Please be patient as the page loads
|
TPD52_MOUSE
|
||||||
| SwissProt Accessions | Q62393 | Gene names | Tpd52 | |||
|
Domain Architecture |
|
|||||
| Description | Tumor protein D52 (mD52). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
TPD52_MOUSE
|
||||||
| θ value | 9.41505e-74 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q62393 | Gene names | Tpd52 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor protein D52 (mD52). | |||||
|
TPD52_HUMAN
|
||||||
| θ value | 8.53533e-67 (rank : 2) | NC score | 0.985032 (rank : 2) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P55327, Q13056 | Gene names | TPD52 | |||
|
Domain Architecture |
|
|||||
| Description | Tumor protein D52 (N8 protein). | |||||
|
TPD53_HUMAN
|
||||||
| θ value | 5.76516e-39 (rank : 3) | NC score | 0.936185 (rank : 3) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q16890 | Gene names | TPD52L1 | |||
|
Domain Architecture |
|
|||||
| Description | Tumor protein D53 (hD53) (Tumor protein D52-like 1). | |||||
|
TPD53_MOUSE
|
||||||
| θ value | 1.57e-36 (rank : 4) | NC score | 0.933408 (rank : 4) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O54818 | Gene names | Tpd52l1 | |||
|
Domain Architecture |
|
|||||
| Description | Tumor protein D53 (mD53) (Tumor protein D52-like 1). | |||||
|
TPD54_HUMAN
|
||||||
| θ value | 9.8567e-31 (rank : 5) | NC score | 0.910600 (rank : 5) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O43399, Q9H3Z6 | Gene names | TPD52L2 | |||
|
Domain Architecture |
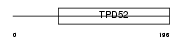
|
|||||
| Description | Tumor protein D54 (hD54) (Tumor protein D52-like 2). | |||||
|
TPD54_MOUSE
|
||||||
| θ value | 1.02001e-27 (rank : 6) | NC score | 0.902988 (rank : 6) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9CYZ2, Q3TVY1 | Gene names | Tpd52l2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor protein D54 (Tumor protein D52-like 2). | |||||
|
CCD21_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 7) | NC score | 0.078429 (rank : 10) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1228 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q6P2H3, Q5VY68, Q5VY70, Q9H6Q1, Q9H828, Q9UF52 | Gene names | CCDC21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
NUF2_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 8) | NC score | 0.095591 (rank : 9) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 585 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q99P69, Q8BTK7, Q8VE05, Q9CST5 | Gene names | Cdca1, Nuf2r | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Nuf2 (Cell division cycle-associated protein 1). | |||||
|
SAS6_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 9) | NC score | 0.108041 (rank : 7) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q80UK7, Q9CYT4 | Gene names | Sass6, Sas6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spindle assembly abnormal protein 6 homolog. | |||||
|
SAS6_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 10) | NC score | 0.106582 (rank : 8) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 791 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q6UVJ0, Q8N3K0 | Gene names | SASS6, SAS6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spindle assembly abnormal protein 6 homolog (HsSAS-6). | |||||
|
SYCP1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 11) | NC score | 0.062434 (rank : 19) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q15431, O14963 | Gene names | SYCP1, SCP1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
FER_HUMAN
|
||||||
| θ value | 0.279714 (rank : 12) | NC score | 0.015166 (rank : 75) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1278 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P16591 | Gene names | FER, TYK3 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase FER (EC 2.7.10.2) (p94-FER) (c- FER). | |||||
|
DYNA_MOUSE
|
||||||
| θ value | 0.365318 (rank : 13) | NC score | 0.069141 (rank : 13) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1084 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O08788 | Gene names | Dctn1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynactin-1 (150 kDa dynein-associated polypeptide) (DP-150) (DAP-150) (p150-glued). | |||||
|
NCOR1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 14) | NC score | 0.034543 (rank : 70) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O75376, Q9UPV5, Q9UQ18 | Gene names | NCOR1, KIAA1047 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR). | |||||
|
NUMA1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 15) | NC score | 0.066664 (rank : 16) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
PCNT_MOUSE
|
||||||
| θ value | 0.47712 (rank : 16) | NC score | 0.067345 (rank : 14) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1207 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P48725 | Gene names | Pcnt, Pcnt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pericentrin. | |||||
|
RB6I2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 17) | NC score | 0.073988 (rank : 12) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1584 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8IUD2, Q6NVK2, Q8IUD3, Q8IUD4, Q8IUD5, Q8NAS1, Q9NXN5, Q9UIK7, Q9UPS1 | Gene names | ERC1, ELKS, KIAA1081, RAB6IP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1). | |||||
|
RB6I2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 18) | NC score | 0.075173 (rank : 11) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q99MI1, Q80TK7, Q8BPL1, Q8C7Y1, Q99MI2 | Gene names | Erc1, Cast2, Kiaa1081, Rab6ip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1) (ERC1) (CAZ-associated structural protein 2) (CAST2). | |||||
|
KIF11_HUMAN
|
||||||
| θ value | 0.62314 (rank : 19) | NC score | 0.045443 (rank : 64) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 583 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P52732, Q15716, Q5VWX0 | Gene names | KIF11, EG5, KNSL1 | |||
|
Domain Architecture |
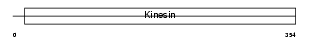
|
|||||
| Description | Kinesin-like protein KIF11 (Kinesin-related motor protein Eg5) (Kinesin-like spindle protein HKSP) (Thyroid receptor-interacting protein 5) (TRIP5) (Kinesin-like protein 1). | |||||
|
MRCKA_HUMAN
|
||||||
