Please be patient as the page loads
|
TANK_HUMAN
|
||||||
| SwissProt Accessions | Q92844, Q92885 | Gene names | TANK, ITRAF | |||
|
Domain Architecture |

|
|||||
| Description | TRAF family member-associated NF-kappa-B activator (TRAF-interacting protein) (I-TRAF). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
TANK_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q92844, Q92885 | Gene names | TANK, ITRAF | |||
|
Domain Architecture |

|
|||||
| Description | TRAF family member-associated NF-kappa-B activator (TRAF-interacting protein) (I-TRAF). | |||||
|
TANK_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.958225 (rank : 2) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P70347, Q61178 | Gene names | Tank, Itraf | |||
|
Domain Architecture |

|
|||||
| Description | TRAF family member-associated NF-kappa-B activator (TRAF-interacting protein) (I-TRAF). | |||||
|
AKAP9_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 3) | NC score | 0.084723 (rank : 4) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
SPAG5_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 4) | NC score | 0.083336 (rank : 5) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 612 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q7TME2, Q8CH61 | Gene names | Spag5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 5 (Mastrin) (Mitotic spindle-associated protein p126) (MAP126). | |||||
|
SLK_HUMAN
|
||||||
| θ value | 0.125558 (rank : 5) | NC score | 0.029053 (rank : 119) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1775 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9H2G2, O00211, Q6P1Z4, Q86WU7, Q86WW1, Q92603, Q9NQL0, Q9NQL1 | Gene names | SLK, KIAA0204, STK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (hSLK) (Serine/threonine-protein kinase 2) (CTCL tumor antigen se20-9). | |||||
|
GUC2F_HUMAN
|
||||||
| θ value | 0.163984 (rank : 6) | NC score | 0.030488 (rank : 117) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 740 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P51841, Q9UJF1 | Gene names | GUCY2F, GUC2F, RETGC2 | |||
|
Domain Architecture |

|
|||||
| Description | Retinal guanylyl cyclase 2 precursor (EC 4.6.1.2) (Guanylate cyclase 2F, retinal) (RETGC-2) (Rod outer segment membrane guanylate cyclase 2) (ROS-GC2) (Guanylate cyclase F) (GC-F). | |||||
|
GCC2_MOUSE
|
||||||
| θ value | 0.279714 (rank : 7) | NC score | 0.070480 (rank : 17) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
MASTL_HUMAN
|
||||||
| θ value | 0.279714 (rank : 8) | NC score | 0.024033 (rank : 123) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 894 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96GX5, Q5T8D5, Q5T8D7, Q8NCD6, Q96SJ5 | Gene names | MASTL, THC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase-like (EC 2.7.11.1). | |||||
|
DMD_HUMAN
|
||||||
| θ value | 0.365318 (rank : 9) | NC score | 0.057196 (rank : 35) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P11532, Q02295, Q14169, Q14170, Q5JYU0, Q7KZ48 | Gene names | DMD | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
GOGA4_MOUSE
|
||||||
| θ value | 0.365318 (rank : 10) | NC score | 0.072555 (rank : 16) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
GUC2D_HUMAN
|
||||||
| θ value | 0.365318 (rank : 11) | NC score | 0.039170 (rank : 108) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 715 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q02846, Q6LEA7 | Gene names | GUCY2D, CORD6, GUC1A4, GUC2D, RETGC, RETGC1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinal guanylyl cyclase 1 precursor (EC 4.6.1.2) (Guanylate cyclase 2D, retinal) (RETGC-1) (Rod outer segment membrane guanylate cyclase) (ROS-GC). | |||||
|
TEX9_HUMAN
|
||||||
| θ value | 0.365318 (rank : 12) | NC score | 0.081517 (rank : 6) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 541 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8N6V9 | Gene names | TEX9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed sequence 9 protein. | |||||
|
MD1L1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 13) | NC score | 0.068222 (rank : 19) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9Y6D9, Q13312, Q75MI0, Q86UM4, Q9UNH0 | Gene names | MAD1L1, MAD1, TXBP181 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1) (Mitotic checkpoint MAD1 protein-homolog) (HsMAD1) (hMAD1) (Tax-binding protein 181). | |||||
|
ZC313_HUMAN
|
||||||
| θ value | 0.47712 (rank : 14) | NC score | 0.086271 (rank : 3) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q5T200, O94936, Q5T1Z9, Q7Z7J3, Q8NDT6, Q9H0L6 | Gene names | ZC3H13, KIAA0853 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 13. | |||||
|
ATN1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 15) | NC score | 0.033955 (rank : 114) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
GLT14_HUMAN
|
||||||
| θ value | 0.62314 (rank : 16) | NC score | 0.016246 (rank : 127) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96FL9, Q8IVI4, Q9BRH1, Q9H827, Q9H9J8 | Gene names | GALNT14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 14 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 14) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 14) (Polypeptide GalNAc transferase 14) (GalNAc-T14) (pp-GaNTase 14). | |||||
|
ERC2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 17) | NC score | 0.076936 (rank : 7) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1156 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O15083, Q86TK4 | Gene names | ERC2, KIAA0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2. | |||||
