976866 (rank : 10) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9Z0S7 | Gene names | Cldn9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Claudin-9. | |||||
|
CLD4_HUMAN
|
||||||
| θ value | 3.26607e-42 (rank : 11) | NC score | 0.976201 (rank : 11) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O14493 | Gene names | CLDN4, CPER, CPETR1, WBSCR8 | |||
|
Domain Architecture |

|
|||||
| Description | Claudin-4 (Clostridium perfringens enterotoxin receptor) (CPE- receptor) (CPE-R) (Williams-Beuren syndrome chromosome region 8 protein). | |||||
|
CLD6_HUMAN
|
||||||
| θ value | 3.26607e-42 (rank : 12) | NC score | 0.973157 (rank : 16) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P56747 | Gene names | CLDN6 | |||
|
Domain Architecture |

|
|||||
| Description | Claudin-6 (Skullin 2). | |||||
|
CLD5_HUMAN
|
||||||
| θ value | 1.62093e-41 (rank : 13) | NC score | 0.971302 (rank : 18) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O00501, Q53XW2, Q8WUW3 | Gene names | CLDN5, TMVCF | |||
|
Domain Architecture |

|
|||||
| Description | Claudin-5 (Transmembrane protein deleted in VCFS) (TMDVCF). | |||||
|
CLD3_MOUSE
|
||||||
| θ value | 3.61106e-41 (rank : 14) | NC score | 0.974030 (rank : 14) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9Z0G9, Q91X40 | Gene names | Cldn3, Cpetr2 | |||
|
Domain Architecture |

|
|||||
| Description | Claudin-3 (Clostridium perfringens enterotoxin receptor 2) (CPE- receptor 2) (CPE-R 2). | |||||
|
CLD6_MOUSE
|
||||||
| θ value | 4.71619e-41 (rank : 15) | NC score | 0.972858 (rank : 17) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9Z262 | Gene names | Cldn6 | |||
|
Domain Architecture |

|
|||||
| Description | Claudin-6 (Skullin). | |||||
|
CLD5_MOUSE
|
||||||
| θ value | 1.05066e-40 (rank : 16) | NC score | 0.970933 (rank : 20) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O54942, O88789 | Gene names | Cldn5, Bec1 | |||
|
Domain Architecture |

|
|||||
| Description | Claudin-5 (Brain endothelial cell clone 1 protein) (Lung-specific membrane protein). | |||||
|
CLD14_MOUSE
|
||||||
| θ value | 1.79215e-40 (rank : 17) | NC score | 0.977886 (rank : 8) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9Z0S3, Q9D284 | Gene names | Cldn14 | |||
|
Domain Architecture |

|
|||||
| Description | Claudin-14. | |||||
|
CLD14_HUMAN
|
||||||
| θ value | 1.16164e-39 (rank : 18) | NC score | 0.979730 (rank : 7) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O95500 | Gene names | CLDN14 | |||
|
Domain Architecture |

|
|||||
| Description | Claudin-14. | |||||
|
CLD10_MOUSE
|
||||||
| θ value | 2.58786e-39 (rank : 19) | NC score | 0.960177 (rank : 27) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9Z0S6 | Gene names | Cldn10 | |||
|
Domain Architecture |

|
|||||
| Description | Claudin-10. | |||||
|
CLD10_HUMAN
|
||||||
| θ value | 4.41421e-39 (rank : 20) | NC score | 0.963707 (rank : 25) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P78369 | Gene names | CLDN10 | |||
|
Domain Architecture |

|
|||||
| Description | Claudin-10 (OSP-like protein). | |||||
|
CLD17_HUMAN
|
||||||
| θ value | 7.79185e-36 (rank : 21) | NC score | 0.965213 (rank : 24) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P56750, Q6UY37 | Gene names | CLDN17 | |||
|
Domain Architecture |

|
|||||
| Description | Claudin-17. | |||||
|
CLD8_HUMAN
|
||||||
| θ value | 1.32909e-35 (rank : 22) | NC score | 0.971168 (rank : 19) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P56748 | Gene names | CLDN8 | |||
|
Domain Architecture |

|
|||||
| Description | Claudin-8. | |||||
|
CLD2_HUMAN
|
||||||
| θ value | 2.26708e-35 (rank : 23) | NC score | 0.968374 (rank : 22) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P57739 | Gene names | CLDN2 | |||
|
Domain Architecture |
|
|||||
| Description | Claudin-2 (SP82). | |||||
|
CLD18_HUMAN
|
||||||
| θ value | 5.0505e-35 (rank : 24) | NC score | 0.956305 (rank : 30) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P56856, Q96PH4 | Gene names | CLDN18 | |||
|
Domain Architecture |

|
|||||
| Description | Claudin-18. | |||||
|
CLD20_HUMAN
|
||||||
| θ value | 6.59618e-35 (rank : 25) | NC score | 0.975961 (rank : 12) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P56880 | Gene names | CLDN20 | |||
|
Domain Architecture |