| θ value | 0.62314 (rank : 20) | NC score | 0.027816 (rank : 73) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1999 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q5VT25, Q59GZ1, Q5H9N9, Q5T797, Q5VT26, Q5VT27, Q86XX2, Q86XX3, Q99646 | Gene names | CDC42BPA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK alpha (EC 2.7.11.1) (CDC42- binding protein kinase alpha) (Myotonic dystrophy kinase-related CDC42-binding kinase alpha) (Myotonic dystrophy protein kinase-like alpha) (MRCK alpha) (DMPK-like alpha). | |||||
|
CUTL1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 21) | NC score | 0.057358 (rank : 27) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P53564, O08994, P70301, Q571L6, Q91ZD2 | Gene names | Cutl1, Cux, Cux1, Kiaa4047 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP) (Homeobox protein Cux). | |||||
|
DYNA_HUMAN
|
||||||
| θ value | 0.813845 (rank : 22) | NC score | 0.067026 (rank : 15) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1194 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q14203, O95296, Q9BRM9, Q9UIU1, Q9UIU2 | Gene names | DCTN1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynactin-1 (150 kDa dynein-associated polypeptide) (DP-150) (DAP-150) (p150-glued) (p135). | |||||
|
GOGA3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 23) | NC score | 0.059630 (rank : 24) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q08378, O43241, Q6P9C7, Q86XW3, Q8TDA9, Q8WZA3 | Gene names | GOLGA3 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Golgi complex-associated protein of 170 kDa) (GCP170). | |||||
|
GOGA1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 24) | NC score | 0.062019 (rank : 21) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1222 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q92805, Q8IYZ9 | Gene names | GOLGA1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
SCAR3_MOUSE
|
||||||
| θ value | 1.06291 (rank : 25) | NC score | 0.032992 (rank : 71) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8C850, Q8C6N1 | Gene names | Scara3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class A member 3. | |||||
|
ZN318_HUMAN
|
||||||
| θ value | 1.06291 (rank : 26) | NC score | 0.049943 (rank : 58) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5VUA4, O94796, Q4G0E4, Q8NEM6, Q9UNU8, Q9Y2W9 | Gene names | ZNF318 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 318 (Endocrine regulatory protein). | |||||
|
KIF11_MOUSE
|
||||||
| θ value | 2.36792 (rank : 27) | NC score | 0.036762 (rank : 69) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 446 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6P9P6, Q9Z1J0 | Gene names | Kif11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin-like protein KIF11 (Kinesin-related motor protein Eg5). | |||||
|
LRSM1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 28) | NC score | 0.036913 (rank : 68) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1078 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q6UWE0, Q5VVV0, Q8NB40, Q96GT5, Q96MX5, Q96MZ7 | Gene names | LRSAM1, TAL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein LRSAM1 (EC 6.3.2.-) (Leucine-rich repeat and sterile alpha motif-containing protein 1) (Tsg101-associated ligase) (hTAL). | |||||
|
MYH4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 29) | NC score | 0.046766 (rank : 61) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
MYH8_MOUSE
|
||||||
| θ value | 3.0926 (rank : 30) | NC score | 0.046246 (rank : 62) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1600 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P13542, Q5SX36 | Gene names | Myh8, Myhsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
ATX3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 31) | NC score | 0.042806 (rank : 67) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P54252, O15284, O15285, O15286, Q8N189, Q96TC3, Q96TC4, Q9H3N0 | Gene names | ATXN3, ATX3, MJD, MJD1, SCA3 | |||
|
Domain Architecture |

|
|||||
| Description | Ataxin-3 (EC 3.4.22.-) (Machado-Joseph disease protein 1) (Spinocerebellar ataxia type 3 protein). | |||||
|
KTN1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 32) | NC score | 0.055469 (rank : 31) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q86UP2, Q13999, Q14707, Q15387, Q86W57 | Gene names | KTN1, CG1, KIAA0004 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin (Kinesin receptor) (CG-1 antigen). | |||||
|
MYH4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 33) | NC score | 0.044859 (rank : 65) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1377 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9Y623 | Gene names | MYH4 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal) (Myosin heavy chain IIb) (MyHC-IIb). | |||||
|
TRIPB_HUMAN
|
||||||
| θ value | 4.03905 (rank : 34) | NC score | 0.053526 (rank : 34) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
ERC2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 35) | NC score | 0.066356 (rank : 18) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1156 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O15083, Q86TK4 | Gene names | ERC2, KIAA0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2. | |||||
|
ERC2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 36) | NC score | 0.066508 (rank : 17) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1166 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q6PH08, Q80U20, Q8CCP1 | Gene names | Erc2, Cast1, D14Ertd171e, Kiaa0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2 (CAZ-associated structural protein 1) (CAST1). | |||||
|
MYH13_HUMAN
|
||||||
| θ value | 5.27518 (rank : 37) | NC score | 0.045546 (rank : 63) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1504 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9UKX3, O95252 | Gene names | MYH13 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-13 (Myosin heavy chain, skeletal muscle, extraocular) (MyHC- eo). | |||||
|
MYH7_HUMAN
|
||||||
| θ value | 5.27518 (rank : 38) | NC score | 0.042975 (rank : 66) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P12883, Q14836, Q14837, Q14904, Q16579, Q92679, Q9H1D5, Q9UMM8 | Gene names | MYH7, MYHCB | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle beta isoform (MyHC-beta). | |||||
|