|
NDE1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 18) | NC score | 0.072986 (rank : 15) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 823 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9NXR1, Q49AQ2 | Gene names | NDE1, NUDE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear distribution protein nudE homolog 1 (NudE). | |||||
|
SLK_MOUSE
|
||||||
| θ value | 1.06291 (rank : 19) | NC score | 0.026948 (rank : 121) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1798 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O54988, Q80U65, Q8CAU2, Q8CDW2, Q9WU41 | Gene names | Slk, Kiaa0204, Stk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (mSLK) (Serine/threonine-protein kinase 2) (STE20-related kinase SMAK) (Etk4). | |||||
|
MYH14_HUMAN
|
||||||
| θ value | 1.38821 (rank : 20) | NC score | 0.043670 (rank : 105) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 995 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q7Z406, Q5CZ75, Q6XYE4, Q76B62, Q8WV23, Q96I22, Q9BT27, Q9BW35, Q9H882 | Gene names | MYH14, KIAA2034 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
NEMO_MOUSE
|
||||||
| θ value | 1.38821 (rank : 21) | NC score | 0.070237 (rank : 18) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O88522 | Gene names | Ikbkg, Nemo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NF-kappa-B essential modulator (NEMO) (NF-kappa-B essential modifier) (Inhibitor of nuclear factor kappa-B kinase subunit gamma) (IkB kinase subunit gamma) (I-kappa-B kinase gamma) (IKK-gamma) (IKKG) (IkB kinase-associated protein 1) (IKKAP1) (mFIP-3). | |||||
|
REST_HUMAN
|
||||||
| θ value | 1.38821 (rank : 22) | NC score | 0.060044 (rank : 29) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1605 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P30622 | Gene names | RSN, CYLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Restin (Cytoplasmic linker protein 170 alpha-2) (CLIP-170) (Reed- Sternberg intermediate filament-associated protein) (Cytoplasmic linker protein 1). | |||||
|
SMC4_MOUSE
|
||||||
| θ value | 1.38821 (rank : 23) | NC score | 0.054933 (rank : 47) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 830 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8CG47, Q8BTS7, Q8BTY9, Q99K21 | Gene names | Smc4l1, Capc, Smc4 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 4-like 1 protein (Chromosome- associated polypeptide C) (XCAP-C homolog). | |||||
|
TEX9_MOUSE
|
||||||
| θ value | 1.38821 (rank : 24) | NC score | 0.076054 (rank : 10) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9D845, O54764 | Gene names | Tex9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed sequence 9 protein (Testis-specifically expressed protein 1) (Tsec-1). | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 25) | NC score | 0.061090 (rank : 28) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
GUC2E_MOUSE
|
||||||
| θ value | 1.81305 (rank : 26) | NC score | 0.034083 (rank : 113) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 692 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P52785 | Gene names | Gucy2e, Guc2e | |||
|
Domain Architecture |

|
|||||
| Description | Guanylyl cyclase GC-E precursor (EC 4.6.1.2) (Guanylate cyclase 2E). | |||||
|
ANR12_HUMAN
|
||||||
| θ value | 2.36792 (rank : 27) | NC score | 0.041358 (rank : 106) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1081 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q6UB98, O94951, Q658K1, Q6QMF7, Q9H231, Q9H784 | Gene names | ANKRD12, ANCO2, KIAA0874 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 12 (Ankyrin repeat-containing cofactor 2) (GAC-1 protein). | |||||
|
CAR10_MOUSE
|
||||||
| θ value | 2.36792 (rank : 28) | NC score | 0.052548 (rank : 73) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 407 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P58660 | Gene names | Card10, Bimp1 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 10 (Bcl10-interacting MAGUK protein 1) (Bimp1). | |||||
|
EEA1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 29) | NC score | 0.062853 (rank : 24) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
ERC2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 30) | NC score | 0.074272 (rank : 12) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1166 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q6PH08, Q80U20, Q8CCP1 | Gene names | Erc2, Cast1, D14Ertd171e, Kiaa0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2 (CAZ-associated structural protein 1) (CAST1). | |||||
|
HOOK3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 31) | NC score | 0.053671 (rank : 60) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1070 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q86VS8, Q8NBH0, Q9BY13 | Gene names | HOOK3 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 3 (hHK3). | |||||
|
MOES_MOUSE
|
||||||
| θ value | 2.36792 (rank : 32) | NC score | 0.037798 (rank : 111) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 923 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P26041, Q3UL28, Q8BSN4 | Gene names | Msn | |||
|
Domain Architecture |

|
|||||
| Description | Moesin (Membrane-organizing extension spike protein). | |||||
|
NDEL1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 33) | NC score | 0.076376 (rank : 8) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9GZM8, Q86T80, Q8TAR7, Q9UH50 | Gene names | NDEL1, EOPA, MITAP1, NUDEL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear distribution protein nudE-like 1 (Protein Nudel) (Mitosin- associated protein 1). | |||||