|
|||||
| Description | Claudin-20. | |||||
|
CLD8_MOUSE
|
||||||
| θ value | 4.27553e-34 (rank : 26) | NC score | 0.970149 (rank : 21) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9Z260 | Gene names | Cldn8 | |||
|
Domain Architecture |

|
|||||
| Description | Claudin-8. | |||||
|
CLD15_HUMAN
|
||||||
| θ value | 9.52487e-34 (rank : 27) | NC score | 0.959337 (rank : 29) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P56746 | Gene names | CLDN15 | |||
|
Domain Architecture |

|
|||||
| Description | Claudin-15. | |||||
|
CLD17_MOUSE
|
||||||
| θ value | 9.52487e-34 (rank : 28) | NC score | 0.962935 (rank : 26) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8BXA6 | Gene names | Cldn17 | |||
|
Domain Architecture |
|
|||||
| Description | Claudin-17. | |||||
|
CLD2_MOUSE
|
||||||
| θ value | 9.52487e-34 (rank : 29) | NC score | 0.967952 (rank : 23) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O88552 | Gene names | Cldn2 | |||
|
Domain Architecture |

|
|||||
| Description | Claudin-2. | |||||
|
CLD15_MOUSE
|
||||||
| θ value | 6.17384e-33 (rank : 30) | NC score | 0.959654 (rank : 28) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9Z0S5, Q9D7Z0, Q9D8A6 | Gene names | Cldn15 | |||
|
Domain Architecture |

|
|||||
| Description | Claudin-15. | |||||
|
CLD18_MOUSE
|
||||||
| θ value | 3.74554e-30 (rank : 31) | NC score | 0.947697 (rank : 31) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P56857, Q91ZY9, Q91ZZ0, Q91ZZ1 | Gene names | Cldn18 | |||
|
Domain Architecture |

|
|||||
| Description | Claudin-18. | |||||
|
CLD22_HUMAN
|
||||||
| θ value | 1.09232e-21 (rank : 32) | NC score | 0.931435 (rank : 33) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8N7P3 | Gene names | CLDN22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Claudin-22. | |||||
|
CLD22_MOUSE
|
||||||
| θ value | 7.82807e-20 (rank : 33) | NC score | 0.932673 (rank : 32) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9D7U6 | Gene names | Cldn22 | |||
|
Domain Architecture |
|
|||||
| Description | Claudin-22. | |||||
|
CLD11_HUMAN
|
||||||
| θ value | 1.47631e-18 (rank : 34) | NC score | 0.845000 (rank : 36) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O75508, Q5U0P3 | Gene names | CLDN11, OSP | |||
|
Domain Architecture |

|
|||||
| Description | Claudin-11 (Oligodendrocyte-specific protein). | |||||
|
CLD11_MOUSE
|
||||||
| θ value | 3.63628e-17 (rank : 35) | NC score | 0.858100 (rank : 35) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q60771, Q545N5, Q9DB65 | Gene names | Cldn11, Osp, Otm | |||
|
Domain Architecture |

|
|||||
| Description | Claudin-11 (Oligodendrocyte transmembrane protein) (Oligodendrocyte- specific protein). | |||||
|
CLD13_MOUSE
|
||||||
| θ value | 3.76295e-14 (rank : 36) | NC score | 0.909384 (rank : 34) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9Z0S4 | Gene names | Cldn13 | |||
|
Domain Architecture |

|
|||||
| Description | Claudin-13. | |||||
|
CLD16_MOUSE
|
||||||
| θ value | 2.36244e-08 (rank : 37) | NC score | 0.732045 (rank : 37) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q925N4, Q542L7 | Gene names | Cldn16 | |||
|
Domain Architecture |
|
|||||
| Description | Claudin-16 (Paracellin-1). | |||||
|
CLD12_MOUSE
|
||||||
| θ value | 5.81887e-07 (rank : 38) | NC score | 0.432054 (rank : 42) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9ET43, Q3TM65, Q8BH13 | Gene names | Cldn12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Claudin-12. | |||||
|
CLD12_HUMAN
|
||||||
| θ value | 1.29631e-06 (rank : 39) | NC score | 0.449209 (rank : 41) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P56749 | Gene names | CLDN12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Claudin-12. | |||||
|
CLD16_HUMAN
|
||||||
| θ value | 2.21117e-06 (rank : 40) | NC score | 0.662371 (rank : 38) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9Y5I7 | Gene names | CLDN16, PCLN1 | |||
|
Domain Architecture |
|
|||||
| Description | Claudin-16 (Paracellin-1) (PCLN-1). | |||||
|
CLD23_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 41) | NC score | 0.603154 (rank : 40) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q96B33 | Gene names | CLDN23 | |||
|
Domain Architecture |
|
|||||
| Description | Claudin-23. | |||||
|
CLD23_MOUSE
|
||||||
| θ value | 3.19293e-05 (rank : 42) | NC score | 0.654093 (rank : 39) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9D7D7 | Gene names | Cldn23 | |||
|
Domain Architecture |