TAXB1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 39) | NC score | 0.056405 (rank : 28) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q3UKC1, Q91YT6, Q9CVF0, Q9DC45 | Gene names | Tax1bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tax1-binding protein 1 homolog. | |||||
|
CK5P2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 40) | NC score | 0.052009 (rank : 44) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1151 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8K389, Q6PCN1, Q6ZPL0 | Gene names | Cdk5rap2, Kiaa1633 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48). | |||||
|
STXB4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 41) | NC score | 0.049177 (rank : 59) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 629 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9WV89, Q5SUA6, Q5SUA8, Q5SUA9, Q8CFL1 | Gene names | Stxbp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Syntaxin-binding protein 4 (Syntaxin 4-interacting protein) (Synip). | |||||
|
TRI39_HUMAN
|
||||||
| θ value | 6.88961 (rank : 42) | NC score | 0.014838 (rank : 77) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9HCM9, Q76BL3, Q8IYT9, Q96IB6 | Gene names | TRIM39, RNF23, TFP | |||
|
Domain Architecture |
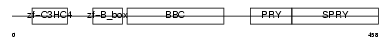
|
|||||
| Description | Tripartite motif-containing protein 39 (RING finger protein 23) (Testis-abundant finger protein). | |||||
|
TRI39_MOUSE
|
||||||
| θ value | 6.88961 (rank : 43) | NC score | 0.015109 (rank : 76) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 691 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9ESN2, Q8BPR5, Q8K0F7 | Gene names | Trim39, Rnf23, Tfp | |||
|
Domain Architecture |
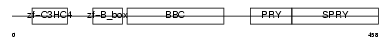
|
|||||
| Description | Tripartite motif-containing protein 39 (RING finger protein 23) (Testis-abundant finger protein). | |||||
|
ZN307_HUMAN
|
||||||
| θ value | 6.88961 (rank : 44) | NC score | -0.001656 (rank : 80) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q969J2 | Gene names | ZNF307 | |||
|
Domain Architecture |
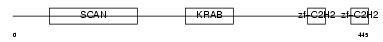
|
|||||
| Description | Zinc finger protein 307 (P373c6.1). | |||||
|
FYCO1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 45) | NC score | 0.050134 (rank : 56) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1149 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9BQS8, Q3MJE6, Q86T41, Q86TB1, Q8TEF9, Q96IV5, Q9H8P9 | Gene names | FYCO1, ZFYVE7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1 (Zinc finger FYVE domain-containing protein 7). | |||||
|
GABP2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 46) | NC score | 0.011202 (rank : 78) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 454 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q06547, Q06545, Q12940, Q12941, Q12942, Q8IYD0 | Gene names | GABPB2, E4TF1B, GABPB, GABPB1 | |||
|
Domain Architecture |
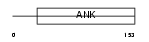
|
|||||
| Description | GA-binding protein beta chain (GABP subunit beta-2) (GABP-2) (GABP subunit beta-1) (GABPB-1) (Transcription factor E4TF1-53) (Transcription factor E4TF1-47) (Nuclear respiratory factor 2). | |||||
|
GABP2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 47) | NC score | 0.010341 (rank : 79) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 429 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q00420, Q00421, Q8BS31, Q91YZ0, Q9QVV2 | Gene names | Gabpb2, Gabpb, Gabpb1 | |||
|
Domain Architecture |
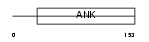
|
|||||
| Description | GA-binding protein beta chain (GABP subunit beta-2) (GABP-2) (GABP subunit beta-1) (GABPB-1). | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 48) | NC score | 0.053251 (rank : 37) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
NIN_MOUSE
|
||||||
| θ value | 8.99809 (rank : 49) | NC score | 0.055657 (rank : 30) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q61043, Q6ZPM7 | Gene names | Nin, Kiaa1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein. | |||||
|
PCNT_HUMAN
|
||||||
| θ value | 8.99809 (rank : 50) | NC score | 0.061592 (rank : 22) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
PFD2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 51) | NC score | 0.030776 (rank : 72) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O70591, Q8R0H2 | Gene names | Pfdn2, Pfd2 | |||
|
Domain Architecture |
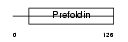
|
|||||
| Description | Prefoldin subunit 2. | |||||
|
RAD50_MOUSE
|
||||||
| θ value | 8.99809 (rank : 52) | NC score | 0.048731 (rank : 60) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1366 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P70388, Q6PEB0, Q8C2T7, Q9CU59 | Gene names | Rad50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (mRad50). | |||||
|
TFAP4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 53) | NC score | 0.019261 (rank : 74) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q01664, O60409 | Gene names | TFAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor AP-4 (Activating enhancer-binding protein 4). | |||||
|
CASP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 54) | NC score | 0.052838 (rank : 39) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q13948, Q53GU9, Q8TBS3 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
CASP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 55) | NC score | 0.053078 (rank : 38) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1152 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P70403, Q7TQJ4, Q91WN6 | Gene names | Cutl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
CCD19_HUMAN
|
||||||
| θ value | θ > 10 (rank : 56) | NC score | 0.051631 (rank : 47) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UL16 | Gene names | CCDC19, NESG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 19 (Nasopharyngeal epithelium- specific protein 1). | |||||
|
CCD21_MOUSE
|
||||||
| θ value | θ > 10 (rank : 57) | NC score | 0.060416 (rank : 23) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8BMK0, Q8BUF1, Q8K0E6 | Gene names | Ccdc21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
CCD39_HUMAN
|
||||||
| θ value | θ > 10 (rank : 58) | NC score | 0.058466 (rank : 25) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 868 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9UFE4 | Gene names | CCDC39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 39. | |||||
