Please be patient as the page loads
|
ACOX3_HUMAN
|
||||||
| SwissProt Accessions | O15254 | Gene names | ACOX3, BRCOX, PRCOX | |||
|
Domain Architecture |

|
|||||
| Description | Acyl-coenzyme A oxidase 3, peroxisomal (EC 1.3.3.6) (Pristanoyl-CoA oxidase) (Branched-chain acyl-CoA oxidase) (BRCACox). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ACOX3_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O15254 | Gene names | ACOX3, BRCOX, PRCOX | |||
|
Domain Architecture |

|
|||||
| Description | Acyl-coenzyme A oxidase 3, peroxisomal (EC 1.3.3.6) (Pristanoyl-CoA oxidase) (Branched-chain acyl-CoA oxidase) (BRCACox). | |||||
|
ACOX3_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.995701 (rank : 2) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9EPL9 | Gene names | Acox3 | |||
|
Domain Architecture |

|
|||||
| Description | Acyl-coenzyme A oxidase 3, peroxisomal (EC 1.3.3.6) (Pristanoyl-CoA oxidase) (Branched-chain acyl-CoA oxidase) (BRCACox). | |||||
|
ACOX1_HUMAN
|
||||||
| θ value | 1.04096e-72 (rank : 3) | NC score | 0.894227 (rank : 4) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q15067, Q12863, Q15068, Q15101, Q16131, Q9UD31 | Gene names | ACOX1, ACOX | |||
|
Domain Architecture |

|
|||||
| Description | Acyl-coenzyme A oxidase 1, peroxisomal (EC 1.3.3.6) (Palmitoyl-CoA oxidase) (AOX) (Straight-chain acyl-CoA oxidase) (SCOX). | |||||
|
ACOX1_MOUSE
|
||||||
| θ value | 1.04096e-72 (rank : 4) | NC score | 0.894783 (rank : 3) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9R0H0, O35616, Q3TDG0 | Gene names | Acox1, Acox, Paox | |||
|
Domain Architecture |

|
|||||
| Description | Acyl-coenzyme A oxidase 1, peroxisomal (EC 1.3.3.6) (Palmitoyl-CoA oxidase) (AOX). | |||||
|
ACOX2_MOUSE
|
||||||
| θ value | 8.81223e-72 (rank : 5) | NC score | 0.874749 (rank : 6) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9QXD1 | Gene names | Acox2 | |||
|
Domain Architecture |

|
|||||
| Description | Acyl-coenzyme A oxidase 2, peroxisomal (EC 1.17.99.3) (3-alpha,7- alpha,12-alpha-trihydroxy-5-beta-cholestanoyl-CoA 24-hydroxylase) (3- alpha,7-alpha,12-alpha-trihydroxy-5-beta-cholestanoyl-CoA oxidase) (Trihydroxycoprostanoyl-CoA oxidase) (THCCox) (THCA-CoA oxidase). | |||||
|
ACOX2_HUMAN
|
||||||
| θ value | 4.52582e-68 (rank : 6) | NC score | 0.878730 (rank : 5) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q99424 | Gene names | ACOX2 | |||
|
Domain Architecture |