|
|||||
| Description | Claudin-23. | |||||
|
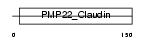
PMP22_MOUSE
|
||||||
| θ value | 0.00035302 (rank : 43) | NC score | 0.228743 (rank : 43) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P16646, Q91XF9 | Gene names | Pmp22, Gas-3, Pmp-22 | |||
|
Domain Architecture |
|
|||||
| Description | Peripheral myelin protein 22 (PMP-22) (Growth-arrest-specific protein 3) (GAS3). | |||||
|
LMIP_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 44) | NC score | 0.137878 (rank : 45) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P56563, Q3TNV8, Q99PA6 | Gene names | Lim2 | |||
|
Domain Architecture |

|
|||||
| Description | Lens fiber membrane intrinsic protein (MP17) (MP18) (MP19) (MP20). | |||||
|
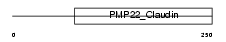
PMP22_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 45) | NC score | 0.152837 (rank : 44) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q01453, Q8WV01 | Gene names | PMP22, GAS3 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral myelin protein 22 (PMP-22). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 0.163984 (rank : 46) | NC score | 0.005642 (rank : 67) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
CLDND_HUMAN
|
||||||
| θ value | 0.365318 (rank : 47) | NC score | 0.076658 (rank : 50) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NY35, Q502Y8, Q6UVX2, Q9BUZ9, Q9NZZ5, Q9Y4S9 | Gene names | CLDND1, C3orf4 | |||
|
Domain Architecture |

|
|||||
| Description | Claudin domain-containing protein 1 (Membrane protein GENX-3745). | |||||
|
GPTC3_MOUSE
|
||||||
| θ value | 0.47712 (rank : 48) | NC score | 0.021017 (rank : 58) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BIY1 | Gene names | Gpatc3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 3. | |||||
|
EMP1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 49) | NC score | 0.106466 (rank : 46) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P54849, O00681, Q13481, Q13834 | Gene names | EMP1, B4B, TMP | |||
|
Domain Architecture |

|
|||||
| Description | Epithelial membrane protein 1 (EMP-1) (Tumor-associated membrane protein) (CL-20) (B4B protein). | |||||
|
CLDND_MOUSE
|
||||||
| θ value | 1.38821 (rank : 50) | NC score | 0.065117 (rank : 55) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9CQX5, Q3U7C4 | Gene names | Cldnd1, D11Moh34 | |||
|
Domain Architecture |

|
|||||
| Description | Claudin domain-containing protein 1. | |||||
|
CCG3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 51) | NC score | 0.071611 (rank : 51) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O60359 | Gene names | CACNG3 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent calcium channel gamma-3 subunit (Neuronal voltage- gated calcium channel gamma-3 subunit). | |||||
|
CO039_HUMAN
|
||||||
| θ value | 4.03905 (rank : 52) | NC score | 0.008213 (rank : 63) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6ZRI6, Q71JB1, Q7L3S0, Q8N3F2, Q96FB6, Q9NTU5 | Gene names | C15orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf39. | |||||
|
MILK2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 53) | NC score | 0.002901 (rank : 69) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IY33, Q7RTP4, Q7Z655, Q8TEQ4, Q9H5F9 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | MICAL-like protein 2. | |||||
|
SOCS7_MOUSE
|
||||||
| θ value | 4.03905 (rank : 54) | NC score | 0.006776 (rank : 66) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VHQ2 | Gene names | Socs7, Cish7, Nap4 | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of cytokine signaling 7 (SOCS-7). | |||||
|
ACL6B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 55) | NC score | 0.008434 (rank : 61) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O94805, O75421 | Gene names | ACTL6B, ACTL6, BAF53B | |||
|
Domain Architecture |
|
|||||
| Description | Actin-like protein 6B (53 kDa BRG1-associated factor B) (Actin-related protein Baf53b) (ArpNalpha). | |||||
|
ACL6B_MOUSE
|
||||||
| θ value | 5.27518 (rank : 56) | NC score | 0.008433 (rank : 62) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99MR0 | Gene names | Actl6b, Actl6, ArpNa, Baf53b | |||
|
Domain Architecture |
|
|||||
| Description | Actin-like protein 6B (53 kDa BRG1-associated factor B) (Actin-related protein Baf53b). | |||||
|
WASIP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 57) | NC score | 0.003309 (rank : 68) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
ACL6A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 58) | NC score | 0.007510 (rank : 65) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O96019, Q8TAE5, Q9BVS8, Q9H0W6 | Gene names | ACTL6A, BAF53, BAF53A | |||
|
Domain Architecture |
|
|||||
| Description | Actin-like protein 6A (53 kDa BRG1-associated factor A) (Actin-related protein Baf53a) (ArpNbeta). | |||||
|
ACL6A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 59) | NC score | 0.007527 (rank : 64) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Z2N8, Q8C1W3, Q99M56 | Gene names | Actl6a, Actl6, Baf53a | |||
|
Domain Architecture |
|
|||||
| Description | Actin-like protein 6A (53 kDa BRG1-associated factor A) (Actin-related protein Baf53a). | |||||
|
CCG3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 60) | NC score | 0.062484 (rank : 56) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9JJV5 | Gene names | Cacng3 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent calcium channel gamma-3 subunit (Neuronal voltage- gated calcium channel gamma-3 subunit). | |||||
|
CF155_HUMAN
|
||||||
| θ value | 8.99809 (rank : 61) | NC score | 0.011170 (rank : 59) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H8W2, Q5TG13 | Gene names | C6orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf155. | |||||
|
FOG1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 62) | NC score | -0.004243 (rank : 71) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1237 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O35615 | Gene names | Zfpm1, Fog, Fog1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
K1802_HUMAN
|
||||||
| θ value | 8.99809 (rank : 63) | NC score | -0.003827 (rank : 70) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
PRR10_HUMAN
|
||||||
| θ value | 8.99809 (rank : 64) | NC score | 0.009767 (rank : 60) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N7Y1 | Gene names | PRR10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 10. | |||||
|
CCG2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.068797 (rank : 52) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y698, Q2M1M1, Q5TGT3, Q9UGZ7 | Gene names | CACNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent calcium channel gamma-2 subunit (Neuronal voltage- gated calcium channel gamma-2 subunit). | |||||
|
CCG2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.068780 (rank : 53) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O88602 | Gene names | Cacng2, Stg | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent calcium channel gamma-2 subunit (Neuronal voltage- gated calcium channel gamma-2 subunit) (Stargazin). | |||||
|
CCG5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.053302 (rank : 57) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UF02, Q8WXS7, Q9UHM3 | Gene names | CACNG5 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent calcium channel gamma-5 subunit (Neuronal voltage- gated calcium channel gamma-5 subunit). | |||||
|
CCG5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.067207 (rank : 54) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8VHW4 | Gene names | Cacng5 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent calcium channel gamma-5 subunit (Neuronal voltage- gated calcium channel gamma-5 subunit). | |||||
|
EMP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.083730 (rank : 48) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P54851 | Gene names | EMP2, XMP | |||
|
Domain Architecture |

