Please be patient as the page loads
|
CAYP2_MOUSE
|
||||||
| SwissProt Accessions | Q8BUG5, Q8C1J6 | Gene names | Caps2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcyphosin-2 (Calcyphosine-2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CAYP2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.923933 (rank : 2) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BXY5, Q6PH84, Q8N242, Q8NAY5 | Gene names | CAPS2 | |||
|
Domain Architecture |

|
|||||
| Description | Calcyphosin-2 (Calcyphosine-2). | |||||
|
CAYP2_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8BUG5, Q8C1J6 | Gene names | Caps2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcyphosin-2 (Calcyphosine-2). | |||||
|
CAYP1_HUMAN
|
||||||
| θ value | 1.6852e-22 (rank : 3) | NC score | 0.473730 (rank : 3) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13938, Q3MJA7, Q8WUZ3 | Gene names | CAPS | |||
|
Domain Architecture |
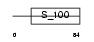
|
|||||
| Description | Calcyphosin (Calcyphosine). | |||||
|
DJBP_MOUSE
|
||||||
| θ value | 0.000461057 (rank : 4) | NC score | 0.209604 (rank : 4) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6P1E8, Q5DTV8, Q6PGK8, Q9D2D8 | Gene names | Djbp, Kiaa1672 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DJ-1-binding protein. | |||||
|
DJBP_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 5) | NC score | 0.206201 (rank : 5) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5THR3, Q5U5T6, Q9BY88, Q9H5C4, Q9NSF5 | Gene names | DJBP, KIAA1672 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DJ-1-binding protein (CAP-binding protein complex-interacting protein 1). | |||||
|
NKTR_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 6) | NC score | 0.036148 (rank : 24) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P30415 | Gene names | Nktr | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
MYL6B_MOUSE
|
||||||
| θ value | 0.62314 (rank : 7) | NC score | 0.058150 (rank : 10) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8CI43 | Gene names | Myl6b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin light polypeptide 6B (Smooth muscle and nonmuscle myosin light chain alkali 6B). | |||||
|
CMC2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 8) | NC score | 0.030296 (rank : 25) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9QXX4, Q9CZF6, Q9DCF5 | Gene names | Slc25a13, Aralar2 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding mitochondrial carrier protein Aralar2 (Mitochondrial aspartate glutamate carrier 2) (Solute carrier family 25 member 13) (Citrin). | |||||
|
MYL6B_HUMAN
|
||||||
| θ value | 0.813845 (rank : 9) | NC score | 0.064133 (rank : 6) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P14649 | Gene names | MYL6B, MLC1SA | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light polypeptide 6B (Smooth muscle and nonmuscle myosin light chain alkali 6B) (Myosin light chain 1 slow-twitch muscle A isoform) (MLC1sa). | |||||
|
NOC3L_HUMAN
|
||||||
| θ value | 0.813845 (rank : 10) | NC score | 0.056060 (rank : 12) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WTT2, Q9H5M6, Q9H9D8 | Gene names | NOC3L, AD24, C10orf117, FAD24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar complex protein 3 homolog (NOC3 protein homolog) (NOC3-like protein) (Nucleolar complex-associated protein 3-like protein) (Factor for adipocyte differentiation 24). | |||||
|
ZN447_HUMAN
|
||||||
| θ value | 0.813845 (rank : 11) | NC score | 0.010046 (rank : 37) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 899 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TBC5, Q9BRK7, Q9H9A0 | Gene names | ZNF447 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 447. | |||||
|
MYL6_HUMAN
|
||||||
| θ value | 1.06291 (rank : 12) | NC score | 0.055876 (rank : 13) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P60660, P16475, P24572, P24573, Q12790, Q6IAZ0, Q6IPY5 | Gene names | MYL6 | |||
|
Domain Architecture |
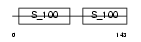
|
|||||
| Description | Myosin light polypeptide 6 (Smooth muscle and nonmuscle myosin light chain alkali 6) (Myosin light chain alkali 3) (Myosin light chain 3) (MLC-3) (LC17). | |||||
|
MYL6_MOUSE
|
||||||
| θ value | 1.06291 (rank : 13) | NC score | 0.055876 (rank : 14) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q60605 | Gene names | Myl6, Myln | |||
|
Domain Architecture |
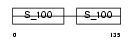
|
|||||
| Description | Myosin light polypeptide 6 (Smooth muscle and nonmuscle myosin light chain alkali 6) (Myosin light chain alkali 3) (Myosin light chain 3) (MLC-3) (LC17). | |||||
|
UBF1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 14) | NC score | 0.041869 (rank : 22) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P25976 | Gene names | Ubtf, Tcfubf, Ubf-1, Ubf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1). | |||||
|
CMC2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 15) | NC score | 0.028617 (rank : 26) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UJS0, O14566, O14575, Q9NZW1, Q9UNI7 | Gene names | SLC25A13, ARALAR2 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding mitochondrial carrier protein Aralar2 (Mitochondrial aspartate glutamate carrier 2) (Solute carrier family 25 member 13) (Citrin). | |||||
|
SYNE1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 16) | NC score | 0.011637 (rank : 35) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
NEST_HUMAN
|
||||||
| θ value | 2.36792 (rank : 17) | NC score | 0.013966 (rank : 32) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 1305 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P48681, O00552 | Gene names | NES | |||
|
Domain Architecture |