|
CCD39_MOUSE
|
||||||
| θ value | θ > 10 (rank : 59) | NC score | 0.050310 (rank : 53) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1026 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9D5Y1, Q8CDG6 | Gene names | Ccdc39, D3Ertd789e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 39. | |||||
|
CCD62_HUMAN
|
||||||
| θ value | θ > 10 (rank : 60) | NC score | 0.054127 (rank : 32) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 631 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6P9F0, Q6ZVF2, Q86VJ0, Q9BYZ5 | Gene names | CCDC62 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 60 (Aaa-protein) (Protein TSP- NY). | |||||
|
CENPF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 61) | NC score | 0.050943 (rank : 50) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
CEP63_HUMAN
|
||||||
| θ value | θ > 10 (rank : 62) | NC score | 0.052069 (rank : 43) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1005 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q96MT8, Q96CR0, Q9H8F5, Q9H8N0 | Gene names | CEP63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 63 kDa (Cep63 protein). | |||||
|
CN145_HUMAN
|
||||||
| θ value | θ > 10 (rank : 63) | NC score | 0.050331 (rank : 52) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1196 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q6ZU80, Q96ML4 | Gene names | C14orf145 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf145. | |||||
|
CN145_MOUSE
|
||||||
| θ value | θ > 10 (rank : 64) | NC score | 0.052216 (rank : 42) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8BI22 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf145 homolog. | |||||
|
GOGA1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.057604 (rank : 26) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 994 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9CW79, Q80YB0 | Gene names | Golga1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
GOGA3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.053522 (rank : 35) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1437 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P55937, Q80VF5, Q8CCK4, Q9QYT2, Q9QYT3 | Gene names | Golga3, Mea2 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Male-enhanced antigen 2) (MEA-2). | |||||
|
GOGA5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.051705 (rank : 46) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1168 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8TBA6, O95287, Q03962, Q9UQQ7 | Gene names | GOLGA5, RETII, RFG5 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (RET-fused gene 5 protein) (Ret-II protein). | |||||
|
GOGA5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.053282 (rank : 36) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1434 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9QYE6, O88317, Q3TGE7, Q3U6S5, Q3UUF9 | Gene names | Golga5, Retii | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (Sumiko protein) (Ret-II protein). | |||||
|
HMMR_HUMAN
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.052527 (rank : 41) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O75330, Q92767 | Gene names | HMMR, IHABP, RHAMM | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility) (CD168 antigen). | |||||
|
HMMR_MOUSE
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.052792 (rank : 40) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q00547 | Gene names | Hmmr, Ihabp, Rhamm | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility). | |||||
|
IFT74_HUMAN
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.051614 (rank : 48) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q96LB3, Q6PGQ8, Q9H643, Q9H8G7 | Gene names | IFT74, CCDC2, CMG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 74 homolog (Coiled-coil domain-containing protein 2) (Capillary morphogenesis protein 1) (CMG-1). | |||||
|
KTN1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.051792 (rank : 45) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1324 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q61595, Q8BG49, Q8BHF4, Q8BHM8, Q8C9Y5, Q8CG51, Q8CG52, Q8CG53, Q8CG54, Q8CG55, Q8CG56, Q8CG57, Q8CG58, Q8CG59, Q8CG60, Q8CG61, Q8CG62, Q8CG63 | Gene names | Ktn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin. | |||||
|
MD1L1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.050500 (rank : 51) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1001 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9WTX8, Q9WTX9 | Gene names | Mad1l1, Mad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1). | |||||
|
NUF2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.062352 (rank : 20) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 661 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9BZD4, Q8WU69, Q96HJ4, Q96Q78 | Gene names | CDCA1, NUF2, NUF2R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Nuf2 (hNuf2) (hNuf2R) (hsNuf2) (Cell division cycle-associated protein 1). | |||||
|
RAD50_HUMAN
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.050205 (rank : 55) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q92878, O43254, Q6GMT7, Q6P5X3, Q9UP86 | Gene names | RAD50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (hRAD50). | |||||
|
SMC4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.053766 (rank : 33) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9NTJ3, O95752, Q8NDL4, Q9UNT9 | Gene names | SMC4L1, CAPC, SMC4 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 4-like 1 protein (Chromosome- associated polypeptide C) (hCAP-C) (XCAP-C homolog). | |||||
|
SYCP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.055777 (rank : 29) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1456 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q62209, O09205, P70192, Q62329 | Gene names | Sycp1, Scp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
T3JAM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.050028 (rank : 57) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 535 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8C0G2, Q8BY50, Q8K1Y9 | Gene names | Traf3ip3, T3jam | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRAF3-interacting JNK-activating modulator (TRAF3-interacting protein 3). | |||||
|