|
|||||
| Description | Acyl-coenzyme A oxidase 2, peroxisomal (EC 1.17.99.3) (3-alpha,7- alpha,12-alpha-trihydroxy-5-beta-cholestanoyl-CoA 24-hydroxylase) (3- alpha,7-alpha,12-alpha-trihydroxy-5-beta-cholestanoyl-CoA oxidase) (Trihydroxycoprostanoyl-CoA oxidase) (THCCox) (THCA-CoA oxidase). | |||||
|
ACADS_HUMAN
|
||||||
| θ value | 4.76016e-09 (rank : 7) | NC score | 0.458208 (rank : 9) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P16219, P78331 | Gene names | ACADS | |||
|
Domain Architecture |
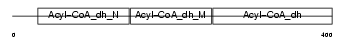
|
|||||
| Description | Short-chain specific acyl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.2) (SCAD) (Butyryl-CoA dehydrogenase). | |||||
|
ACADV_MOUSE
|
||||||
| θ value | 1.06045e-08 (rank : 8) | NC score | 0.447230 (rank : 11) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P50544, O35289, O55133 | Gene names | Acadvl, Vlcad | |||
|
Domain Architecture |
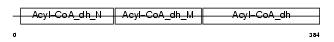
|
|||||
| Description | Very-long-chain specific acyl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.-) (VLCAD) (MVLCAD). | |||||
|
ACAD9_HUMAN
|
||||||
| θ value | 1.38499e-08 (rank : 9) | NC score | 0.449613 (rank : 10) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9H845, Q8WXX3 | Gene names | ACAD9 | |||
|
Domain Architecture |
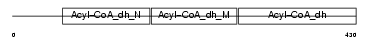
|
|||||
| Description | Acyl-CoA dehydrogenase family member 9, mitochondrial precursor (EC 1.3.99.-) (ACAD-9). | |||||
|
ACDSB_HUMAN
|
||||||
| θ value | 1.38499e-08 (rank : 10) | NC score | 0.424105 (rank : 14) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P45954, Q96CX7 | Gene names | ACADSB | |||
|
Domain Architecture |
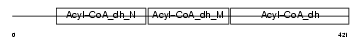
|
|||||
| Description | Short/branched chain specific acyl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.-) (SBCAD) (2-methyl branched chain acyl-CoA dehydrogenase) (2-MEBCAD) (2-methylbutyryl-coenzyme A dehydrogenase) (2-methylbutyryl-CoA dehydrogenase). | |||||
|
ACAD9_MOUSE
|
||||||
| θ value | 2.36244e-08 (rank : 11) | NC score | 0.460744 (rank : 8) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8JZN5, Q8BK76, Q8C0B5 | Gene names | Acad9 | |||
|
Domain Architecture |
|
|||||
| Description | Acyl-CoA dehydrogenase family member 9, mitochondrial precursor (EC 1.3.99.-) (ACAD-9). | |||||
|
ACADS_MOUSE
|
||||||
| θ value | 4.0297e-08 (rank : 12) | NC score | 0.462471 (rank : 7) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q07417 | Gene names | Acads | |||
|
Domain Architecture |
|
|||||
| Description | Short-chain specific acyl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.2) (SCAD) (Butyryl-CoA dehydrogenase). | |||||
|
ACADV_HUMAN
|
||||||
| θ value | 4.0297e-08 (rank : 13) | NC score | 0.441780 (rank : 12) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P49748, O76056, Q8WUL0 | Gene names | ACADVL, VLCAD | |||
|
Domain Architecture |
|
|||||
| Description | Very-long-chain specific acyl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.-) (VLCAD). | |||||
|
ACDSB_MOUSE
|
||||||
| θ value | 1.99992e-07 (rank : 14) | NC score | 0.396080 (rank : 17) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9DBL1 | Gene names | Acadsb | |||
|
Domain Architecture |
|
|||||
| Description | Short/branched chain specific acyl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.-) (SBCAD) (2-methyl branched chain acyl-CoA dehydrogenase) (2-MEBCAD) (2-methylbutyryl-coenzyme A dehydrogenase) (2-methylbutyryl-CoA dehydrogenase). | |||||
|
GCDH_MOUSE
|
||||||
| θ value | 1.29631e-06 (rank : 15) | NC score | 0.440595 (rank : 13) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q60759 | Gene names | Gcdh | |||
|
Domain Architecture |
|
|||||
| Description | Glutaryl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.7) (GCD). | |||||
|
GCDH_HUMAN
|
||||||
| θ value | 3.19293e-05 (rank : 16) | NC score | 0.423140 (rank : 15) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q92947, O14719 | Gene names | GCDH | |||
|
Domain Architecture |
|
|||||
| Description | Glutaryl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.7) (GCD). | |||||
|
ACADM_MOUSE
|
||||||
| θ value | 0.000121331 (rank : 17) | NC score | 0.384836 (rank : 18) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P45952, Q64235 | Gene names | Acadm | |||
|
Domain Architecture |
|
|||||
| Description | Medium-chain specific acyl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.3) (MCAD). | |||||
|
ACADM_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 18) | NC score | 0.396297 (rank : 16) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P11310, Q9NYF1 | Gene names | ACADM | |||
|
Domain Architecture |
|
|||||
| Description | Medium-chain specific acyl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.3) (MCAD). | |||||
|
ACAD8_MOUSE
|
||||||
| θ value | 0.000786445 (rank : 19) | NC score | 0.351919 (rank : 23) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9D7B6, Q8BK36, Q8BKQ8, Q8CFY9, Q9D2L8 | Gene names | Acad8 | |||
|
Domain Architecture |
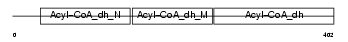
|
|||||
| Description | Acyl-CoA dehydrogenase family member 8, mitochondrial precursor (EC 1.3.99.-) (ACAD-8) (Isobutyryl-CoA dehydrogenase). | |||||
|
IVD_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 20) | NC score | 0.359289 (rank : 21) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9JHI5, Q9CYI3, Q9DBD7 | Gene names | Ivd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Isovaleryl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.10) (IVD). | |||||
|
ACAD8_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 21) | NC score | 0.343125 (rank : 24) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UKU7, Q9BUS8 | Gene names | ACAD8, ARC42 | |||
|
Domain Architecture |
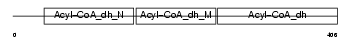
|
|||||
| Description | Acyl-CoA dehydrogenase family member 8, mitochondrial precursor (EC 1.3.99.-) (ACAD-8) (Isobutyryl-CoA dehydrogenase) (Activator- recruited cofactor 42 kDa component) (ARC42). | |||||
|
ACADL_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 22) | NC score | 0.365181 (rank : 19) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P28330 | Gene names | ACADL | |||
|
Domain Architecture |
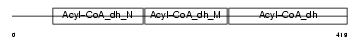
|
|||||
| Description | Long-chain specific acyl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.13) (LCAD). | |||||
|
IVD_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 23) | NC score | 0.352554 (rank : 22) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P26440, Q96AF6 | Gene names | IVD | |||
|
Domain Architecture |
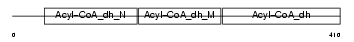
|
|||||
| Description | Isovaleryl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.10) (IVD). | |||||
|
ACADL_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 24) | NC score | 0.363765 (rank : 20) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P51174, O35302, Q8QZR6, Q9CU29, Q9DB83 | Gene names | Acadl | |||
|
Domain Architecture |
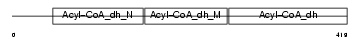
|
|||||
| Description | Long-chain specific acyl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.13) (LCAD). | |||||
|
PIGO_HUMAN
|
||||||
| θ value | 1.38821 (rank : 25) | NC score | 0.024647 (rank : 25) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TEQ8, Q6P154, Q6UX80, Q8TDS8, Q96CS9, Q9BVN9, Q9Y4B0 | Gene names | PIGO | |||
|
Domain Architecture |