|
|||||
| Description | Nestin. | |||||
|
UBF1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 18) | NC score | 0.038760 (rank : 23) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P17480 | Gene names | UBTF, UBF, UBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1) (Autoantigen NOR-90). | |||||
|
CHLE_HUMAN
|
||||||
| θ value | 4.03905 (rank : 19) | NC score | 0.011159 (rank : 36) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P06276 | Gene names | BCHE, CHE1 | |||
|
Domain Architecture |

|
|||||
| Description | Cholinesterase precursor (EC 3.1.1.8) (Acylcholine acylhydrolase) (Choline esterase II) (Butyrylcholine esterase) (Pseudocholinesterase). | |||||
|
ELOA1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 20) | NC score | 0.020476 (rank : 28) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CB77, Q80VB2, Q9R0Q5 | Gene names | Tceb3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription elongation factor B polypeptide 3 (RNA polymerase II transcription factor SIII subunit A) (SIII p110) (Elongin-A) (EloA) (Elongin 110 kDa subunit). | |||||
|
AHTF1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 21) | NC score | 0.019092 (rank : 29) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WYP5, Q7Z4E3, Q8IZA4, Q96EH9, Q9Y4Q6 | Gene names | AHCTF1, ELYS, TMBS62 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-hook-containing transcription factor 1 (Embryonic large molecule derived from yolk sac). | |||||
|
KCNH5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 22) | NC score | 0.009115 (rank : 39) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NCM2 | Gene names | KCNH5, EAG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 5 (Voltage-gated potassium channel subunit Kv10.2) (Ether-a-go-go potassium channel 2) (hEAG2). | |||||
|
ZEP2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 23) | NC score | 0.005339 (rank : 42) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3UHF7, O55140, Q3UVD4, Q3UVH5 | Gene names | Hivep2, Mibp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Human immunodeficiency virus type I enhancer-binding protein 2 homolog (Myc intron-binding protein 1) (MIBP-1). | |||||
|
4ET_MOUSE
|
||||||
| θ value | 6.88961 (rank : 24) | NC score | 0.017675 (rank : 30) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9EST3, Q9CSS3 | Gene names | Eif4enif1, Clast4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4E transporter (eIF4E transporter) (4E-T) (Eukaryotic translation initiation factor 4E nuclear import factor 1). | |||||
|
CCHL_MOUSE
|
||||||
| θ value | 6.88961 (rank : 25) | NC score | 0.021851 (rank : 27) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P53702 | Gene names | Hccs, Cchl | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome c-type heme lyase (EC 4.4.1.17) (CCHL) (Holocytochrome c- type synthase). | |||||
|
CCNB3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 26) | NC score | 0.012928 (rank : 33) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8WWL7, Q96SB5, Q96SB6, Q96SB7, Q9NT38 | Gene names | CCNB3, CYCB3 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B3. | |||||
|
MLE1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 27) | NC score | 0.047241 (rank : 19) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P05977 | Gene names | Myl1, Mylf | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light chain 1, skeletal muscle isoform (MLC1F) (A1 catalytic) (Alkali myosin light chain 1). | |||||
|
MLE3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 28) | NC score | 0.046848 (rank : 20) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P05978 | Gene names | Myl1, Mylf | |||
|
Domain Architecture |
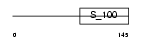
|
|||||
| Description | Myosin light chain 3, skeletal muscle isoform (A2 catalytic) (Alkali myosin light chain 3) (MLC3F). | |||||
|
ADA19_MOUSE
|
||||||
| θ value | 8.99809 (rank : 29) | NC score | 0.003711 (rank : 43) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O35674 | Gene names | Adam19, Mltnb | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 19 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 19) (Meltrin beta). | |||||
|
CN155_HUMAN
|
||||||
| θ value | 8.99809 (rank : 30) | NC score | 0.014056 (rank : 31) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
FSTL1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 31) | NC score | 0.012656 (rank : 34) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q62356, Q99JI9 | Gene names | Fstl1, Frp, Fstl, Tsc36 | |||
|
Domain Architecture |
|
|||||
| Description | Follistatin-related protein 1 precursor (Follistatin-like 1) (TGF- beta-inducible protein TSC-36). | |||||
|
LRRF2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 32) | NC score | 0.007789 (rank : 40) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91WK0, Q8BVD1, Q9CU89 | Gene names | Lrrfip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 2 (LRR FLII- interacting protein 2). | |||||
|
NOC3L_MOUSE
|
||||||
| θ value | 8.99809 (rank : 33) | NC score | 0.042118 (rank : 21) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8VI84, Q9JKM3 | Gene names | Noc3l, Ad24, Fad24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar complex protein 3 homolog (NOC3 protein homolog) (NOC3-like protein) (Nucleolar complex-associated protein 3-like protein) (Factor for adipocyte differentiation 24). | |||||
|
OPHN1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 34) | NC score | 0.007429 (rank : 41) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99J31, Q544K7 | Gene names | Ophn1 | |||
|
Domain Architecture |