TAXB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.051533 (rank : 49) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 829 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q86VP1, O60398, O95770, Q13311, Q9BQG5, Q9UI88 | Gene names | TAX1BP1, T6BP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tax1-binding protein 1 (TRAF6-binding protein). | |||||
|
TMF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.050283 (rank : 54) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 987 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P82094 | Gene names | TMF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TATA element modulatory factor (TMF). | |||||
|
TPD52_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 9.41505e-74 (rank : 1) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q62393 | Gene names | Tpd52 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor protein D52 (mD52). | |||||
|
TPD52_HUMAN
|
||||||
| NC score | 0.985032 (rank : 2) | θ value | 8.53533e-67 (rank : 2) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P55327, Q13056 | Gene names | TPD52 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor protein D52 (N8 protein). | |||||
|
TPD53_HUMAN
|
||||||
| NC score | 0.936185 (rank : 3) | θ value | 5.76516e-39 (rank : 3) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q16890 | Gene names | TPD52L1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor protein D53 (hD53) (Tumor protein D52-like 1). | |||||
|
TPD53_MOUSE
|
||||||
| NC score | 0.933408 (rank : 4) | θ value | 1.57e-36 (rank : 4) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O54818 | Gene names | Tpd52l1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor protein D53 (mD53) (Tumor protein D52-like 1). | |||||
|
TPD54_HUMAN
|
||||||
| NC score | 0.910600 (rank : 5) | θ value | 9.8567e-31 (rank : 5) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O43399, Q9H3Z6 | Gene names | TPD52L2 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor protein D54 (hD54) (Tumor protein D52-like 2). | |||||
|
TPD54_MOUSE
|
||||||
| NC score | 0.902988 (rank : 6) | θ value | 1.02001e-27 (rank : 6) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9CYZ2, Q3TVY1 | Gene names | Tpd52l2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor protein D54 (Tumor protein D52-like 2). | |||||
|
SAS6_MOUSE
|
||||||
| NC score | 0.108041 (rank : 7) | θ value | 0.0330416 (rank : 9) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q80UK7, Q9CYT4 | Gene names | Sass6, Sas6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spindle assembly abnormal protein 6 homolog. | |||||
|
SAS6_HUMAN
|
||||||
| NC score | 0.106582 (rank : 8) | θ value | 0.0563607 (rank : 10) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 791 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q6UVJ0, Q8N3K0 | Gene names | SASS6, SAS6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spindle assembly abnormal protein 6 homolog (HsSAS-6). | |||||
|
NUF2_MOUSE
|
||||||
| NC score | 0.095591 (rank : 9) | θ value | 0.0330416 (rank : 8) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 585 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q99P69, Q8BTK7, Q8VE05, Q9CST5 | Gene names | Cdca1, Nuf2r | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Nuf2 (Cell division cycle-associated protein 1). | |||||
|
CCD21_HUMAN
|
||||||
| NC score | 0.078429 (rank : 10) | θ value | 0.0330416 (rank : 7) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1228 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q6P2H3, Q5VY68, Q5VY70, Q9H6Q1, Q9H828, Q9UF52 | Gene names | CCDC21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
RB6I2_MOUSE
|
||||||
| NC score | 0.075173 (rank : 11) | θ value | 0.47712 (rank : 18) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q99MI1, Q80TK7, Q8BPL1, Q8C7Y1, Q99MI2 | Gene names | Erc1, Cast2, Kiaa1081, Rab6ip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1) (ERC1) (CAZ-associated structural protein 2) (CAST2). | |||||
|
RB6I2_HUMAN
|
||||||
| NC score | 0.073988 (rank : 12) | θ value | 0.47712 (rank : 17) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1584 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8IUD2, Q6NVK2, Q8IUD3, Q8IUD4, Q8IUD5, Q8NAS1, Q9NXN5, Q9UIK7, Q9UPS1 | Gene names | ERC1, ELKS, KIAA1081, RAB6IP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1). | |||||
|
DYNA_MOUSE
|
||||||
| NC score | 0.069141 (rank : 13) | θ value | 0.365318 (rank : 13) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1084 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O08788 | Gene names | Dctn1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynactin-1 (150 kDa dynein-associated polypeptide) (DP-150) (DAP-150) (p150-glued). | |||||
|
PCNT_MOUSE
|
||||||
| NC score | 0.067345 (rank : 14) | θ value | 0.47712 (rank : 16) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1207 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P48725 | Gene names | Pcnt, Pcnt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pericentrin. | |||||
|
DYNA_HUMAN
|
||||||
| NC score | 0.067026 (rank : 15) | θ value | 0.813845 (rank : 22) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1194 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q14203, O95296, Q9BRM9, Q9UIU1, Q9UIU2 | Gene names | DCTN1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynactin-1 (150 kDa dynein-associated polypeptide) (DP-150) (DAP-150) (p150-glued) (p135). | |||||
|
NUMA1_HUMAN
|
||||||
| NC score | 0.066664 (rank : 16) | θ value | 0.47712 (rank : 15) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
ERC2_MOUSE
|
||||||
| NC score | 0.066508 (rank : 17) | θ value | 5.27518 (rank : 36) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1166 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q6PH08, Q80U20, Q8CCP1 | Gene names | Erc2, Cast1, D14Ertd171e, Kiaa0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2 (CAZ-associated structural protein 1) (CAST1). | |||||
|
ERC2_HUMAN
|
||||||
| NC score | 0.066356 (rank : 18) | θ value | 5.27518 (rank : 35) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1156 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O15083, Q86TK4 | Gene names | ERC2, KIAA0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2. | |||||
|
SYCP1_HUMAN
|
||||||