|
|||||
| Description | GPI ethanolamine phosphate transferase 3 (EC 2.-.-.-) (Phosphatidylinositol-glycan biosynthesis class O protein) (PIG-O). | |||||
|
DUS18_MOUSE
|
||||||
| θ value | 2.36792 (rank : 26) | NC score | 0.012295 (rank : 30) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VE01 | Gene names | Dusp18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 18 (EC 3.1.3.48) (EC 3.1.3.16). | |||||
|
ADA15_HUMAN
|
||||||
| θ value | 3.0926 (rank : 27) | NC score | 0.005182 (rank : 31) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q13444, Q13493, Q96C78 | Gene names | ADAM15, MDC15 | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 15 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 15) (Metalloproteinase-like, disintegrin-like, and cysteine- rich protein 15) (MDC-15) (Metalloprotease RGD disintegrin protein) (Metargidin). | |||||
|
MYH11_HUMAN
|
||||||
| θ value | 3.0926 (rank : 28) | NC score | 0.002360 (rank : 33) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
MYH11_MOUSE
|
||||||
| θ value | 3.0926 (rank : 29) | NC score | 0.002465 (rank : 32) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
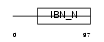
XPO7_HUMAN
|
||||||
| θ value | 4.03905 (rank : 30) | NC score | 0.018415 (rank : 27) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UIA9, O94846, Q6PJK9, Q8NEK7 | Gene names | XPO7, KIAA0745, RANBP16 | |||
|
Domain Architecture |
|
|||||
| Description | Exportin-7 (Exp7) (Ran-binding protein 16). | |||||
|
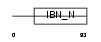
XPO7_MOUSE
|
||||||
| θ value | 4.03905 (rank : 31) | NC score | 0.018433 (rank : 26) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9EPK7, Q3TP94, Q80TS9, Q8BSK5, Q8C9M7, Q8CB42, Q8CBL8, Q8CEF5 | Gene names | Xpo7, Kiaa0745, Ranbp16 | |||
|
Domain Architecture |
|
|||||
| Description | Exportin-7 (Exp7) (Ran-binding protein 16). | |||||
|
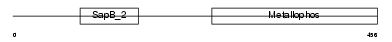
ASM_HUMAN
|
||||||
| θ value | 5.27518 (rank : 32) | NC score | 0.012887 (rank : 28) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P17405, P17406, Q13811, Q16837, Q16841 | Gene names | SMPD1, ASM | |||
|
Domain Architecture |
|
|||||
| Description | Sphingomyelin phosphodiesterase precursor (EC 3.1.4.12) (Acid sphingomyelinase) (aSMase). | |||||
|
MOCOS_MOUSE
|
||||||
| θ value | 6.88961 (rank : 33) | NC score | 0.012833 (rank : 29) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14CH1 | Gene names | Mocos | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Molybdenum cofactor sulfurase (EC 4.4.-.-) (MoCo sulfurase) (MOS). | |||||
|
ACOX3_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O15254 | Gene names | ACOX3, BRCOX, PRCOX | |||
|
Domain Architecture |