|
|||||
| Description | Oligophrenin 1. | |||||
|
RABE2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 35) | NC score | 0.009494 (rank : 38) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 545 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H5N1 | Gene names | RABEP2, RABPT5B | |||
|
Domain Architecture |
|
|||||
| Description | Rab GTPase-binding effector protein 2 (Rabaptin-5beta). | |||||
|
CALL3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 36) | NC score | 0.053003 (rank : 15) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P27482 | Gene names | CALML3 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-like protein 3 (Calmodulin-related protein NB-1) (CaM-like protein) (CLP). | |||||
|
CALL5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 37) | NC score | 0.050898 (rank : 16) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9NZT1, Q8IXU8 | Gene names | CALML5, CLSP | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-like protein 5 (Calmodulin-like skin protein). | |||||
|
CALM4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 38) | NC score | 0.060556 (rank : 8) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9JM83, Q9CR31, Q9D1E9 | Gene names | Calm4, Dd112 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-4 (Calcium-binding protein Dd112). | |||||
|
CALN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 39) | NC score | 0.050355 (rank : 18) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BXU9 | Gene names | CALN1, CABP8 | |||
|
Domain Architecture |
|
|||||
| Description | Calneuron-1 (Calcium-binding protein CaBP8). | |||||
|
CALN1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 40) | NC score | 0.050603 (rank : 17) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9JJG7 | Gene names | Caln1 | |||
|
Domain Architecture |
|
|||||
| Description | Calneuron-1. | |||||
|
CETN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 41) | NC score | 0.059320 (rank : 9) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q12798 | Gene names | CETN1, CEN1, CETN | |||
|
Domain Architecture |
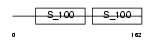
|
|||||
| Description | Centrin-1 (Caltractin isoform 2). | |||||
|
CETN2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 42) | NC score | 0.061869 (rank : 7) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P41208, Q53XW1 | Gene names | CETN2, CALT, CEN2 | |||
|
Domain Architecture |
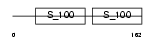
|
|||||
| Description | Centrin-2 (Caltractin isoform 1). | |||||
|
CETN2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 43) | NC score | 0.057951 (rank : 11) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9R1K9, Q3UBB4, Q9CWM0 | Gene names | Cetn2, Calt | |||
|
Domain Architecture |

|
|||||
| Description | Centrin-2 (Caltractin isoform 1). | |||||
|
CAYP2_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8BUG5, Q8C1J6 | Gene names | Caps2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcyphosin-2 (Calcyphosine-2). | |||||
|
CAYP2_HUMAN
|
||||||
| NC score | 0.923933 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BXY5, Q6PH84, Q8N242, Q8NAY5 | Gene names | CAPS2 | |||
|
Domain Architecture |