|
NDEL1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 34) | NC score | 0.076272 (rank : 9) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9ERR1, Q9D0Q4, Q9EPT6 | Gene names | Ndel1, Nudel | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear distribution protein nudE-like 1 (Protein Nudel) (Protein mNudE-like) (mNudE-L). | |||||
|
RB6I2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 35) | NC score | 0.073625 (rank : 14) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1584 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8IUD2, Q6NVK2, Q8IUD3, Q8IUD4, Q8IUD5, Q8NAS1, Q9NXN5, Q9UIK7, Q9UPS1 | Gene names | ERC1, ELKS, KIAA1081, RAB6IP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1). | |||||
|
RB6I2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 36) | NC score | 0.075196 (rank : 11) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q99MI1, Q80TK7, Q8BPL1, Q8C7Y1, Q99MI2 | Gene names | Erc1, Cast2, Kiaa1081, Rab6ip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1) (ERC1) (CAZ-associated structural protein 2) (CAST2). | |||||
|
TMCC3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 37) | NC score | 0.028844 (rank : 120) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8R310 | Gene names | Tmcc3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane and coiled-coil domains protein 3. | |||||
|
TRHY_HUMAN
|
||||||
| θ value | 2.36792 (rank : 38) | NC score | 0.064391 (rank : 21) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
UACA_MOUSE
|
||||||
| θ value | 2.36792 (rank : 39) | NC score | 0.052373 (rank : 74) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8CGB3, Q69ZG3, Q7TN77, Q8BJC8 | Gene names | Uaca, Kiaa1561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uveal autoantigen with coiled-coil domains and ankyrin repeats protein (Nucling) (Nuclear membrane-binding protein). | |||||
|
K0831_MOUSE
|
||||||
| θ value | 3.0926 (rank : 40) | NC score | 0.054917 (rank : 49) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8CDJ3, Q69ZY1, Q6PFY6, Q8C6N0, Q8R3M3 | Gene names | Kiaa0831, D14Ertd436e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0831. | |||||
|
KIF11_MOUSE
|
||||||
| θ value | 3.0926 (rank : 41) | NC score | 0.029131 (rank : 118) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 446 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6P9P6, Q9Z1J0 | Gene names | Kif11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin-like protein KIF11 (Kinesin-related motor protein Eg5). | |||||
|
NDE1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 42) | NC score | 0.074096 (rank : 13) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 602 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9CZA6, Q3UBS6, Q3UIC1, Q9ERR0 | Gene names | Nde1, Nude | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear distribution protein nudE homolog 1 (NudE) (mNudE). | |||||
|
RSF1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 43) | NC score | 0.036294 (rank : 112) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q96T23, Q86X86, Q9H3L8, Q9NVZ8, Q9NYU0 | Gene names | RSF1, HBXAP, XAP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Remodeling and spacing factor 1 (Rsf-1) (Hepatitis B virus X- associated protein) (HBV pX-associated protein 8) (p325 subunit of RSF chromatin remodelling complex). | |||||
|
SYNE1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 44) | NC score | 0.057570 (rank : 33) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
TRAF5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 45) | NC score | 0.032740 (rank : 115) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O00463 | Gene names | TRAF5, RNF84 | |||
|
Domain Architecture |

|
|||||
| Description | TNF receptor-associated factor 5 (RING finger protein 84). | |||||
|
ANLN_HUMAN
|
||||||
| θ value | 4.03905 (rank : 46) | NC score | 0.026051 (rank : 122) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NQW6, Q5CZ78, Q6NSK5, Q9H8Y4, Q9NVN9, Q9NVP0 | Gene names | ANLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding protein anillin. | |||||
|
REST_MOUSE
|
||||||
| θ value | 4.03905 (rank : 47) | NC score | 0.058865 (rank : 30) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1506 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q922J3, Q571L7 | Gene names | Rsn, Kiaa4046 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin. | |||||
|
KINH_MOUSE
|
||||||
| θ value | 5.27518 (rank : 48) | NC score | 0.040027 (rank : 107) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q61768, O08711, Q61580 | Gene names | Kif5b, Khcs, Kns1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain (Ubiquitous kinesin heavy chain) (UKHC). | |||||
|
MYH10_MOUSE
|
||||||
| θ value | 5.27518 (rank : 49) | NC score | 0.046559 (rank : 103) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1612 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q61879, Q5SV63 | Gene names | Myh10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
PER3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 50) | NC score | 0.017291 (rank : 125) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O70361 | Gene names | Per3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (mPER3). | |||||
|
SDCG8_MOUSE
|
||||||