|
|||||
| Description | Epithelial membrane protein 2 (EMP-2) (Protein XMP). | |||||
|
EMP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.085217 (rank : 47) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O88662 | Gene names | Emp2, Xmp | |||
|
Domain Architecture |

|
|||||
| Description | Epithelial membrane protein 2 (EMP-2) (Protein XMP). | |||||
|
LMIP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.077759 (rank : 49) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P55344, Q9BXD0, Q9HAR5 | Gene names | LIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Lens fiber membrane intrinsic protein (MP18) (MP19) (MP20). | |||||
|
CLD19_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 1.2549e-118 (rank : 2) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q8N6F1, Q8N8X0 | Gene names | CLDN19 | |||
|
Domain Architecture |

|
|||||
| Description | Claudin-19. | |||||
|
CLD19_MOUSE
|
||||||
| NC score | 0.996180 (rank : 2) | θ value | 2.06847e-121 (rank : 1) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9ET38, Q8R4B7 | Gene names | Cldn19 | |||
|
Domain Architecture |

|
|||||
| Description | Claudin-19. | |||||
|
CLD1_HUMAN
|
||||||
| NC score | 0.991304 (rank : 3) | θ value | 1.07972e-61 (rank : 4) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O95832 | Gene names | CLDN1, CLD1, SEMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Claudin-1 (Senescence-associated epithelial membrane protein). | |||||
|
CLD1_MOUSE
|
||||||
| NC score | 0.990988 (rank : 4) | θ value | 3.47335e-60 (rank : 5) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O88551 | Gene names | Cldn1 | |||
|
Domain Architecture |

|
|||||
| Description | Claudin-1. | |||||
|
CLD7_MOUSE
|
||||||
| NC score | 0.988385 (rank : 5) | θ value | 5.0155e-59 (rank : 6) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9Z261, Q3TJX4 | Gene names | Cldn7 | |||
|
Domain Architecture |

|
|||||
| Description | Claudin-7. | |||||
|
CLD7_HUMAN
|
||||||
| NC score | 0.985818 (rank : 6) | θ value | 6.11685e-65 (rank : 3) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O95471, Q6IPN3, Q7Z4Y7, Q9BVN0 | Gene names | CLDN7 | |||
|
Domain Architecture |

|
|||||
| Description | Claudin-7 (CLDN-7). | |||||
|
CLD14_HUMAN
|
||||||
| NC score | 0.979730 (rank : 7) | θ value | 1.16164e-39 (rank : 18) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O95500 | Gene names | CLDN14 | |||
|
Domain Architecture |