|
|||||
| Description | Calcyphosin-2 (Calcyphosine-2). | |||||
|
CAYP1_HUMAN
|
||||||
| NC score | 0.473730 (rank : 3) | θ value | 1.6852e-22 (rank : 3) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13938, Q3MJA7, Q8WUZ3 | Gene names | CAPS | |||
|
Domain Architecture |

|
|||||
| Description | Calcyphosin (Calcyphosine). | |||||
|
DJBP_MOUSE
|
||||||
| NC score | 0.209604 (rank : 4) | θ value | 0.000461057 (rank : 4) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6P1E8, Q5DTV8, Q6PGK8, Q9D2D8 | Gene names | Djbp, Kiaa1672 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DJ-1-binding protein. | |||||
|
DJBP_HUMAN
|
||||||
| NC score | 0.206201 (rank : 5) | θ value | 0.00298849 (rank : 5) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5THR3, Q5U5T6, Q9BY88, Q9H5C4, Q9NSF5 | Gene names | DJBP, KIAA1672 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DJ-1-binding protein (CAP-binding protein complex-interacting protein 1). | |||||
|
MYL6B_HUMAN
|
||||||
| NC score | 0.064133 (rank : 6) | θ value | 0.813845 (rank : 9) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P14649 | Gene names | MYL6B, MLC1SA | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light polypeptide 6B (Smooth muscle and nonmuscle myosin light chain alkali 6B) (Myosin light chain 1 slow-twitch muscle A isoform) (MLC1sa). | |||||
|
CETN2_HUMAN
|
||||||
| NC score | 0.061869 (rank : 7) | θ value | θ > 10 (rank : 42) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P41208, Q53XW1 | Gene names | CETN2, CALT, CEN2 | |||
|
Domain Architecture |

|
|||||
| Description | Centrin-2 (Caltractin isoform 1). | |||||
|
CALM4_MOUSE
|
||||||
| NC score | 0.060556 (rank : 8) | θ value | θ > 10 (rank : 38) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9JM83, Q9CR31, Q9D1E9 | Gene names | Calm4, Dd112 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-4 (Calcium-binding protein Dd112). | |||||
|
CETN1_HUMAN
|
||||||
| NC score | 0.059320 (rank : 9) | θ value | θ > 10 (rank : 41) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q12798 | Gene names | CETN1, CEN1, CETN | |||
|
Domain Architecture |

|
|||||
| Description | Centrin-1 (Caltractin isoform 2). | |||||
|
MYL6B_MOUSE
|
||||||
| NC score | 0.058150 (rank : 10) | θ value | 0.62314 (rank : 7) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8CI43 | Gene names | Myl6b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin light polypeptide 6B (Smooth muscle and nonmuscle myosin light chain alkali 6B). | |||||
|
CETN2_MOUSE
|
||||||
| NC score | 0.057951 (rank : 11) | θ value | θ > 10 (rank : 43) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9R1K9, Q3UBB4, Q9CWM0 | Gene names | Cetn2, Calt | |||
|
Domain Architecture |

|
|||||
| Description | Centrin-2 (Caltractin isoform 1). | |||||
|
NOC3L_HUMAN
|
||||||
| NC score | 0.056060 (rank : 12) | θ value | 0.813845 (rank : 10) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WTT2, Q9H5M6, Q9H9D8 | Gene names | NOC3L, AD24, C10orf117, FAD24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar complex protein 3 homolog (NOC3 protein homolog) (NOC3-like protein) (Nucleolar complex-associated protein 3-like protein) (Factor for adipocyte differentiation 24). | |||||
|
MYL6_HUMAN
|
||||||
| NC score | 0.055876 (rank : 13) | θ value | 1.06291 (rank : 12) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P60660, P16475, P24572, P24573, Q12790, Q6IAZ0, Q6IPY5 | Gene names | MYL6 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light polypeptide 6 (Smooth muscle and nonmuscle myosin light chain alkali 6) (Myosin light chain alkali 3) (Myosin light chain 3) (MLC-3) (LC17). | |||||
|
MYL6_MOUSE
|
||||||
| NC score | 0.055876 (rank : 14) | θ value | 1.06291 (rank : 13) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q60605 | Gene names | Myl6, Myln | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light polypeptide 6 (Smooth muscle and nonmuscle myosin light chain alkali 6) (Myosin light chain alkali 3) (Myosin light chain 3) (MLC-3) (LC17). | |||||
|
CALL3_HUMAN
|
||||||
| NC score | 0.053003 (rank : 15) | θ value | θ > 10 (rank : 36) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P27482 | Gene names | CALML3 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-like protein 3 (Calmodulin-related protein NB-1) (CaM-like protein) (CLP). | |||||
|
CALL5_HUMAN
|
||||||
| NC score | 0.050898 (rank : 16) | θ value | θ > 10 (rank : 37) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9NZT1, Q8IXU8 | Gene names | CALML5, CLSP | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-like protein 5 (Calmodulin-like skin protein). | |||||
|
CALN1_MOUSE
|
||||||
| NC score | 0.050603 (rank : 17) | θ value | θ > 10 (rank : 40) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9JJG7 | Gene names | Caln1 | |||
|
Domain Architecture |