| θ value | 5.27518 (rank : 51) | NC score | 0.055777 (rank : 43) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q80UF4 | Gene names | Sdccag8, Cccap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serologically defined colon cancer antigen 8 (Centrosomal colon cancer autoantigen protein) (mCCCAP). | |||||
|
CE290_HUMAN
|
||||||
| θ value | 6.88961 (rank : 52) | NC score | 0.061508 (rank : 27) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
CLOCK_MOUSE
|
||||||
| θ value | 6.88961 (rank : 53) | NC score | 0.011725 (rank : 129) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O08785 | Gene names | Clock | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (mCLOCK). | |||||
|
CP250_HUMAN
|
||||||
| θ value | 6.88961 (rank : 54) | NC score | 0.056076 (rank : 41) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
EEA1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 55) | NC score | 0.061902 (rank : 26) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1637 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8BL66, Q6DIC2 | Gene names | Eea1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early endosome antigen 1. | |||||
|
MYH10_HUMAN
|
||||||
| θ value | 6.88961 (rank : 56) | NC score | 0.046075 (rank : 104) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P35580, Q16087 | Gene names | MYH10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
SNPC4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 57) | NC score | 0.017019 (rank : 126) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BP86, Q6PGG7, Q80UG9, Q810L1 | Gene names | Snapc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 4 (SNAPc subunit 4) (snRNA- activating protein complex 190 kDa subunit) (SNAPc 190 kDa subunit). | |||||
|
TAXB1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 58) | NC score | 0.066247 (rank : 20) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q3UKC1, Q91YT6, Q9CVF0, Q9DC45 | Gene names | Tax1bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tax1-binding protein 1 homolog. | |||||
|
TBCD2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 59) | NC score | 0.031953 (rank : 116) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9BYX2 | Gene names | TBC1D2, PARIS1 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 2 (Prostate antigen recognized and identified by SEREX) (PARIS-1). | |||||
|
EP15_HUMAN
|
||||||
| θ value | 8.99809 (rank : 60) | NC score | 0.038517 (rank : 110) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1021 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P42566 | Gene names | EPS15, AF1P | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15 (Protein Eps15) (AF-1p protein). | |||||
|
EVI2B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 61) | NC score | 0.016101 (rank : 128) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P34910 | Gene names | EVI2B, EVDB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EVI2B protein precursor (Ecotropic viral integration site 2B protein). | |||||
|
GOGA4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 62) | NC score | 0.064260 (rank : 22) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
IRAK1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 63) | NC score | 0.008671 (rank : 130) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P51617, Q7Z5V4, Q96RL2 | Gene names | IRAK1, IRAK | |||
|
Domain Architecture |
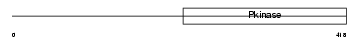
|
|||||
| Description | Interleukin-1 receptor-associated kinase 1 (EC 2.7.11.1) (IRAK-1). | |||||
|
MPP8_HUMAN
|
||||||
| θ value | 8.99809 (rank : 64) | NC score | 0.022912 (rank : 124) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 948 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q99549, Q86TK3, Q9BTP1 | Gene names | MPHOSPH8, MPP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | M-phase phosphoprotein 8. | |||||
|
TXLNG_HUMAN
|
||||||
| θ value | 8.99809 (rank : 65) | NC score | 0.038883 (rank : 109) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 568 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9NUQ3, Q9P0X1 | Gene names | TXLNG, CXorf15, LSR5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gamma-taxilin (Lipopolysaccharide-specific response protein 5). | |||||
|
ANR26_MOUSE
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.050071 (rank : 100) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1554 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q811D2, Q69ZS2, Q9CS61 | Gene names | Ankrd26, Kiaa1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
CAR10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.051222 (rank : 88) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 569 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9BWT7, Q14CQ8, Q5TFG6, Q9UGR5, Q9UGR6, Q9Y3H0 | Gene names | CARD10, CARMA3 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 10 (CARD-containing MAGUK protein 3) (Carma 3). | |||||
|
CARD9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.050367 (rank : 98) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 602 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9H257, Q5SXM6, Q9H854 | Gene names | CARD9 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 9 (hCARD9). | |||||
|
CCD39_HUMAN
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.054910 (rank : 50) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 868 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9UFE4 | Gene names | CCDC39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 39. | |||||
|
CCD39_MOUSE