|
|||||
| Description | Acyl-coenzyme A oxidase 3, peroxisomal (EC 1.3.3.6) (Pristanoyl-CoA oxidase) (Branched-chain acyl-CoA oxidase) (BRCACox). | |||||
|
ACOX3_MOUSE
|
||||||
| NC score | 0.995701 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9EPL9 | Gene names | Acox3 | |||
|
Domain Architecture |

|
|||||
| Description | Acyl-coenzyme A oxidase 3, peroxisomal (EC 1.3.3.6) (Pristanoyl-CoA oxidase) (Branched-chain acyl-CoA oxidase) (BRCACox). | |||||
|
ACOX1_MOUSE
|
||||||
| NC score | 0.894783 (rank : 3) | θ value | 1.04096e-72 (rank : 4) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9R0H0, O35616, Q3TDG0 | Gene names | Acox1, Acox, Paox | |||
|
Domain Architecture |

|
|||||
| Description | Acyl-coenzyme A oxidase 1, peroxisomal (EC 1.3.3.6) (Palmitoyl-CoA oxidase) (AOX). | |||||
|
ACOX1_HUMAN
|
||||||
| NC score | 0.894227 (rank : 4) | θ value | 1.04096e-72 (rank : 3) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q15067, Q12863, Q15068, Q15101, Q16131, Q9UD31 | Gene names | ACOX1, ACOX | |||
|
Domain Architecture |

|
|||||
| Description | Acyl-coenzyme A oxidase 1, peroxisomal (EC 1.3.3.6) (Palmitoyl-CoA oxidase) (AOX) (Straight-chain acyl-CoA oxidase) (SCOX). | |||||
|
ACOX2_HUMAN
|
||||||
| NC score | 0.878730 (rank : 5) | θ value | 4.52582e-68 (rank : 6) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q99424 | Gene names | ACOX2 | |||
|
Domain Architecture |

