Please be patient as the page loads
|
CB013_HUMAN
|
||||||
| SwissProt Accessions | Q8IW19, Q53P47, Q53PB9, Q53QU0 | Gene names | C2orf13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C2orf13. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CB013_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8IW19, Q53P47, Q53PB9, Q53QU0 | Gene names | C2orf13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C2orf13. | |||||
|
CB013_MOUSE
|
||||||
| θ value | 8.61018e-152 (rank : 2) | NC score | 0.954211 (rank : 2) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9D842, Q8BZL5, Q99LX6 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C2orf13 homolog. | |||||
|
PNKP_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 3) | NC score | 0.130889 (rank : 3) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JLV6, Q6PFA3 | Gene names | Pnkp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional polynucleotide phosphatase/kinase (Polynucleotide kinase- 3'-phosphatase) (DNA 5'-kinase/3'-phosphatase) [Includes: Polynucleotide 3'-phosphatase (EC 3.1.3.32) (2'(3')-polynucleotidase); Polynucleotide 5'-hydroxyl-kinase (EC 2.7.1.78)]. | |||||
|
LEO1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 4) | NC score | 0.061813 (rank : 9) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
ZF106_HUMAN
|
||||||
| θ value | 0.125558 (rank : 5) | NC score | 0.045861 (rank : 15) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 470 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9H2Y7, Q6NSD9, Q6PEK1, Q86T43, Q86T45, Q86T50, Q86T58, Q86TA9, Q96M37, Q9H7B8 | Gene names | ZFP106, ZNF474 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 106 homolog (Zfp-106) (Zinc finger protein 474). | |||||
|
OXR1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 6) | NC score | 0.058977 (rank : 11) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q4KMM3, Q5FWW1, Q99L06, Q99MK1, Q99MP4 | Gene names | Oxr1, C7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oxidation resistance protein 1 (Protein C7). | |||||
|
CD2L1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 7) | NC score | 0.019195 (rank : 37) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 1467 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P21127, O95265, Q12817, Q12818, Q12819, Q12820, Q12822, Q8N530, Q9NZS5, Q9UBJ0, Q9UBQ1, Q9UBR0, Q9UNY2, Q9UP57, Q9UP58, Q9UP59 | Gene names | CDC2L1, PITSLREA, PK58 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1) (CLK-1) (CDK11) (p58 CLK-1). | |||||
|
PNKP_HUMAN
|
||||||
| θ value | 0.279714 (rank : 8) | NC score | 0.116334 (rank : 4) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96T60, Q9P1V2, Q9UKU8, Q9UNF8 | Gene names | PNKP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional polynucleotide phosphatase/kinase (Polynucleotide kinase- 3'-phosphatase) (DNA 5'-kinase/3'-phosphatase) [Includes: Polynucleotide 3'-phosphatase (EC 3.1.3.32) (2'(3')-polynucleotidase); Polynucleotide 5'-hydroxyl-kinase (EC 2.7.1.78)]. | |||||
|
SFRIP_HUMAN
|
||||||
| θ value | 0.279714 (rank : 9) | NC score | 0.071939 (rank : 7) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99590, Q8IW59 | Gene names | SFRS2IP, CASP11, SIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SFRS2-interacting protein (Splicing factor, arginine/serine-rich 2- interacting protein) (SC35-interacting protein 1) (CTD-associated SR protein 11) (Splicing regulatory protein 129) (SRrp129) (NY-REN-40 antigen). | |||||
|
CD2L1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 10) | NC score | 0.018535 (rank : 39) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 1463 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P24788, Q3UI03, Q61399, Q7TST4, Q8BP53 | Gene names | Cdc2l1 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1). | |||||
|
APTX_HUMAN
|
||||||
| θ value | 0.47712 (rank : 11) | NC score | 0.098911 (rank : 5) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7Z2E3, Q5T781, Q7Z2F3, Q7Z336, Q7Z5R5, Q7Z6V7, Q7Z6V8, Q9NXM5 | Gene names | APTX | |||
|
Domain Architecture |

|
|||||
| Description | Aprataxin (Forkhead-associated domain histidine triad-like protein) (FHA-HIT). | |||||
|
CAS1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 12) | NC score | 0.088373 (rank : 6) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P19228 | Gene names | Csn1s1, Csn1, Csna | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-s1-casein precursor. | |||||