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.053248 (rank : 64) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1026 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9D5Y1, Q8CDG6 | Gene names | Ccdc39, D3Ertd789e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 39. | |||||
|
CCD41_HUMAN
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.052704 (rank : 71) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1139 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9Y592 | Gene names | CCDC41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41 (NY-REN-58 antigen). | |||||
|
CCD41_MOUSE
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.051342 (rank : 86) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9D5R3, Q3U7X7, Q3UX57, Q80VF0 | Gene names | Ccdc41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41. | |||||
|
CCD46_HUMAN
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.056603 (rank : 40) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1430 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8N8E3, Q6PIB5, Q8NCR4, Q8NFR4 | Gene names | CCDC46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 46. | |||||
|
CCD46_MOUSE
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.054919 (rank : 48) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q5PR68, Q80ZN6, Q99JS4, Q9D9T1 | Gene names | Ccdc46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled coil domain-containing protein 46. | |||||
|
CE152_HUMAN
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.056873 (rank : 37) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 976 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O94986, Q6NTA0 | Gene names | CEP152, KIAA0912 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 152 kDa (Cep152 protein). | |||||
|
CENPE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.050984 (rank : 91) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
CENPF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.056971 (rank : 36) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
CEP63_HUMAN
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.054590 (rank : 52) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1005 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q96MT8, Q96CR0, Q9H8F5, Q9H8N0 | Gene names | CEP63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 63 kDa (Cep63 protein). | |||||
|
CF060_HUMAN
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.063118 (rank : 23) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1270 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8NB25, Q96GY8, Q9H851 | Gene names | C6orf60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf60. | |||||
|
CI039_HUMAN
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.053435 (rank : 61) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1171 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9NXG0, Q5VYJ0, Q9HAJ5 | Gene names | C9orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf39. | |||||
|
CI093_HUMAN
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.055125 (rank : 46) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q6TFL3, Q5SU58, Q6P1W1, Q6ZR13, Q8N1Z4, Q8N8L3, Q8TBR2 | Gene names | C9orf93 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf93. | |||||
|
CJ118_HUMAN
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.052933 (rank : 70) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q7Z3E2, Q2M2V6, Q6NS91, Q7RTP1, Q8N117, Q8N3G3, Q8N6C2, Q9NWA3 | Gene names | C10orf118 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf118 (CTCL tumor antigen HD-CL-01/L14-2). | |||||
|
CJ118_MOUSE
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.053782 (rank : 59) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1132 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8C9S4, Q6P3D6, Q8CJC0 | Gene names | Otg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf118 homolog (Oocyte-testis gene 1 protein). | |||||
|
CK5P2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.050331 (rank : 99) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1151 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8K389, Q6PCN1, Q6ZPL0 | Gene names | Cdk5rap2, Kiaa1633 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48). | |||||
|
CN145_HUMAN
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.053263 (rank : 63) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1196 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q6ZU80, Q96ML4 | Gene names | C14orf145 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf145. | |||||
|
CP135_HUMAN
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.053894 (rank : 57) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1290 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q66GS9, O75130, Q9H8H7 | Gene names | CEP135, CEP4, KIAA0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
CP135_MOUSE
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.054582 (rank : 54) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1278 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q6P5D4, Q6A030 | Gene names | Cep135, Cep4, Kiaa0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
CP250_MOUSE
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.057680 (rank : 32) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
CROCC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.050732 (rank : 95) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1409 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q5TZA2, Q2VHY3, Q66GT7, Q7Z2L4, Q7Z5D7 | Gene names | CROCC, KIAA0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