|
|||||
| Description | Claudin-14. | |||||
|
CLD14_MOUSE
|
||||||
| NC score | 0.977886 (rank : 8) | θ value | 1.79215e-40 (rank : 17) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9Z0S3, Q9D284 | Gene names | Cldn14 | |||
|
Domain Architecture |

|
|||||
| Description | Claudin-14. | |||||
|
CLD9_HUMAN
|
||||||
| NC score | 0.977097 (rank : 9) | θ value | 3.48949e-44 (rank : 8) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O95484 | Gene names | CLDN9 | |||
|
Domain Architecture |

|
|||||
| Description | Claudin-9. | |||||
|
CLD9_MOUSE
|
||||||
| NC score | 0.976866 (rank : 10) | θ value | 2.50074e-42 (rank : 10) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9Z0S7 | Gene names | Cldn9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Claudin-9. | |||||
|
CLD4_HUMAN
|
||||||
| NC score | 0.976201 (rank : 11) | θ value | 3.26607e-42 (rank : 11) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O14493 | Gene names | CLDN4, CPER, CPETR1, WBSCR8 | |||
|
Domain Architecture |

|
|||||
| Description | Claudin-4 (Clostridium perfringens enterotoxin receptor) (CPE- receptor) (CPE-R) (Williams-Beuren syndrome chromosome region 8 protein). | |||||
|
CLD20_HUMAN
|
||||||
| NC score | 0.975961 (rank : 12) | θ value | 6.59618e-35 (rank : 25) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P56880 | Gene names | CLDN20 | |||
|
Domain Architecture |

|
|||||
| Description | Claudin-20. | |||||
|
CLD3_HUMAN
|
||||||
| NC score | 0.975814 (rank : 13) | θ value | 1.73182e-43 (rank : 9) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O15551 | Gene names | CLDN3, C7orf1, CPETR2 | |||
|
Domain Architecture |

|
|||||
| Description | Claudin-3 (Clostridium perfringens enterotoxin receptor 2) (CPE- receptor 2) (CPE-R 2) (Ventral prostate.1 protein homolog) (HRVP1). | |||||
|
CLD3_MOUSE
|
||||||
| NC score | 0.974030 (rank : 14) | θ value | 3.61106e-41 (rank : 14) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9Z0G9, Q91X40 | Gene names | Cldn3, Cpetr2 | |||
|
Domain Architecture |

|
|||||
| Description | Claudin-3 (Clostridium perfringens enterotoxin receptor 2) (CPE- receptor 2) (CPE-R 2). | |||||
|
CLD4_MOUSE
|
||||||
| NC score | 0.973589 (rank : 15) | θ value | 8.87376e-48 (rank : 7) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O35054 | Gene names | Cldn4, Cper, Cpetr1 | |||
|
Domain Architecture |

|
|||||
| Description | Claudin-4 (Clostridium perfringens enterotoxin receptor) (CPE- receptor) (CPE-R). | |||||
|
CLD6_HUMAN
|
||||||
| NC score | 0.973157 (rank : 16) | θ value | 3.26607e-42 (rank : 12) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P56747 | Gene names | CLDN6 | |||
|
Domain Architecture |

|
|||||
| Description | Claudin-6 (Skullin 2). | |||||
|
CLD6_MOUSE
|
||||||
| NC score | 0.972858 (rank : 17) | θ value | 4.71619e-41 (rank : 15) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9Z262 | Gene names | Cldn6 | |||
|
Domain Architecture |

|
|||||
| Description | Claudin-6 (Skullin). | |||||
|
CLD5_HUMAN
|
||||||
| NC score | 0.971302 (rank : 18) | θ value | 1.62093e-41 (rank : 13) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O00501, Q53XW2, Q8WUW3 | Gene names | CLDN5, TMVCF | |||
|
Domain Architecture |

|
|||||
| Description | Claudin-5 (Transmembrane protein deleted in VCFS) (TMDVCF). | |||||
|
CLD8_HUMAN
|
||||||
| NC score | 0.971168 (rank : 19) | θ value | 1.32909e-35 (rank : 22) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P56748 | Gene names | CLDN8 | |||
|
Domain Architecture |

|
|||||
| Description | Claudin-8. | |||||
|
CLD5_MOUSE
|
||||||
| NC score | 0.970933 (rank : 20) | θ value | 1.05066e-40 (rank : 16) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O54942, O88789 | Gene names | Cldn5, Bec1 | |||
|
Domain Architecture |

|
|||||
| Description | Claudin-5 (Brain endothelial cell clone 1 protein) (Lung-specific membrane protein). | |||||
|
CLD8_MOUSE
|
||||||
| NC score | 0.970149 (rank : 21) | θ value | 4.27553e-34 (rank : 26) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9Z260 | Gene names | Cldn8 | |||
|
Domain Architecture |