|
|||||
| Description | Acyl-coenzyme A oxidase 2, peroxisomal (EC 1.17.99.3) (3-alpha,7- alpha,12-alpha-trihydroxy-5-beta-cholestanoyl-CoA 24-hydroxylase) (3- alpha,7-alpha,12-alpha-trihydroxy-5-beta-cholestanoyl-CoA oxidase) (Trihydroxycoprostanoyl-CoA oxidase) (THCCox) (THCA-CoA oxidase). | |||||
|
ACOX2_MOUSE
|
||||||
| NC score | 0.874749 (rank : 6) | θ value | 8.81223e-72 (rank : 5) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9QXD1 | Gene names | Acox2 | |||
|
Domain Architecture |

|
|||||
| Description | Acyl-coenzyme A oxidase 2, peroxisomal (EC 1.17.99.3) (3-alpha,7- alpha,12-alpha-trihydroxy-5-beta-cholestanoyl-CoA 24-hydroxylase) (3- alpha,7-alpha,12-alpha-trihydroxy-5-beta-cholestanoyl-CoA oxidase) (Trihydroxycoprostanoyl-CoA oxidase) (THCCox) (THCA-CoA oxidase). | |||||
|
ACADS_MOUSE
|
||||||
| NC score | 0.462471 (rank : 7) | θ value | 4.0297e-08 (rank : 12) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q07417 | Gene names | Acads | |||
|
Domain Architecture |
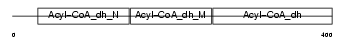
|
|||||
| Description | Short-chain specific acyl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.2) (SCAD) (Butyryl-CoA dehydrogenase). | |||||
|
ACAD9_MOUSE
|
||||||
| NC score | 0.460744 (rank : 8) | θ value | 2.36244e-08 (rank : 11) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8JZN5, Q8BK76, Q8C0B5 | Gene names | Acad9 | |||
|
Domain Architecture |
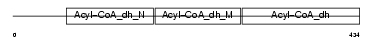
|
|||||
| Description | Acyl-CoA dehydrogenase family member 9, mitochondrial precursor (EC 1.3.99.-) (ACAD-9). | |||||
|
ACADS_HUMAN
|
||||||
| NC score | 0.458208 (rank : 9) | θ value | 4.76016e-09 (rank : 7) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P16219, P78331 | Gene names | ACADS | |||
|
Domain Architecture |
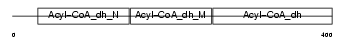
|
|||||
| Description | Short-chain specific acyl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.2) (SCAD) (Butyryl-CoA dehydrogenase). | |||||
|
ACAD9_HUMAN
|
||||||
| NC score | 0.449613 (rank : 10) | θ value | 1.38499e-08 (rank : 9) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9H845, Q8WXX3 | Gene names | ACAD9 | |||
|
Domain Architecture |
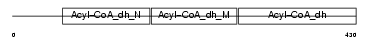
|
|||||
| Description | Acyl-CoA dehydrogenase family member 9, mitochondrial precursor (EC 1.3.99.-) (ACAD-9). | |||||
|
ACADV_MOUSE
|
||||||
| NC score | 0.447230 (rank : 11) | θ value | 1.06045e-08 (rank : 8) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P50544, O35289, O55133 | Gene names | Acadvl, Vlcad | |||
|
Domain Architecture |
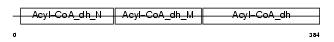
|
|||||
| Description | Very-long-chain specific acyl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.-) (VLCAD) (MVLCAD). | |||||
|
ACADV_HUMAN
|
||||||
| NC score | 0.441780 (rank : 12) | θ value | 4.0297e-08 (rank : 13) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P49748, O76056, Q8WUL0 | Gene names | ACADVL, VLCAD | |||
|
Domain Architecture |