CYLN2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.051376 (rank : 84) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1004 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9UDT6, O14527, O43611 | Gene names | CYLN2, KIAA0291, WBSCR4, WSCR4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic linker protein 2 (Cytoplasmic linker protein 115) (CLIP- 115) (Williams-Beuren syndrome chromosome region 4 protein). | |||||
|
CYLN2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.054120 (rank : 56) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1002 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9Z0H8, Q7TSI9, Q8CHU1, Q9EP81 | Gene names | Cyln2, Kiaa0291 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic linker protein 2 (Cytoplasmic linker protein 115) (CLIP- 115). | |||||
|
FLIP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.050826 (rank : 94) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q7Z7B0, Q5VUL6, Q86TC3, Q8N8B9, Q96SK6, Q9NVI8, Q9ULE5 | Gene names | FILIP1, KIAA1275 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
FYCO1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.053040 (rank : 67) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1149 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9BQS8, Q3MJE6, Q86T41, Q86TB1, Q8TEF9, Q96IV5, Q9H8P9 | Gene names | FYCO1, ZFYVE7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1 (Zinc finger FYVE domain-containing protein 7). | |||||
|
GCC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.056617 (rank : 39) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
GOGA1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.050003 (rank : 102) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 994 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9CW79, Q80YB0 | Gene names | Golga1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
GOGA3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.050573 (rank : 96) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q08378, O43241, Q6P9C7, Q86XW3, Q8TDA9, Q8WZA3 | Gene names | GOLGA3 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Golgi complex-associated protein of 170 kDa) (GCP170). | |||||
|
GOGA3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.051904 (rank : 80) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1437 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P55937, Q80VF5, Q8CCK4, Q9QYT2, Q9QYT3 | Gene names | Golga3, Mea2 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Male-enhanced antigen 2) (MEA-2). | |||||
|
HMMR_HUMAN
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.051735 (rank : 82) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O75330, Q92767 | Gene names | HMMR, IHABP, RHAMM | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility) (CD168 antigen). | |||||
|
HMMR_MOUSE
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.052000 (rank : 78) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q00547 | Gene names | Hmmr, Ihabp, Rhamm | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility). | |||||
|
IFT74_HUMAN
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.055568 (rank : 44) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q96LB3, Q6PGQ8, Q9H643, Q9H8G7 | Gene names | IFT74, CCDC2, CMG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 74 homolog (Coiled-coil domain-containing protein 2) (Capillary morphogenesis protein 1) (CMG-1). | |||||
|
IFT74_MOUSE
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.051035 (rank : 90) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 939 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8BKE9, Q80ZI3, Q9CSY1, Q9CUS0, Q9D9T5 | Gene names | Ift74, Ccdc2, Cmg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 74 homolog (Coiled-coil domain-containing protein 2) (Capillary morphogenesis protein 1) (CMG-1). | |||||
|
IFT81_MOUSE
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.052148 (rank : 76) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O35594, Q8R4W2, Q9CSA1, Q9EQM1 | Gene names | Ift81, Cdv-1, Cdv1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 81 (Carnitine deficiency-associated protein expressed in ventricle 1) (CDV-1 protein). | |||||
|
K0555_HUMAN
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.051447 (rank : 83) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 880 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q96AA8, O60302 | Gene names | KIAA0555 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0555. | |||||
|
KMHN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.052262 (rank : 75) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 950 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8TBZ0, Q86YI9, Q8N7W0 | Gene names | KMHN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cancer/testis antigen KM-HN-1. | |||||
|
KTN1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.054168 (rank : 55) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1324 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q61595, Q8BG49, Q8BHF4, Q8BHM8, Q8C9Y5, Q8CG51, Q8CG52, Q8CG53, Q8CG54, Q8CG55, Q8CG56, Q8CG57, Q8CG58, Q8CG59, Q8CG60, Q8CG61, Q8CG62, Q8CG63 | Gene names | Ktn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin. | |||||
|
LRRF2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.054587 (rank : 53) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q91WK0, Q8BVD1, Q9CU89 | Gene names | Lrrfip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 2 (LRR FLII- interacting protein 2). | |||||