|
|||||
| Description | Claudin-8. | |||||
|
CLD2_HUMAN
|
||||||
| NC score | 0.968374 (rank : 22) | θ value | 2.26708e-35 (rank : 23) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P57739 | Gene names | CLDN2 | |||
|
Domain Architecture |

|
|||||
| Description | Claudin-2 (SP82). | |||||
|
CLD2_MOUSE
|
||||||
| NC score | 0.967952 (rank : 23) | θ value | 9.52487e-34 (rank : 29) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O88552 | Gene names | Cldn2 | |||
|
Domain Architecture |

|
|||||
| Description | Claudin-2. | |||||
|
CLD17_HUMAN
|
||||||
| NC score | 0.965213 (rank : 24) | θ value | 7.79185e-36 (rank : 21) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P56750, Q6UY37 | Gene names | CLDN17 | |||
|
Domain Architecture |

|
|||||
| Description | Claudin-17. | |||||
|
CLD10_HUMAN
|
||||||
| NC score | 0.963707 (rank : 25) | θ value | 4.41421e-39 (rank : 20) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P78369 | Gene names | CLDN10 | |||
|
Domain Architecture |

|
|||||
| Description | Claudin-10 (OSP-like protein). | |||||
|
CLD17_MOUSE
|
||||||
| NC score | 0.962935 (rank : 26) | θ value | 9.52487e-34 (rank : 28) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8BXA6 | Gene names | Cldn17 | |||
|
Domain Architecture |

|
|||||
| Description | Claudin-17. | |||||
|
CLD10_MOUSE
|
||||||
| NC score | 0.960177 (rank : 27) | θ value | 2.58786e-39 (rank : 19) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9Z0S6 | Gene names | Cldn10 | |||
|
Domain Architecture |

|
|||||
| Description | Claudin-10. | |||||
|
CLD15_MOUSE
|
||||||
| NC score | 0.959654 (rank : 28) | θ value | 6.17384e-33 (rank : 30) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9Z0S5, Q9D7Z0, Q9D8A6 | Gene names | Cldn15 | |||
|
Domain Architecture |

|
|||||
| Description | Claudin-15. | |||||
|
CLD15_HUMAN
|
||||||
| NC score | 0.959337 (rank : 29) | θ value | 9.52487e-34 (rank : 27) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P56746 | Gene names | CLDN15 | |||
|
Domain Architecture |

|
|||||
| Description | Claudin-15. | |||||
|
CLD18_HUMAN
|
||||||
| NC score | 0.956305 (rank : 30) | θ value | 5.0505e-35 (rank : 24) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P56856, Q96PH4 | Gene names | CLDN18 | |||
|
Domain Architecture |

|
|||||
| Description | Claudin-18. | |||||
|
CLD18_MOUSE
|
||||||
| NC score | 0.947697 (rank : 31) | θ value | 3.74554e-30 (rank : 31) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P56857, Q91ZY9, Q91ZZ0, Q91ZZ1 | Gene names | Cldn18 | |||
|
Domain Architecture |

|
|||||
| Description | Claudin-18. | |||||
|
CLD22_MOUSE
|
||||||
| NC score | 0.932673 (rank : 32) | θ value | 7.82807e-20 (rank : 33) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9D7U6 | Gene names | Cldn22 | |||
|
Domain Architecture |

|
|||||
| Description | Claudin-22. | |||||
|
CLD22_HUMAN
|
||||||
| NC score | 0.931435 (rank : 33) | θ value | 1.09232e-21 (rank : 32) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8N7P3 | Gene names | CLDN22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Claudin-22. | |||||
|
CLD13_MOUSE
|
||||||
| NC score | 0.909384 (rank : 34) | θ value | 3.76295e-14 (rank : 36) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9Z0S4 | Gene names | Cldn13 | |||
|
Domain Architecture |

|
|||||
| Description | Claudin-13. | |||||
|
CLD11_MOUSE
|
||||||
| NC score | 0.858100 (rank : 35) | θ value | 3.63628e-17 (rank : 35) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q60771, Q545N5, Q9DB65 | Gene names | Cldn11, Osp, Otm | |||
|
Domain Architecture |

|
|||||
| Description | Claudin-11 (Oligodendrocyte transmembrane protein) (Oligodendrocyte- specific protein). | |||||
|
CLD11_HUMAN
|
||||||
| NC score | 0.845000 (rank : 36) | θ value | 1.47631e-18 (rank : 34) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O75508, Q5U0P3 | Gene names | CLDN11, OSP | |||
|
Domain Architecture |

|
|||||
| Description | Claudin-11 (Oligodendrocyte-specific protein). | |||||
|
CLD16_MOUSE
|
||||||
| NC score | 0.732045 (rank : 37) | θ value | 2.36244e-08 (rank : 37) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q925N4, Q542L7 | Gene names | Cldn16 | |||
|
Domain Architecture |