|
NGEF_HUMAN
|
||||||
| θ value | 0.47712 (rank : 13) | NC score | 0.028510 (rank : 32) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N5V2, Q53QQ4, Q53ST7, Q6GMQ5, Q9NQD6 | Gene names | NGEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ephexin-1 (Eph-interacting exchange protein) (Neuronal guanine nucleotide exchange factor). | |||||
|
NOG2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 14) | NC score | 0.042885 (rank : 18) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13823, Q9BWN7 | Gene names | GNL2, NGP1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar GTP-binding protein 2 (Autoantigen NGP-1). | |||||
|
ARI5B_MOUSE
|
||||||
| θ value | 1.06291 (rank : 15) | NC score | 0.039156 (rank : 22) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BM75, Q8C0E0, Q8K4G8, Q8K4L9, Q9CU78, Q9JIX4 | Gene names | Arid5b, Desrt, Mrf2 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 5B (ARID domain- containing protein 5B) (MRF1-like) (Modulator recognition factor protein 2) (MRF-2) (Developmentally and sexually retarded with transient immune abnormalities protein). | |||||
|
FHOD1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 16) | NC score | 0.035449 (rank : 25) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6P9Q4, Q8BMK2 | Gene names | Fhod1, Fhos1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FH1/FH2 domain-containing protein (Formin homolog overexpressed in spleen) (FHOS) (Formin homology 2 domain-containing protein 1). | |||||
|
GOGA4_MOUSE
|
||||||
| θ value | 1.38821 (rank : 17) | NC score | 0.012969 (rank : 50) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
PDE1C_MOUSE
|
||||||
| θ value | 1.38821 (rank : 18) | NC score | 0.018234 (rank : 40) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q64338, Q62045, Q8BSV6 | Gene names | Pde1c | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1C (EC 3.1.4.17) (Cam-PDE 1C). | |||||
|
TTF1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 19) | NC score | 0.045176 (rank : 16) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q62187, Q9JKK5 | Gene names | Ttf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 1 (TTF-1) (TTF-I) (mTFF-I) (RNA polymerase I termination factor). | |||||
|
CD2L2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 20) | NC score | 0.016810 (rank : 44) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 1439 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UQ88, O95227, O95228, O96012, Q12821, Q12853, Q12854, Q5QPR0, Q5QPR1, Q5QPR2, Q9UBC4, Q9UBI3, Q9UEI1, Q9UEI2, Q9UP53, Q9UP54, Q9UP55, Q9UP56, Q9UQ86, Q9UQ87, Q9UQ89 | Gene names | CDC2L2, PITSLREB | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L2 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 2) (CDK11). | |||||
|
IRS1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 21) | NC score | 0.040089 (rank : 20) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P35569 | Gene names | Irs1, Irs-1 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin receptor substrate 1 (IRS-1). | |||||
|
GRIN1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 22) | NC score | 0.022487 (rank : 34) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7Z2K8, Q8ND74, Q96PZ4 | Gene names | GPRIN1, KIAA1893 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 23) | NC score | 0.040424 (rank : 19) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
MASP2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 24) | NC score | 0.007329 (rank : 53) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O00187, O75754, Q5TEQ5, Q5TER0, Q96QG4, Q9BZH0, Q9H498, Q9H499, Q9UBP3, Q9ULC7, Q9UMV3, Q9Y270 | Gene names | MASP2 | |||
|
Domain Architecture |

|
|||||
| Description | Mannan-binding lectin serine protease 2 precursor (EC 3.4.21.104) (Mannose-binding protein-associated serine protease 2) (MASP-2) (MBL- associated serine protease 2) [Contains: Mannan-binding lectin serine protease 2 A chain; Mannan-binding lectin serine protease 2 B chain]. | |||||
|
NP1L3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 25) | NC score | 0.032775 (rank : 28) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q794H2, O54802 | Gene names | Nap1l3, Mb20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome assembly protein 1-like 3 (Brain-specific protein MB20). | |||||
|