|
|||||
| Description | Very-long-chain specific acyl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.-) (VLCAD). | |||||
|
GCDH_MOUSE
|
||||||
| NC score | 0.440595 (rank : 13) | θ value | 1.29631e-06 (rank : 15) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q60759 | Gene names | Gcdh | |||
|
Domain Architecture |
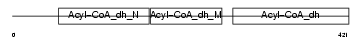
|
|||||
| Description | Glutaryl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.7) (GCD). | |||||
|
ACDSB_HUMAN
|
||||||
| NC score | 0.424105 (rank : 14) | θ value | 1.38499e-08 (rank : 10) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P45954, Q96CX7 | Gene names | ACADSB | |||
|
Domain Architecture |
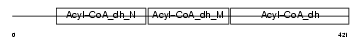
|
|||||
| Description | Short/branched chain specific acyl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.-) (SBCAD) (2-methyl branched chain acyl-CoA dehydrogenase) (2-MEBCAD) (2-methylbutyryl-coenzyme A dehydrogenase) (2-methylbutyryl-CoA dehydrogenase). | |||||
|
GCDH_HUMAN
|
||||||
| NC score | 0.423140 (rank : 15) | θ value | 3.19293e-05 (rank : 16) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q92947, O14719 | Gene names | GCDH | |||
|
Domain Architecture |
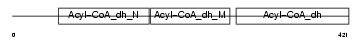
|
|||||
| Description | Glutaryl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.7) (GCD). | |||||
|
ACADM_HUMAN
|
||||||
| NC score | 0.396297 (rank : 16) | θ value | 0.000461057 (rank : 18) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P11310, Q9NYF1 | Gene names | ACADM | |||
|
Domain Architecture |
|
|||||
| Description | Medium-chain specific acyl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.3) (MCAD). | |||||
|
ACDSB_MOUSE
|
||||||
| NC score | 0.396080 (rank : 17) | θ value | 1.99992e-07 (rank : 14) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9DBL1 | Gene names | Acadsb | |||
|
Domain Architecture |
|
|||||
| Description | Short/branched chain specific acyl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.-) (SBCAD) (2-methyl branched chain acyl-CoA dehydrogenase) (2-MEBCAD) (2-methylbutyryl-coenzyme A dehydrogenase) (2-methylbutyryl-CoA dehydrogenase). | |||||
|
ACADM_MOUSE
|
||||||
| NC score | 0.384836 (rank : 18) | θ value | 0.000121331 (rank : 17) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P45952, Q64235 | Gene names | Acadm | |||
|
Domain Architecture |
|
|||||
| Description | Medium-chain specific acyl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.3) (MCAD). | |||||
|
ACADL_HUMAN
|
||||||
| NC score | 0.365181 (rank : 19) | θ value | 0.00665767 (rank : 22) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P28330 | Gene names | ACADL | |||
|
Domain Architecture |
|
|||||
| Description | Long-chain specific acyl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.13) (LCAD). | |||||
|
ACADL_MOUSE
|
||||||
| NC score | 0.363765 (rank : 20) | θ value | 0.0961366 (rank : 24) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P51174, O35302, Q8QZR6, Q9CU29, Q9DB83 | Gene names | Acadl | |||
|
Domain Architecture |
|
|||||
| Description | Long-chain specific acyl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.13) (LCAD). | |||||
|
IVD_MOUSE
|
||||||
| NC score | 0.359289 (rank : 21) | θ value | 0.00175202 (rank : 20) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9JHI5, Q9CYI3, Q9DBD7 | Gene names | Ivd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Isovaleryl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.10) (IVD). | |||||
|
IVD_HUMAN
|
||||||
| NC score | 0.352554 (rank : 22) | θ value | 0.0148317 (rank : 23) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P26440, Q96AF6 | Gene names | IVD | |||
|
Domain Architecture |
|
|||||
| Description | Isovaleryl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.10) (IVD). | |||||
|
ACAD8_MOUSE
|
||||||
| NC score | 0.351919 (rank : 23) | θ value | 0.000786445 (rank : 19) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9D7B6, Q8BK36, Q8BKQ8, Q8CFY9, Q9D2L8 | Gene names | Acad8 | |||
|
Domain Architecture |
|
|||||
| Description | Acyl-CoA dehydrogenase family member 8, mitochondrial precursor (EC 1.3.99.-) (ACAD-8) (Isobutyryl-CoA dehydrogenase). | |||||
|
ACAD8_HUMAN
|
||||||
| NC score | 0.343125 (rank : 24) | θ value | 0.00390308 (rank : 21) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UKU7, Q9BUS8 | Gene names | ACAD8, ARC42 | |||
|
Domain Architecture |
|
|||||
| Description | Acyl-CoA dehydrogenase family member 8, mitochondrial precursor (EC 1.3.99.-) (ACAD-8) (Isobutyryl-CoA dehydrogenase) (Activator- recruited cofactor 42 kDa component) (ARC42). | |||||
|
PIGO_HUMAN
|
||||||
| NC score | 0.024647 (rank : 25) | θ value | 1.38821 (rank : 25) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TEQ8, Q6P154, Q6UX80, Q8TDS8, Q96CS9, Q9BVN9, Q9Y4B0 | Gene names | PIGO | |||
|
Domain Architecture |