| NC score | 0.062434 (rank : 19) | θ value | 0.125558 (rank : 11) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q15431, O14963 | Gene names | SYCP1, SCP1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
NUF2_HUMAN
|
||||||
| NC score | 0.062352 (rank : 20) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 661 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9BZD4, Q8WU69, Q96HJ4, Q96Q78 | Gene names | CDCA1, NUF2, NUF2R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Nuf2 (hNuf2) (hNuf2R) (hsNuf2) (Cell division cycle-associated protein 1). | |||||
|
GOGA1_HUMAN
|
||||||
| NC score | 0.062019 (rank : 21) | θ value | 1.06291 (rank : 24) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1222 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q92805, Q8IYZ9 | Gene names | GOLGA1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
PCNT_HUMAN
|
||||||
| NC score | 0.061592 (rank : 22) | θ value | 8.99809 (rank : 50) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
CCD21_MOUSE
|
||||||
| NC score | 0.060416 (rank : 23) | θ value | θ > 10 (rank : 57) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8BMK0, Q8BUF1, Q8K0E6 | Gene names | Ccdc21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
GOGA3_HUMAN
|
||||||
| NC score | 0.059630 (rank : 24) | θ value | 0.813845 (rank : 23) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q08378, O43241, Q6P9C7, Q86XW3, Q8TDA9, Q8WZA3 | Gene names | GOLGA3 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Golgi complex-associated protein of 170 kDa) (GCP170). | |||||
|
CCD39_HUMAN
|
||||||
| NC score | 0.058466 (rank : 25) | θ value | θ > 10 (rank : 58) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 868 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9UFE4 | Gene names | CCDC39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 39. | |||||
|
GOGA1_MOUSE
|
||||||
| NC score | 0.057604 (rank : 26) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 994 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9CW79, Q80YB0 | Gene names | Golga1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
CUTL1_MOUSE
|
||||||
| NC score | 0.057358 (rank : 27) | θ value | 0.813845 (rank : 21) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P53564, O08994, P70301, Q571L6, Q91ZD2 | Gene names | Cutl1, Cux, Cux1, Kiaa4047 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP) (Homeobox protein Cux). | |||||
|
TAXB1_MOUSE
|
||||||
| NC score | 0.056405 (rank : 28) | θ value | 5.27518 (rank : 39) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q3UKC1, Q91YT6, Q9CVF0, Q9DC45 | Gene names | Tax1bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tax1-binding protein 1 homolog. | |||||
|
SYCP1_MOUSE
|
||||||
| NC score | 0.055777 (rank : 29) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1456 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q62209, O09205, P70192, Q62329 | Gene names | Sycp1, Scp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
NIN_MOUSE
|
||||||
| NC score | 0.055657 (rank : 30) | θ value | 8.99809 (rank : 49) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q61043, Q6ZPM7 | Gene names | Nin, Kiaa1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein. | |||||
|
KTN1_HUMAN
|
||||||
| NC score | 0.055469 (rank : 31) | θ value | 4.03905 (rank : 32) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q86UP2, Q13999, Q14707, Q15387, Q86W57 | Gene names | KTN1, CG1, KIAA0004 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin (Kinesin receptor) (CG-1 antigen). | |||||
|
CCD62_HUMAN
|
||||||
| NC score | 0.054127 (rank : 32) | θ value | θ > 10 (rank : 60) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 631 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6P9F0, Q6ZVF2, Q86VJ0, Q9BYZ5 | Gene names | CCDC62 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 60 (Aaa-protein) (Protein TSP- NY). | |||||
|
SMC4_HUMAN
|
||||||
| NC score | 0.053766 (rank : 33) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9NTJ3, O95752, Q8NDL4, Q9UNT9 | Gene names | SMC4L1, CAPC, SMC4 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 4-like 1 protein (Chromosome- associated polypeptide C) (hCAP-C) (XCAP-C homolog). | |||||
|
TRIPB_HUMAN
|
||||||
| NC score | 0.053526 (rank : 34) | θ value | 4.03905 (rank : 34) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
GOGA3_MOUSE
|
||||||
| NC score | 0.053522 (rank : 35) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1437 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P55937, Q80VF5, Q8CCK4, Q9QYT2, Q9QYT3 | Gene names | Golga3, Mea2 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Male-enhanced antigen 2) (MEA-2). | |||||
|
GOGA5_MOUSE
|
||||||
| NC score | 0.053282 (rank : 36) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1434 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9QYE6, O88317, Q3TGE7, Q3U6S5, Q3UUF9 | Gene names | Golga5, Retii | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (Sumiko protein) (Ret-II protein). | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.053251 (rank : 37) | θ value | 8.99809 (rank : 48) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
CASP_MOUSE
|
||||||
| NC score | 0.053078 (rank : 38) | θ value | θ > 10 (rank : 55) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1152 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P70403, Q7TQJ4, Q91WN6 | Gene names | Cutl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
CASP_HUMAN
|
||||||
| NC score | 0.052838 (rank : 39) | θ value | θ > 10 (rank : 54) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q13948, Q53GU9, Q8TBS3 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
HMMR_MOUSE
|
||||||
| NC score | 0.052792 (rank : 40) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q00547 | Gene names | Hmmr, Ihabp, Rhamm | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility). | |||||
|
HMMR_HUMAN
|
||||||
| NC score | 0.052527 (rank : 41) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O75330, Q92767 | Gene names | HMMR, IHABP, RHAMM | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility) (CD168 antigen). | |||||
|
CN145_MOUSE
|
||||||
| NC score | 0.052216 (rank : 42) | θ value | θ > 10 (rank : 64) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8BI22 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf145 homolog. | |||||
|
CEP63_HUMAN
|