|
|||||
| Description | Claudin-16 (Paracellin-1). | |||||
|
CLD16_HUMAN
|
||||||
| NC score | 0.662371 (rank : 38) | θ value | 2.21117e-06 (rank : 40) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9Y5I7 | Gene names | CLDN16, PCLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Claudin-16 (Paracellin-1) (PCLN-1). | |||||
|
CLD23_MOUSE
|
||||||
| NC score | 0.654093 (rank : 39) | θ value | 3.19293e-05 (rank : 42) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9D7D7 | Gene names | Cldn23 | |||
|
Domain Architecture |

|
|||||
| Description | Claudin-23. | |||||
|
CLD23_HUMAN
|
||||||
| NC score | 0.603154 (rank : 40) | θ value | 2.44474e-05 (rank : 41) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q96B33 | Gene names | CLDN23 | |||
|
Domain Architecture |

|
|||||
| Description | Claudin-23. | |||||
|
CLD12_HUMAN
|
||||||
| NC score | 0.449209 (rank : 41) | θ value | 1.29631e-06 (rank : 39) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P56749 | Gene names | CLDN12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Claudin-12. | |||||
|
CLD12_MOUSE
|
||||||
| NC score | 0.432054 (rank : 42) | θ value | 5.81887e-07 (rank : 38) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9ET43, Q3TM65, Q8BH13 | Gene names | Cldn12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Claudin-12. | |||||
|
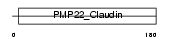
PMP22_MOUSE
|
||||||
| NC score | 0.228743 (rank : 43) | θ value | 0.00035302 (rank : 43) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P16646, Q91XF9 | Gene names | Pmp22, Gas-3, Pmp-22 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral myelin protein 22 (PMP-22) (Growth-arrest-specific protein 3) (GAS3). | |||||
|
PMP22_HUMAN
|
||||||
| NC score | 0.152837 (rank : 44) | θ value | 0.0563607 (rank : 45) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q01453, Q8WV01 | Gene names | PMP22, GAS3 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral myelin protein 22 (PMP-22). | |||||
|
LMIP_MOUSE
|
||||||
| NC score | 0.137878 (rank : 45) | θ value | 0.0113563 (rank : 44) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P56563, Q3TNV8, Q99PA6 | Gene names | Lim2 | |||
|
Domain Architecture |

|
|||||
| Description | Lens fiber membrane intrinsic protein (MP17) (MP18) (MP19) (MP20). | |||||
|
EMP1_HUMAN
|
||||||
| NC score | 0.106466 (rank : 46) | θ value | 0.62314 (rank : 49) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P54849, O00681, Q13481, Q13834 | Gene names | EMP1, B4B, TMP | |||
|
Domain Architecture |

|
|||||
| Description | Epithelial membrane protein 1 (EMP-1) (Tumor-associated membrane protein) (CL-20) (B4B protein). | |||||
|
EMP2_MOUSE
|
||||||
| NC score | 0.085217 (rank : 47) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O88662 | Gene names | Emp2, Xmp | |||
|
Domain Architecture |

|
|||||
| Description | Epithelial membrane protein 2 (EMP-2) (Protein XMP). | |||||
|
EMP2_HUMAN
|
||||||
| NC score | 0.083730 (rank : 48) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P54851 | Gene names | EMP2, XMP | |||
|
Domain Architecture |

|
|||||
| Description | Epithelial membrane protein 2 (EMP-2) (Protein XMP). | |||||
|
LMIP_HUMAN
|
||||||
| NC score | 0.077759 (rank : 49) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P55344, Q9BXD0, Q9HAR5 | Gene names | LIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Lens fiber membrane intrinsic protein (MP18) (MP19) (MP20). | |||||
|
CLDND_HUMAN
|
||||||
| NC score | 0.076658 (rank : 50) | θ value | 0.365318 (rank : 47) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NY35, Q502Y8, Q6UVX2, Q9BUZ9, Q9NZZ5, Q9Y4S9 | Gene names | CLDND1, C3orf4 | |||
|
Domain Architecture |

|
|||||
| Description | Claudin domain-containing protein 1 (Membrane protein GENX-3745). | |||||
|
CCG3_HUMAN
|
||||||
| NC score | 0.071611 (rank : 51) | θ value | 4.03905 (rank : 51) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O60359 | Gene names | CACNG3 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent calcium channel gamma-3 subunit (Neuronal voltage- gated calcium channel gamma-3 subunit). | |||||
|
CCG2_HUMAN
|
||||||
| NC score | 0.068797 (rank : 52) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y698, Q2M1M1, Q5TGT3, Q9UGZ7 | Gene names | CACNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent calcium channel gamma-2 subunit (Neuronal voltage- gated calcium channel gamma-2 subunit). | |||||
|
CCG2_MOUSE
|
||||||
| NC score | 0.068780 (rank : 53) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O88602 | Gene names | Cacng2, Stg | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent calcium channel gamma-2 subunit (Neuronal voltage- gated calcium channel gamma-2 subunit) (Stargazin). | |||||
|
CCG5_MOUSE
|
||||||
| NC score | 0.067207 (rank : 54) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8VHW4 | Gene names | Cacng5 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent calcium channel gamma-5 subunit (Neuronal voltage- gated calcium channel gamma-5 subunit). | |||||
|
CLDND_MOUSE
|
||||||
| NC score | 0.065117 (rank : 55) | θ value | 1.38821 (rank : 50) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9CQX5, Q3U7C4 | Gene names | Cldnd1, D11Moh34 | |||
|
Domain Architecture |