PRGC2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 26) | NC score | 0.060322 (rank : 10) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VHJ7, Q8C1C0 | Gene names | Ppargc1b, Errl1, Pgc1, Pgc1b, Ppargc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor gamma coactivator 1-beta (PPAR gamma coactivator-1beta protein) (PGC-1beta) (ERR ligand 1) (PPARGC-1-beta) (PGC-1-beta). | |||||
|
AKA12_HUMAN
|
||||||
| θ value | 3.0926 (rank : 27) | NC score | 0.044643 (rank : 17) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
APBA1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 28) | NC score | 0.046674 (rank : 14) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q02410, O14914, O60570, Q5VYR8 | Gene names | APBA1, MINT1, X11 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 1 (Neuron- specific X11 protein) (Neuronal Munc18-1-interacting protein 1) (Mint- 1) (Adapter protein X11alpha). | |||||
|
E41L2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 29) | NC score | 0.014049 (rank : 49) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O43491 | Gene names | EPB41L2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 2 (Generally expressed protein 4.1) (4.1G). | |||||
|
NSBP1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 30) | NC score | 0.056959 (rank : 12) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
AFF4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 31) | NC score | 0.039206 (rank : 21) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UHB7, Q498B2, Q59FB3, Q6P592, Q8TDR1, Q9P0E4 | Gene names | AFF4, AF5Q31, MCEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4 (ALL1-fused gene from chromosome 5q31) (Major CDK9 elongation factor-associated protein). | |||||
|
IRS1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 32) | NC score | 0.036959 (rank : 23) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P35568 | Gene names | IRS1 | |||
|
Domain Architecture |
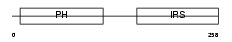
|
|||||
| Description | Insulin receptor substrate 1 (IRS-1). | |||||
|
NBN_HUMAN
|
||||||
| θ value | 4.03905 (rank : 33) | NC score | 0.028805 (rank : 31) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O60934, O60672, Q32NF7, Q53FM6, Q63HR6, Q7LDM2 | Gene names | NBN, NBS, NBS1, P95 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nibrin (Nijmegen breakage syndrome protein 1) (Cell cycle regulatory protein p95). | |||||
|
NFAT5_MOUSE
|
||||||
| θ value | 4.03905 (rank : 34) | NC score | 0.018007 (rank : 41) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9WV30 | Gene names | Nfat5 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells 5 (T cell transcription factor NFAT5) (NF-AT5) (Rel domain-containing transcription factor NFAT5). | |||||
|
RCOR3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 35) | NC score | 0.016867 (rank : 43) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9P2K3, Q5VT47, Q7L9I5, Q8N5U3, Q9NV83 | Gene names | RCOR3, KIAA1343 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 3. | |||||
|
AP3B2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 36) | NC score | 0.021817 (rank : 36) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9JME5, Q6QR53, Q8R1E5 | Gene names | Ap3b2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain). | |||||
|
ATRX_HUMAN
|
||||||
| θ value | 5.27518 (rank : 37) | NC score | 0.030842 (rank : 30) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P46100, P51068, Q15886, Q59FB5, Q59H31, Q5H9A2, Q5JWI4, Q7Z2J1, Q9H0Z1, Q9NTS3 | Gene names | ATRX, RAD54L, XH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked helicase II) (X-linked nuclear protein) (XNP) (Znf- HX). | |||||
|
DSCL1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 38) | NC score | 0.005413 (rank : 54) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TD84, Q76MU9, Q8IZY3, Q8IZY4, Q8WXU7, Q9ULT7 | Gene names | DSCAML1, DSCAM2, KIAA1132 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Down syndrome cell adhesion molecule-like protein 1 precursor (Down syndrome cell adhesion molecule 2). | |||||
|
GCP5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 39) | NC score | 0.027424 (rank : 33) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96RT8, Q96PY8 | Gene names | TUBGCP5, GCP5, KIAA1899 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-tubulin complex component 5 (GCP-5). | |||||
|
KI21A_MOUSE
|