||||||
| NC score | 0.052069 (rank : 43) | θ value | θ > 10 (rank : 62) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1005 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q96MT8, Q96CR0, Q9H8F5, Q9H8N0 | Gene names | CEP63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 63 kDa (Cep63 protein). | |||||
|
CK5P2_MOUSE
|
||||||
| NC score | 0.052009 (rank : 44) | θ value | 6.88961 (rank : 40) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1151 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8K389, Q6PCN1, Q6ZPL0 | Gene names | Cdk5rap2, Kiaa1633 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48). | |||||
|
KTN1_MOUSE
|
||||||
| NC score | 0.051792 (rank : 45) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1324 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q61595, Q8BG49, Q8BHF4, Q8BHM8, Q8C9Y5, Q8CG51, Q8CG52, Q8CG53, Q8CG54, Q8CG55, Q8CG56, Q8CG57, Q8CG58, Q8CG59, Q8CG60, Q8CG61, Q8CG62, Q8CG63 | Gene names | Ktn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin. | |||||
|
GOGA5_HUMAN
|
||||||
| NC score | 0.051705 (rank : 46) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1168 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8TBA6, O95287, Q03962, Q9UQQ7 | Gene names | GOLGA5, RETII, RFG5 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (RET-fused gene 5 protein) (Ret-II protein). | |||||
|
CCD19_HUMAN
|
||||||
| NC score | 0.051631 (rank : 47) | θ value | θ > 10 (rank : 56) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UL16 | Gene names | CCDC19, NESG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 19 (Nasopharyngeal epithelium- specific protein 1). | |||||
|
IFT74_HUMAN
|
||||||
| NC score | 0.051614 (rank : 48) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q96LB3, Q6PGQ8, Q9H643, Q9H8G7 | Gene names | IFT74, CCDC2, CMG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 74 homolog (Coiled-coil domain-containing protein 2) (Capillary morphogenesis protein 1) (CMG-1). | |||||
|
TAXB1_HUMAN
|
||||||
| NC score | 0.051533 (rank : 49) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 829 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q86VP1, O60398, O95770, Q13311, Q9BQG5, Q9UI88 | Gene names | TAX1BP1, T6BP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tax1-binding protein 1 (TRAF6-binding protein). | |||||
|
CENPF_HUMAN
|
||||||
| NC score | 0.050943 (rank : 50) | θ value | θ > 10 (rank : 61) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
MD1L1_MOUSE
|
||||||
| NC score | 0.050500 (rank : 51) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1001 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9WTX8, Q9WTX9 | Gene names | Mad1l1, Mad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1). | |||||
|
CN145_HUMAN
|
||||||
| NC score | 0.050331 (rank : 52) | θ value | θ > 10 (rank : 63) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1196 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q6ZU80, Q96ML4 | Gene names | C14orf145 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf145. | |||||
|
CCD39_MOUSE
|
||||||
| NC score | 0.050310 (rank : 53) | θ value | θ > 10 (rank : 59) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1026 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9D5Y1, Q8CDG6 | Gene names | Ccdc39, D3Ertd789e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 39. | |||||
|
TMF1_HUMAN
|
||||||
| NC score | 0.050283 (rank : 54) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 987 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P82094 | Gene names | TMF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TATA element modulatory factor (TMF). | |||||
|
RAD50_HUMAN
|
||||||
| NC score | 0.050205 (rank : 55) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q92878, O43254, Q6GMT7, Q6P5X3, Q9UP86 | Gene names | RAD50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (hRAD50). | |||||
|
FYCO1_HUMAN
|
||||||
| NC score | 0.050134 (rank : 56) | θ value | 8.99809 (rank : 45) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1149 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9BQS8, Q3MJE6, Q86T41, Q86TB1, Q8TEF9, Q96IV5, Q9H8P9 | Gene names | FYCO1, ZFYVE7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1 (Zinc finger FYVE domain-containing protein 7). | |||||
|
T3JAM_MOUSE
|
||||||
| NC score | 0.050028 (rank : 57) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 535 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8C0G2, Q8BY50, Q8K1Y9 | Gene names | Traf3ip3, T3jam | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRAF3-interacting JNK-activating modulator (TRAF3-interacting protein 3). | |||||
|
ZN318_HUMAN
|
||||||
| NC score | 0.049943 (rank : 58) | θ value | 1.06291 (rank : 26) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5VUA4, O94796, Q4G0E4, Q8NEM6, Q9UNU8, Q9Y2W9 | Gene names | ZNF318 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 318 (Endocrine regulatory protein). | |||||
|
STXB4_MOUSE
|
||||||
| NC score | 0.049177 (rank : 59) | θ value | 6.88961 (rank : 41) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 629 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9WV89, Q5SUA6, Q5SUA8, Q5SUA9, Q8CFL1 | Gene names | Stxbp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Syntaxin-binding protein 4 (Syntaxin 4-interacting protein) (Synip). | |||||
|
RAD50_MOUSE
|
||||||
| NC score | 0.048731 (rank : 60) | θ value | 8.99809 (rank : 52) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1366 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P70388, Q6PEB0, Q8C2T7, Q9CU59 | Gene names | Rad50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (mRad50). | |||||
|
MYH4_MOUSE
|
||||||
| NC score | 0.046766 (rank : 61) | θ value | 3.0926 (rank : 29) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
MYH8_MOUSE
|
||||||
| NC score | 0.046246 (rank : 62) | θ value | 3.0926 (rank : 30) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1600 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P13542, Q5SX36 | Gene names | Myh8, Myhsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
MYH13_HUMAN
|
||||||
| NC score | 0.045546 (rank : 63) | θ value | 5.27518 (rank : 37) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1504 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9UKX3, O95252 | Gene names | MYH13 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-13 (Myosin heavy chain, skeletal muscle, extraocular) (MyHC- eo). | |||||