|
|||||
| Description | Claudin domain-containing protein 1. | |||||
|
CCG3_MOUSE
|
||||||
| NC score | 0.062484 (rank : 56) | θ value | 8.99809 (rank : 60) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9JJV5 | Gene names | Cacng3 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent calcium channel gamma-3 subunit (Neuronal voltage- gated calcium channel gamma-3 subunit). | |||||
|
CCG5_HUMAN
|
||||||
| NC score | 0.053302 (rank : 57) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UF02, Q8WXS7, Q9UHM3 | Gene names | CACNG5 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent calcium channel gamma-5 subunit (Neuronal voltage- gated calcium channel gamma-5 subunit). | |||||
|
GPTC3_MOUSE
|
||||||
| NC score | 0.021017 (rank : 58) | θ value | 0.47712 (rank : 48) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BIY1 | Gene names | Gpatc3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 3. | |||||
|
CF155_HUMAN
|
||||||
| NC score | 0.011170 (rank : 59) | θ value | 8.99809 (rank : 61) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H8W2, Q5TG13 | Gene names | C6orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf155. | |||||
|
PRR10_HUMAN
|
||||||
| NC score | 0.009767 (rank : 60) | θ value | 8.99809 (rank : 64) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N7Y1 | Gene names | PRR10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 10. | |||||
|
ACL6B_HUMAN
|
||||||
| NC score | 0.008434 (rank : 61) | θ value | 5.27518 (rank : 55) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O94805, O75421 | Gene names | ACTL6B, ACTL6, BAF53B | |||
|
Domain Architecture |
|
|||||
| Description | Actin-like protein 6B (53 kDa BRG1-associated factor B) (Actin-related protein Baf53b) (ArpNalpha). | |||||
|
ACL6B_MOUSE
|
||||||
| NC score | 0.008433 (rank : 62) | θ value | 5.27518 (rank : 56) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99MR0 | Gene names | Actl6b, Actl6, ArpNa, Baf53b | |||
|
Domain Architecture |
|
|||||
| Description | Actin-like protein 6B (53 kDa BRG1-associated factor B) (Actin-related protein Baf53b). | |||||
|
CO039_HUMAN
|
||||||
| NC score | 0.008213 (rank : 63) | θ value | 4.03905 (rank : 52) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6ZRI6, Q71JB1, Q7L3S0, Q8N3F2, Q96FB6, Q9NTU5 | Gene names | C15orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf39. | |||||
|
ACL6A_MOUSE
|
||||||
| NC score | 0.007527 (rank : 64) | θ value | 8.99809 (rank : 59) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Z2N8, Q8C1W3, Q99M56 | Gene names | Actl6a, Actl6, Baf53a | |||
|
Domain Architecture |
|
|||||
| Description | Actin-like protein 6A (53 kDa BRG1-associated factor A) (Actin-related protein Baf53a). | |||||
|
ACL6A_HUMAN
|
||||||
| NC score | 0.007510 (rank : 65) | θ value | 8.99809 (rank : 58) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O96019, Q8TAE5, Q9BVS8, Q9H0W6 | Gene names | ACTL6A, BAF53, BAF53A | |||
|
Domain Architecture |
|
|||||
| Description | Actin-like protein 6A (53 kDa BRG1-associated factor A) (Actin-related protein Baf53a) (ArpNbeta). | |||||
|
SOCS7_MOUSE
|
||||||
| NC score | 0.006776 (rank : 66) | θ value | 4.03905 (rank : 54) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VHQ2 | Gene names | Socs7, Cish7, Nap4 | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of cytokine signaling 7 (SOCS-7). | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.005642 (rank : 67) | θ value | 0.163984 (rank : 46) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
WASIP_HUMAN
|
||||||
| NC score | 0.003309 (rank : 68) | θ value | 5.27518 (rank : 57) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
MILK2_HUMAN
|
||||||
| NC score | 0.002901 (rank : 69) | θ value | 4.03905 (rank : 53) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IY33, Q7RTP4, Q7Z655, Q8TEQ4, Q9H5F9 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | MICAL-like protein 2. | |||||
|
K1802_HUMAN
|
||||||
| NC score | -0.003827 (rank : 70) | θ value | 8.99809 (rank : 63) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
FOG1_MOUSE
|
||||||
| NC score | -0.004243 (rank : 71) | θ value | 8.99809 (rank : 62) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1237 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O35615 | Gene names | Zfpm1, Fog, Fog1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||