||||||
| θ value | 5.27518 (rank : 40) | NC score | 0.015103 (rank : 48) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9QXL2, Q6P5H1, Q6ZPJ8, Q8BWZ9, Q8BXF1 | Gene names | Kif21a, Kiaa1708 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A. | |||||
|
KIF3B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 41) | NC score | 0.011538 (rank : 51) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 540 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O15066 | Gene names | KIF3B, KIAA0359 | |||
|
Domain Architecture |
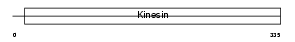
|
|||||
| Description | Kinesin-like protein KIF3B (Microtubule plus end-directed kinesin motor 3B) (HH0048). | |||||
|
LEO1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 42) | NC score | 0.056377 (rank : 13) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
ATRX_MOUSE
|
||||||
| θ value | 6.88961 (rank : 43) | NC score | 0.031455 (rank : 29) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q61687 | Gene names | Atrx, Hp1bp2, Xnp | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked nuclear protein) (Heterochromatin protein 2) (HP1 alpha-interacting protein) (HP1-BP38 protein). | |||||
|
DLG7_HUMAN
|
||||||
| θ value | 6.88961 (rank : 44) | NC score | 0.018600 (rank : 38) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15398, Q8NG58 | Gene names | DLG7, KIAA0008 | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 7 (Hepatoma up-regulated protein) (HURP). | |||||
|
FA13A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 45) | NC score | 0.016644 (rank : 45) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O94988, Q8NBA3 | Gene names | FAM13A1, KIAA0914 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM13A1. | |||||
|
KIF3B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 46) | NC score | 0.011308 (rank : 52) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 533 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61771 | Gene names | Kif3b | |||
|
Domain Architecture |
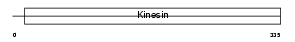
|
|||||
| Description | Kinesin-like protein KIF3B (Microtubule plus end-directed kinesin motor 3B). | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | 6.88961 (rank : 47) | NC score | 0.033301 (rank : 26) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
OR4M2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 48) | NC score | -0.000094 (rank : 55) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 703 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NGB6 | Gene names | OR4M2 | |||
|
Domain Architecture |
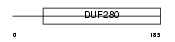
|
|||||
| Description | Olfactory receptor 4M2. | |||||
|
AFF4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 49) | NC score | 0.032820 (rank : 27) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
APTX_MOUSE
|
||||||
| θ value | 8.99809 (rank : 50) | NC score | 0.062518 (rank : 8) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7TQC5, Q8BPA7, Q8C2B5, Q8K3D1, Q9CQ59 | Gene names | Aptx | |||
|
Domain Architecture |

|
|||||
| Description | Aprataxin homolog (Forkhead-associated domain histidine triad-like protein) (FHA-HIT). | |||||
|
CCD94_MOUSE
|
||||||
| θ value | 8.99809 (rank : 51) | NC score | 0.022194 (rank : 35) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9D6J3 | Gene names | Ccdc94 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 94. | |||||
|
CYLC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 52) | NC score | 0.036072 (rank : 24) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P35663, Q5JQQ9 | Gene names | CYLC1, CYL, CYL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-1 (Cylicin I) (Multiple-band polypeptide I). | |||||
|
LCOR_MOUSE
|
||||||
| θ value | 8.99809 (rank : 53) | NC score | 0.017097 (rank : 42) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6ZPI3, Q3U302, Q5CZW7, Q80VA8, Q8BGT2, Q8C9Q0 | Gene names | Lcor, Kiaa1795, Mlr2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ligand-dependent corepressor (LCoR) (Mblk1-related protein 2). | |||||
|
PALB2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 54) | NC score | 0.015727 (rank : 47) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3U0P1, Q6NZG9, Q7TMQ4, Q8CEA9 | Gene names | Palb2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Partner and localizer of BRCA2. | |||||
|