|
KIF11_HUMAN
|
||||||
| NC score | 0.045443 (rank : 64) | θ value | 0.62314 (rank : 19) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 583 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P52732, Q15716, Q5VWX0 | Gene names | KIF11, EG5, KNSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF11 (Kinesin-related motor protein Eg5) (Kinesin-like spindle protein HKSP) (Thyroid receptor-interacting protein 5) (TRIP5) (Kinesin-like protein 1). | |||||
|
MYH4_HUMAN
|
||||||
| NC score | 0.044859 (rank : 65) | θ value | 4.03905 (rank : 33) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1377 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9Y623 | Gene names | MYH4 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal) (Myosin heavy chain IIb) (MyHC-IIb). | |||||
|
MYH7_HUMAN
|
||||||
| NC score | 0.042975 (rank : 66) | θ value | 5.27518 (rank : 38) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P12883, Q14836, Q14837, Q14904, Q16579, Q92679, Q9H1D5, Q9UMM8 | Gene names | MYH7, MYHCB | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle beta isoform (MyHC-beta). | |||||
|
ATX3_HUMAN
|
||||||
| NC score | 0.042806 (rank : 67) | θ value | 4.03905 (rank : 31) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P54252, O15284, O15285, O15286, Q8N189, Q96TC3, Q96TC4, Q9H3N0 | Gene names | ATXN3, ATX3, MJD, MJD1, SCA3 | |||
|
Domain Architecture |

|
|||||
| Description | Ataxin-3 (EC 3.4.22.-) (Machado-Joseph disease protein 1) (Spinocerebellar ataxia type 3 protein). | |||||
|
LRSM1_HUMAN
|
||||||
| NC score | 0.036913 (rank : 68) | θ value | 2.36792 (rank : 28) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1078 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q6UWE0, Q5VVV0, Q8NB40, Q96GT5, Q96MX5, Q96MZ7 | Gene names | LRSAM1, TAL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein LRSAM1 (EC 6.3.2.-) (Leucine-rich repeat and sterile alpha motif-containing protein 1) (Tsg101-associated ligase) (hTAL). | |||||
|
KIF11_MOUSE
|
||||||
| NC score | 0.036762 (rank : 69) | θ value | 2.36792 (rank : 27) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 446 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6P9P6, Q9Z1J0 | Gene names | Kif11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin-like protein KIF11 (Kinesin-related motor protein Eg5). | |||||
|
NCOR1_HUMAN
|
||||||
| NC score | 0.034543 (rank : 70) | θ value | 0.47712 (rank : 14) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O75376, Q9UPV5, Q9UQ18 | Gene names | NCOR1, KIAA1047 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR). | |||||
|
SCAR3_MOUSE
|
||||||
| NC score | 0.032992 (rank : 71) | θ value | 1.06291 (rank : 25) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8C850, Q8C6N1 | Gene names | Scara3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class A member 3. | |||||
|
PFD2_MOUSE
|
||||||
| NC score | 0.030776 (rank : 72) | θ value | 8.99809 (rank : 51) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O70591, Q8R0H2 | Gene names | Pfdn2, Pfd2 | |||
|
Domain Architecture |

|
|||||
| Description | Prefoldin subunit 2. | |||||
|
MRCKA_HUMAN
|
||||||
| NC score | 0.027816 (rank : 73) | θ value | 0.62314 (rank : 20) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1999 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q5VT25, Q59GZ1, Q5H9N9, Q5T797, Q5VT26, Q5VT27, Q86XX2, Q86XX3, Q99646 | Gene names | CDC42BPA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK alpha (EC 2.7.11.1) (CDC42- binding protein kinase alpha) (Myotonic dystrophy kinase-related CDC42-binding kinase alpha) (Myotonic dystrophy protein kinase-like alpha) (MRCK alpha) (DMPK-like alpha). | |||||
|
TFAP4_HUMAN
|
||||||
| NC score | 0.019261 (rank : 74) | θ value | 8.99809 (rank : 53) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q01664, O60409 | Gene names | TFAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor AP-4 (Activating enhancer-binding protein 4). | |||||
|
FER_HUMAN
|
||||||
| NC score | 0.015166 (rank : 75) | θ value | 0.279714 (rank : 12) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1278 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P16591 | Gene names | FER, TYK3 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase FER (EC 2.7.10.2) (p94-FER) (c- FER). | |||||
|
TRI39_MOUSE
|
||||||
| NC score | 0.015109 (rank : 76) | θ value | 6.88961 (rank : 43) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 691 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9ESN2, Q8BPR5, Q8K0F7 | Gene names | Trim39, Rnf23, Tfp | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 39 (RING finger protein 23) (Testis-abundant finger protein). | |||||
|
TRI39_HUMAN
|
||||||
| NC score | 0.014838 (rank : 77) | θ value | 6.88961 (rank : 42) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9HCM9, Q76BL3, Q8IYT9, Q96IB6 | Gene names | TRIM39, RNF23, TFP | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 39 (RING finger protein 23) (Testis-abundant finger protein). | |||||
|
GABP2_HUMAN
|
||||||
| NC score | 0.011202 (rank : 78) | θ value | 8.99809 (rank : 46) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 454 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q06547, Q06545, Q12940, Q12941, Q12942, Q8IYD0 | Gene names | GABPB2, E4TF1B, GABPB, GABPB1 | |||
|
Domain Architecture |

|
|||||
| Description | GA-binding protein beta chain (GABP subunit beta-2) (GABP-2) (GABP subunit beta-1) (GABPB-1) (Transcription factor E4TF1-53) (Transcription factor E4TF1-47) (Nuclear respiratory factor 2). | |||||
|
GABP2_MOUSE
|
||||||
| NC score | 0.010341 (rank : 79) | θ value | 8.99809 (rank : 47) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 429 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q00420, Q00421, Q8BS31, Q91YZ0, Q9QVV2 | Gene names | Gabpb2, Gabpb, Gabpb1 | |||
|
Domain Architecture |

|
|||||
| Description | GA-binding protein beta chain (GABP subunit beta-2) (GABP-2) (GABP subunit beta-1) (GABPB-1). | |||||
|
ZN307_HUMAN
|
||||||
| NC score | -0.001656 (rank : 80) | θ value | 6.88961 (rank : 44) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q969J2 | Gene names | ZNF307 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 307 (P373c6.1). | |||||