|
|||||
| Description | GPI ethanolamine phosphate transferase 3 (EC 2.-.-.-) (Phosphatidylinositol-glycan biosynthesis class O protein) (PIG-O). | |||||
|
XPO7_MOUSE
|
||||||
| NC score | 0.018433 (rank : 26) | θ value | 4.03905 (rank : 31) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9EPK7, Q3TP94, Q80TS9, Q8BSK5, Q8C9M7, Q8CB42, Q8CBL8, Q8CEF5 | Gene names | Xpo7, Kiaa0745, Ranbp16 | |||
|
Domain Architecture |

|
|||||
| Description | Exportin-7 (Exp7) (Ran-binding protein 16). | |||||
|
XPO7_HUMAN
|
||||||
| NC score | 0.018415 (rank : 27) | θ value | 4.03905 (rank : 30) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UIA9, O94846, Q6PJK9, Q8NEK7 | Gene names | XPO7, KIAA0745, RANBP16 | |||
|
Domain Architecture |

|
|||||
| Description | Exportin-7 (Exp7) (Ran-binding protein 16). | |||||
|
ASM_HUMAN
|
||||||
| NC score | 0.012887 (rank : 28) | θ value | 5.27518 (rank : 32) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P17405, P17406, Q13811, Q16837, Q16841 | Gene names | SMPD1, ASM | |||
|
Domain Architecture |

|
|||||
| Description | Sphingomyelin phosphodiesterase precursor (EC 3.1.4.12) (Acid sphingomyelinase) (aSMase). | |||||
|
MOCOS_MOUSE
|
||||||
| NC score | 0.012833 (rank : 29) | θ value | 6.88961 (rank : 33) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14CH1 | Gene names | Mocos | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Molybdenum cofactor sulfurase (EC 4.4.-.-) (MoCo sulfurase) (MOS). | |||||
|
DUS18_MOUSE
|
||||||
| NC score | 0.012295 (rank : 30) | θ value | 2.36792 (rank : 26) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VE01 | Gene names | Dusp18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 18 (EC 3.1.3.48) (EC 3.1.3.16). | |||||
|
ADA15_HUMAN
|
||||||
| NC score | 0.005182 (rank : 31) | θ value | 3.0926 (rank : 27) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q13444, Q13493, Q96C78 | Gene names | ADAM15, MDC15 | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 15 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 15) (Metalloproteinase-like, disintegrin-like, and cysteine- rich protein 15) (MDC-15) (Metalloprotease RGD disintegrin protein) (Metargidin). | |||||
|
MYH11_MOUSE
|
||||||
| NC score | 0.002465 (rank : 32) | θ value | 3.0926 (rank : 29) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
MYH11_HUMAN
|
||||||
| NC score | 0.002360 (rank : 33) | θ value | 3.0926 (rank : 28) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||