PPR3A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 55) | NC score | 0.015946 (rank : 46) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99MR9, Q8BUJ4, Q8BUL0 | Gene names | Ppp1r3a, Pp1g | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase 1 regulatory subunit 3A (Protein phosphatase 1 glycogen-associated regulatory subunit) (Protein phosphatase type-1 glycogen targeting subunit) (RGL). | |||||
|
CB013_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8IW19, Q53P47, Q53PB9, Q53QU0 | Gene names | C2orf13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C2orf13. | |||||
|
CB013_MOUSE
|
||||||
| NC score | 0.954211 (rank : 2) | θ value | 8.61018e-152 (rank : 2) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9D842, Q8BZL5, Q99LX6 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C2orf13 homolog. | |||||
|
PNKP_MOUSE
|
||||||
| NC score | 0.130889 (rank : 3) | θ value | 0.0736092 (rank : 3) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JLV6, Q6PFA3 | Gene names | Pnkp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional polynucleotide phosphatase/kinase (Polynucleotide kinase- 3'-phosphatase) (DNA 5'-kinase/3'-phosphatase) [Includes: Polynucleotide 3'-phosphatase (EC 3.1.3.32) (2'(3')-polynucleotidase); Polynucleotide 5'-hydroxyl-kinase (EC 2.7.1.78)]. | |||||
|
PNKP_HUMAN
|
||||||
| NC score | 0.116334 (rank : 4) | θ value | 0.279714 (rank : 8) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96T60, Q9P1V2, Q9UKU8, Q9UNF8 | Gene names | PNKP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional polynucleotide phosphatase/kinase (Polynucleotide kinase- 3'-phosphatase) (DNA 5'-kinase/3'-phosphatase) [Includes: Polynucleotide 3'-phosphatase (EC 3.1.3.32) (2'(3')-polynucleotidase); Polynucleotide 5'-hydroxyl-kinase (EC 2.7.1.78)]. | |||||
|
APTX_HUMAN
|
||||||
| NC score | 0.098911 (rank : 5) | θ value | 0.47712 (rank : 11) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7Z2E3, Q5T781, Q7Z2F3, Q7Z336, Q7Z5R5, Q7Z6V7, Q7Z6V8, Q9NXM5 | Gene names | APTX | |||
|
Domain Architecture |

|
|||||
| Description | Aprataxin (Forkhead-associated domain histidine triad-like protein) (FHA-HIT). | |||||
|
CAS1_MOUSE
|
||||||
| NC score | 0.088373 (rank : 6) | θ value | 0.47712 (rank : 12) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P19228 | Gene names | Csn1s1, Csn1, Csna | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-s1-casein precursor. | |||||
|
SFRIP_HUMAN
|
||||||
| NC score | 0.071939 (rank : 7) | θ value | 0.279714 (rank : 9) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99590, Q8IW59 | Gene names | SFRS2IP, CASP11, SIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SFRS2-interacting protein (Splicing factor, arginine/serine-rich 2- interacting protein) (SC35-interacting protein 1) (CTD-associated SR protein 11) (Splicing regulatory protein 129) (SRrp129) (NY-REN-40 antigen). | |||||
|
APTX_MOUSE
|
||||||
| NC score | 0.062518 (rank : 8) | θ value | 8.99809 (rank : 50) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7TQC5, Q8BPA7, Q8C2B5, Q8K3D1, Q9CQ59 | Gene names | Aptx | |||
|
Domain Architecture |

|
|||||
| Description | Aprataxin homolog (Forkhead-associated domain histidine triad-like protein) (FHA-HIT). | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.061813 (rank : 9) | θ value | 0.0961366 (rank : 4) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
PRGC2_MOUSE
|
||||||
| NC score | 0.060322 (rank : 10) | θ value | 2.36792 (rank : 26) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VHJ7, Q8C1C0 | Gene names | Ppargc1b, Errl1, Pgc1, Pgc1b, Ppargc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor gamma coactivator 1-beta (PPAR gamma coactivator-1beta protein) (PGC-1beta) (ERR ligand 1) (PPARGC-1-beta) (PGC-1-beta). | |||||
|
OXR1_MOUSE
|
||||||
| NC score | 0.058977 (rank : 11) | θ value | 0.163984 (rank : 6) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q4KMM3, Q5FWW1, Q99L06, Q99MK1, Q99MP4 | Gene names | Oxr1, C7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oxidation resistance protein 1 (Protein C7). | |||||
|
NSBP1_MOUSE
|
||||||
| NC score | 0.056959 (rank : 12) | θ value | 3.0926 (rank : 30) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.056377 (rank : 13) | θ value | 5.27518 (rank : 42) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
APBA1_HUMAN
|
||||||