|
|||||
| Description | Calneuron-1. | |||||
|
CALN1_HUMAN
|
||||||
| NC score | 0.050355 (rank : 18) | θ value | θ > 10 (rank : 39) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BXU9 | Gene names | CALN1, CABP8 | |||
|
Domain Architecture |

|
|||||
| Description | Calneuron-1 (Calcium-binding protein CaBP8). | |||||
|
MLE1_MOUSE
|
||||||
| NC score | 0.047241 (rank : 19) | θ value | 6.88961 (rank : 27) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P05977 | Gene names | Myl1, Mylf | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light chain 1, skeletal muscle isoform (MLC1F) (A1 catalytic) (Alkali myosin light chain 1). | |||||
|
MLE3_MOUSE
|
||||||
| NC score | 0.046848 (rank : 20) | θ value | 6.88961 (rank : 28) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P05978 | Gene names | Myl1, Mylf | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light chain 3, skeletal muscle isoform (A2 catalytic) (Alkali myosin light chain 3) (MLC3F). | |||||
|
NOC3L_MOUSE
|
||||||
| NC score | 0.042118 (rank : 21) | θ value | 8.99809 (rank : 33) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8VI84, Q9JKM3 | Gene names | Noc3l, Ad24, Fad24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar complex protein 3 homolog (NOC3 protein homolog) (NOC3-like protein) (Nucleolar complex-associated protein 3-like protein) (Factor for adipocyte differentiation 24). | |||||
|
UBF1_MOUSE
|
||||||
| NC score | 0.041869 (rank : 22) | θ value | 1.06291 (rank : 14) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P25976 | Gene names | Ubtf, Tcfubf, Ubf-1, Ubf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1). | |||||
|
UBF1_HUMAN
|
||||||
| NC score | 0.038760 (rank : 23) | θ value | 2.36792 (rank : 18) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P17480 | Gene names | UBTF, UBF, UBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1) (Autoantigen NOR-90). | |||||
|
NKTR_MOUSE
|
||||||
| NC score | 0.036148 (rank : 24) | θ value | 0.0961366 (rank : 6) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P30415 | Gene names | Nktr | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
CMC2_MOUSE
|
||||||
| NC score | 0.030296 (rank : 25) | θ value | 0.813845 (rank : 8) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9QXX4, Q9CZF6, Q9DCF5 | Gene names | Slc25a13, Aralar2 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding mitochondrial carrier protein Aralar2 (Mitochondrial aspartate glutamate carrier 2) (Solute carrier family 25 member 13) (Citrin). | |||||
|
CMC2_HUMAN
|
||||||
| NC score | 0.028617 (rank : 26) | θ value | 1.38821 (rank : 15) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UJS0, O14566, O14575, Q9NZW1, Q9UNI7 | Gene names | SLC25A13, ARALAR2 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding mitochondrial carrier protein Aralar2 (Mitochondrial aspartate glutamate carrier 2) (Solute carrier family 25 member 13) (Citrin). | |||||
|
CCHL_MOUSE
|
||||||
| NC score | 0.021851 (rank : 27) | θ value | 6.88961 (rank : 25) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P53702 | Gene names | Hccs, Cchl | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome c-type heme lyase (EC 4.4.1.17) (CCHL) (Holocytochrome c- type synthase). | |||||
|
ELOA1_MOUSE
|
||||||
| NC score | 0.020476 (rank : 28) | θ value | 4.03905 (rank : 20) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CB77, Q80VB2, Q9R0Q5 | Gene names | Tceb3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription elongation factor B polypeptide 3 (RNA polymerase II transcription factor SIII subunit A) (SIII p110) (Elongin-A) (EloA) (Elongin 110 kDa subunit). | |||||
|
AHTF1_HUMAN
|
||||||
| NC score | 0.019092 (rank : 29) | θ value | 5.27518 (rank : 21) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WYP5, Q7Z4E3, Q8IZA4, Q96EH9, Q9Y4Q6 | Gene names | AHCTF1, ELYS, TMBS62 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-hook-containing transcription factor 1 (Embryonic large molecule derived from yolk sac). | |||||
|
4ET_MOUSE
|
||||||
| NC score | 0.017675 (rank : 30) | θ value | 6.88961 (rank : 24) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9EST3, Q9CSS3 | Gene names | Eif4enif1, Clast4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4E transporter (eIF4E transporter) (4E-T) (Eukaryotic translation initiation factor 4E nuclear import factor 1). | |||||
|
CN155_HUMAN
|
||||||
| NC score | 0.014056 (rank : 31) | θ value | 8.99809 (rank : 30) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
NEST_HUMAN
|
||||||
| NC score | 0.013966 (rank : 32) | θ value | 2.36792 (rank : 17) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 1305 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P48681, O00552 | Gene names | NES | |||
|
Domain Architecture |