| NC score | 0.046674 (rank : 14) | θ value | 3.0926 (rank : 28) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q02410, O14914, O60570, Q5VYR8 | Gene names | APBA1, MINT1, X11 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 1 (Neuron- specific X11 protein) (Neuronal Munc18-1-interacting protein 1) (Mint- 1) (Adapter protein X11alpha). | |||||
|
ZF106_HUMAN
|
||||||
| NC score | 0.045861 (rank : 15) | θ value | 0.125558 (rank : 5) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 470 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9H2Y7, Q6NSD9, Q6PEK1, Q86T43, Q86T45, Q86T50, Q86T58, Q86TA9, Q96M37, Q9H7B8 | Gene names | ZFP106, ZNF474 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 106 homolog (Zfp-106) (Zinc finger protein 474). | |||||
|
TTF1_MOUSE
|
||||||
| NC score | 0.045176 (rank : 16) | θ value | 1.38821 (rank : 19) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q62187, Q9JKK5 | Gene names | Ttf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 1 (TTF-1) (TTF-I) (mTFF-I) (RNA polymerase I termination factor). | |||||
|
AKA12_HUMAN
|
||||||
| NC score | 0.044643 (rank : 17) | θ value | 3.0926 (rank : 27) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
NOG2_HUMAN
|
||||||
| NC score | 0.042885 (rank : 18) | θ value | 0.813845 (rank : 14) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13823, Q9BWN7 | Gene names | GNL2, NGP1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar GTP-binding protein 2 (Autoantigen NGP-1). | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.040424 (rank : 19) | θ value | 2.36792 (rank : 23) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
IRS1_MOUSE
|
||||||
| NC score | 0.040089 (rank : 20) | θ value | 1.81305 (rank : 21) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P35569 | Gene names | Irs1, Irs-1 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin receptor substrate 1 (IRS-1). | |||||
|
AFF4_HUMAN
|
||||||
| NC score | 0.039206 (rank : 21) | θ value | 4.03905 (rank : 31) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UHB7, Q498B2, Q59FB3, Q6P592, Q8TDR1, Q9P0E4 | Gene names | AFF4, AF5Q31, MCEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4 (ALL1-fused gene from chromosome 5q31) (Major CDK9 elongation factor-associated protein). | |||||
|
ARI5B_MOUSE
|
||||||
| NC score | 0.039156 (rank : 22) | θ value | 1.06291 (rank : 15) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BM75, Q8C0E0, Q8K4G8, Q8K4L9, Q9CU78, Q9JIX4 | Gene names | Arid5b, Desrt, Mrf2 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 5B (ARID domain- containing protein 5B) (MRF1-like) (Modulator recognition factor protein 2) (MRF-2) (Developmentally and sexually retarded with transient immune abnormalities protein). | |||||
|
IRS1_HUMAN
|
||||||
| NC score | 0.036959 (rank : 23) | θ value | 4.03905 (rank : 32) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P35568 | Gene names | IRS1 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin receptor substrate 1 (IRS-1). | |||||
|
CYLC1_HUMAN
|
||||||
| NC score | 0.036072 (rank : 24) | θ value | 8.99809 (rank : 52) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P35663, Q5JQQ9 | Gene names | CYLC1, CYL, CYL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-1 (Cylicin I) (Multiple-band polypeptide I). | |||||
|
FHOD1_MOUSE
|
||||||
| NC score | 0.035449 (rank : 25) | θ value | 1.06291 (rank : 16) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6P9Q4, Q8BMK2 | Gene names | Fhod1, Fhos1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FH1/FH2 domain-containing protein (Formin homolog overexpressed in spleen) (FHOS) (Formin homology 2 domain-containing protein 1). | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.033301 (rank : 26) | θ value | 6.88961 (rank : 47) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
AFF4_MOUSE
|
||||||
| NC score | 0.032820 (rank : 27) | θ value | 8.99809 (rank : 49) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
NP1L3_MOUSE
|
||||||
| NC score | 0.032775 (rank : 28) | θ value | 2.36792 (rank : 25) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q794H2, O54802 | Gene names | Nap1l3, Mb20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome assembly protein 1-like 3 (Brain-specific protein MB20). | |||||
|
ATRX_MOUSE
|
||||||
| NC score | 0.031455 (rank : 29) | θ value | 6.88961 (rank : 43) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q61687 | Gene names | Atrx, Hp1bp2, Xnp | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked nuclear protein) (Heterochromatin protein 2) (HP1 alpha-interacting protein) (HP1-BP38 protein). | |||||
|
ATRX_HUMAN
|
||||||
| NC score | 0.030842 (rank : 30) | θ value | 5.27518 (rank : 37) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P46100, P51068, Q15886, Q59FB5, Q59H31, Q5H9A2, Q5JWI4, Q7Z2J1, Q9H0Z1, Q9NTS3 | Gene names | ATRX, RAD54L, XH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked helicase II) (X-linked nuclear protein) (XNP) (Znf- HX). | |||||
|
NBN_HUMAN
|
||||||
| NC score | 0.028805 (rank : 31) | θ value | 4.03905 (rank : 33) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O60934, O60672, Q32NF7, Q53FM6, Q63HR6, Q7LDM2 | Gene names | NBN, NBS, NBS1, P95 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nibrin (Nijmegen breakage syndrome protein 1) (Cell cycle regulatory protein p95). | |||||
|
NGEF_HUMAN
|
||||||
| NC score | 0.028510 (rank : 32) | θ value | 0.47712 (rank : 13) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N5V2, Q53QQ4, Q53ST7, Q6GMQ5, Q9NQD6 | Gene names | NGEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ephexin-1 (Eph-interacting exchange protein) (Neuronal guanine nucleotide exchange factor). | |||||
|
GCP5_HUMAN
|
||||||
| NC score | 0.027424 (rank : 33) | θ value | 5.27518 (rank : 39) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96RT8, Q96PY8 | Gene names | TUBGCP5, GCP5, KIAA1899 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-tubulin complex component 5 (GCP-5). | |||||
|
GRIN1_HUMAN
|
||||||
| NC score | 0.022487 (rank : 34) | θ value | 2.36792 (rank : 22) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7Z2K8, Q8ND74, Q96PZ4 | Gene names | GPRIN1, KIAA1893 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
CCD94_MOUSE
|
||||||
| NC score | 0.022194 (rank : 35) | θ value | 8.99809 (rank : 51) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9D6J3 | Gene names | Ccdc94 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 94. | |||||
|
AP3B2_MOUSE
|
||||||
| NC score | 0.021817 (rank : 36) | θ value | 5.27518 (rank : 36) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9JME5, Q6QR53, Q8R1E5 | Gene names | Ap3b2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain). | |||||
|
CD2L1_HUMAN
|
||||||
| NC score | 0.019195 (rank : 37) | θ value | 0.21417 (rank : 7) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 1467 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P21127, O95265, Q12817, Q12818, Q12819, Q12820, Q12822, Q8N530, Q9NZS5, Q9UBJ0, Q9UBQ1, Q9UBR0, Q9UNY2, Q9UP57, Q9UP58, Q9UP59 | Gene names | CDC2L1, PITSLREA, PK58 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1) (CLK-1) (CDK11) (p58 CLK-1). | |||||
|
DLG7_HUMAN
|
||||||
| NC score | 0.018600 (rank : 38) | θ value | 6.88961 (rank : 44) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15398, Q8NG58 | Gene names | DLG7, KIAA0008 | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 7 (Hepatoma up-regulated protein) (HURP). | |||||
|
CD2L1_MOUSE
|
||||||
| NC score | 0.018535 (rank : 39) | θ value | 0.365318 (rank : 10) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 1463 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P24788, Q3UI03, Q61399, Q7TST4, Q8BP53 | Gene names | Cdc2l1 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1). | |||||
|
PDE1C_MOUSE
|
||||||
| NC score | 0.018234 (rank : 40) | θ value | 1.38821 (rank : 18) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q64338, Q62045, Q8BSV6 | Gene names | Pde1c | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1C (EC 3.1.4.17) (Cam-PDE 1C). | |||||
|
NFAT5_MOUSE
|
||||||
| NC score | 0.018007 (rank : 41) | θ value | 4.03905 (rank : 34) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9WV30 | Gene names | Nfat5 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells 5 (T cell transcription factor NFAT5) (NF-AT5) (Rel domain-containing transcription factor NFAT5). | |||||
|
LCOR_MOUSE
|
||||||
| NC score | 0.017097 (rank : 42) | θ value | 8.99809 (rank : 53) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6ZPI3, Q3U302, Q5CZW7, Q80VA8, Q8BGT2, Q8C9Q0 | Gene names | Lcor, Kiaa1795, Mlr2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ligand-dependent corepressor (LCoR) (Mblk1-related protein 2). | |||||
|
RCOR3_HUMAN
|
||||||