|
|||||
| Description | Nestin. | |||||
|
CCNB3_HUMAN
|
||||||
| NC score | 0.012928 (rank : 33) | θ value | 6.88961 (rank : 26) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8WWL7, Q96SB5, Q96SB6, Q96SB7, Q9NT38 | Gene names | CCNB3, CYCB3 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B3. | |||||
|
FSTL1_MOUSE
|
||||||
| NC score | 0.012656 (rank : 34) | θ value | 8.99809 (rank : 31) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q62356, Q99JI9 | Gene names | Fstl1, Frp, Fstl, Tsc36 | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin-related protein 1 precursor (Follistatin-like 1) (TGF- beta-inducible protein TSC-36). | |||||
|
SYNE1_HUMAN
|
||||||
| NC score | 0.011637 (rank : 35) | θ value | 1.81305 (rank : 16) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
CHLE_HUMAN
|
||||||
| NC score | 0.011159 (rank : 36) | θ value | 4.03905 (rank : 19) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P06276 | Gene names | BCHE, CHE1 | |||
|
Domain Architecture |

|
|||||
| Description | Cholinesterase precursor (EC 3.1.1.8) (Acylcholine acylhydrolase) (Choline esterase II) (Butyrylcholine esterase) (Pseudocholinesterase). | |||||
|
ZN447_HUMAN
|
||||||
| NC score | 0.010046 (rank : 37) | θ value | 0.813845 (rank : 11) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 899 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TBC5, Q9BRK7, Q9H9A0 | Gene names | ZNF447 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 447. | |||||
|
RABE2_HUMAN
|
||||||
| NC score | 0.009494 (rank : 38) | θ value | 8.99809 (rank : 35) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 545 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H5N1 | Gene names | RABEP2, RABPT5B | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 2 (Rabaptin-5beta). | |||||
|
KCNH5_HUMAN
|
||||||
| NC score | 0.009115 (rank : 39) | θ value | 5.27518 (rank : 22) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NCM2 | Gene names | KCNH5, EAG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 5 (Voltage-gated potassium channel subunit Kv10.2) (Ether-a-go-go potassium channel 2) (hEAG2). | |||||
|
LRRF2_MOUSE
|
||||||
| NC score | 0.007789 (rank : 40) | θ value | 8.99809 (rank : 32) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91WK0, Q8BVD1, Q9CU89 | Gene names | Lrrfip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 2 (LRR FLII- interacting protein 2). | |||||
|
OPHN1_MOUSE
|
||||||
| NC score | 0.007429 (rank : 41) | θ value | 8.99809 (rank : 34) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99J31, Q544K7 | Gene names | Ophn1 | |||
|
Domain Architecture |

|
|||||
| Description | Oligophrenin 1. | |||||
|
ZEP2_MOUSE
|
||||||
| NC score | 0.005339 (rank : 42) | θ value | 5.27518 (rank : 23) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3UHF7, O55140, Q3UVD4, Q3UVH5 | Gene names | Hivep2, Mibp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Human immunodeficiency virus type I enhancer-binding protein 2 homolog (Myc intron-binding protein 1) (MIBP-1). | |||||
|
ADA19_MOUSE
|
||||||
| NC score | 0.003711 (rank : 43) | θ value | 8.99809 (rank : 29) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O35674 | Gene names | Adam19, Mltnb | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 19 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 19) (Meltrin beta). | |||||