| NC score | 0.016867 (rank : 43) | θ value | 4.03905 (rank : 35) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9P2K3, Q5VT47, Q7L9I5, Q8N5U3, Q9NV83 | Gene names | RCOR3, KIAA1343 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 3. | |||||
|
CD2L2_HUMAN
|
||||||
| NC score | 0.016810 (rank : 44) | θ value | 1.81305 (rank : 20) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 1439 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UQ88, O95227, O95228, O96012, Q12821, Q12853, Q12854, Q5QPR0, Q5QPR1, Q5QPR2, Q9UBC4, Q9UBI3, Q9UEI1, Q9UEI2, Q9UP53, Q9UP54, Q9UP55, Q9UP56, Q9UQ86, Q9UQ87, Q9UQ89 | Gene names | CDC2L2, PITSLREB | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L2 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 2) (CDK11). | |||||
|
FA13A_HUMAN
|
||||||
| NC score | 0.016644 (rank : 45) | θ value | 6.88961 (rank : 45) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O94988, Q8NBA3 | Gene names | FAM13A1, KIAA0914 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM13A1. | |||||
|
PPR3A_MOUSE
|
||||||
| NC score | 0.015946 (rank : 46) | θ value | 8.99809 (rank : 55) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99MR9, Q8BUJ4, Q8BUL0 | Gene names | Ppp1r3a, Pp1g | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase 1 regulatory subunit 3A (Protein phosphatase 1 glycogen-associated regulatory subunit) (Protein phosphatase type-1 glycogen targeting subunit) (RGL). | |||||
|
PALB2_MOUSE
|
||||||
| NC score | 0.015727 (rank : 47) | θ value | 8.99809 (rank : 54) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3U0P1, Q6NZG9, Q7TMQ4, Q8CEA9 | Gene names | Palb2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Partner and localizer of BRCA2. | |||||
|
KI21A_MOUSE
|
||||||
| NC score | 0.015103 (rank : 48) | θ value | 5.27518 (rank : 40) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9QXL2, Q6P5H1, Q6ZPJ8, Q8BWZ9, Q8BXF1 | Gene names | Kif21a, Kiaa1708 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A. | |||||
|
E41L2_HUMAN
|
||||||
| NC score | 0.014049 (rank : 49) | θ value | 3.0926 (rank : 29) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O43491 | Gene names | EPB41L2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 2 (Generally expressed protein 4.1) (4.1G). | |||||
|
GOGA4_MOUSE
|
||||||
| NC score | 0.012969 (rank : 50) | θ value | 1.38821 (rank : 17) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
KIF3B_HUMAN
|
||||||
| NC score | 0.011538 (rank : 51) | θ value | 5.27518 (rank : 41) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 540 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O15066 | Gene names | KIF3B, KIAA0359 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF3B (Microtubule plus end-directed kinesin motor 3B) (HH0048). | |||||
|
KIF3B_MOUSE
|
||||||
| NC score | 0.011308 (rank : 52) | θ value | 6.88961 (rank : 46) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 533 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61771 | Gene names | Kif3b | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF3B (Microtubule plus end-directed kinesin motor 3B). | |||||
|
MASP2_HUMAN
|
||||||
| NC score | 0.007329 (rank : 53) | θ value | 2.36792 (rank : 24) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O00187, O75754, Q5TEQ5, Q5TER0, Q96QG4, Q9BZH0, Q9H498, Q9H499, Q9UBP3, Q9ULC7, Q9UMV3, Q9Y270 | Gene names | MASP2 | |||
|
Domain Architecture |

|
|||||
| Description | Mannan-binding lectin serine protease 2 precursor (EC 3.4.21.104) (Mannose-binding protein-associated serine protease 2) (MASP-2) (MBL- associated serine protease 2) [Contains: Mannan-binding lectin serine protease 2 A chain; Mannan-binding lectin serine protease 2 B chain]. | |||||
|
DSCL1_HUMAN
|
||||||
| NC score | 0.005413 (rank : 54) | θ value | 5.27518 (rank : 38) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TD84, Q76MU9, Q8IZY3, Q8IZY4, Q8WXU7, Q9ULT7 | Gene names | DSCAML1, DSCAM2, KIAA1132 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Down syndrome cell adhesion molecule-like protein 1 precursor (Down syndrome cell adhesion molecule 2). | |||||
|
OR4M2_HUMAN
|
||||||
| NC score | -0.000094 (rank : 55) | θ value | 6.88961 (rank : 48) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 703 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NGB6 | Gene names | OR4M2 | |||
|
Domain Architecture |

|
|||||
| Description | Olfactory receptor 4M2. | |||||