|
MD1L1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.057809 (rank : 31) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1001 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9WTX8, Q9WTX9 | Gene names | Mad1l1, Mad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1). | |||||
|
MPP9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.051365 (rank : 85) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q99550, Q9H976 | Gene names | MPHOSPH9, MPP9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | M-phase phosphoprotein 9. | |||||
|
NIN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.053106 (rank : 66) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1416 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8N4C6, Q6P0P6, Q9BWU6, Q9C012, Q9C013, Q9C014, Q9H5I6, Q9HAT7, Q9HBY5, Q9HCK7, Q9UH61 | Gene names | NIN, KIAA1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein (hNinein) (Glycogen synthase kinase 3 beta-interacting protein) (GSK3B-interacting protein). | |||||
|
NIN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.052962 (rank : 69) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q61043, Q6ZPM7 | Gene names | Nin, Kiaa1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein. | |||||
|
NUF2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.050940 (rank : 93) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 661 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9BZD4, Q8WU69, Q96HJ4, Q96Q78 | Gene names | CDCA1, NUF2, NUF2R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Nuf2 (hNuf2) (hNuf2R) (hsNuf2) (Cell division cycle-associated protein 1). | |||||
|
NUMA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.052139 (rank : 77) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
OPTN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.053798 (rank : 58) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1120 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8K3K8 | Gene names | Optn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin. | |||||
|
PCNT_HUMAN
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.055211 (rank : 45) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
PCNT_MOUSE
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.051068 (rank : 89) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1207 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P48725 | Gene names | Pcnt, Pcnt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pericentrin. | |||||
|
PIBF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.054787 (rank : 51) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8WXW3, O95664, Q6U9V2, Q6UG50, Q86V07, Q96SF4 | Gene names | PIBF1, C13orf24, PIBF | |||
|
Domain Architecture |

|
|||||
| Description | Progesterone-induced-blocking factor 1. | |||||
|
RABE1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.051949 (rank : 79) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q15276, O95369, Q8IVX3 | Gene names | RABEP1, RABPT5, RABPT5A | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha) (Rabaptin-4) (NY-REN-17 antigen). | |||||
|
RABE1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.051783 (rank : 81) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1105 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O35551, Q99L08, Q9CRP3, Q9EQF9, Q9JI94 | Gene names | Rabep1, Rab5ep, Rabpt5, Rabpt5a | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha). | |||||
|
RAD50_HUMAN
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.051294 (rank : 87) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q92878, O43254, Q6GMT7, Q6P5X3, Q9UP86 | Gene names | RAD50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (hRAD50). | |||||
|
RAD50_MOUSE
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.050963 (rank : 92) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1366 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P70388, Q6PEB0, Q8C2T7, Q9CU59 | Gene names | Rad50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (mRad50). | |||||
|
RBP24_HUMAN
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.056724 (rank : 38) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
SAS6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.050484 (rank : 97) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q80UK7, Q9CYT4 | Gene names | Sass6, Sas6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spindle assembly abnormal protein 6 homolog. | |||||
|
SDCG8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.052693 (rank : 72) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1002 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q86SQ7, O60527, Q3ZCR6, Q8N5F2, Q9P0F1 | Gene names | SDCCAG8, CCCAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serologically defined colon cancer antigen 8 (Centrosomal colon cancer autoantigen protein) (hCCCAP) (Antigen NY-CO-8). | |||||
|
SM1L2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.050040 (rank : 101) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 918 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q920F6 | Gene names | Smc1l2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 1-like 2 protein (SMC1beta protein). | |||||
|
SPAG5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.062188 (rank : 25) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 625 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q96R06, O95213, Q9BWE8, Q9NT17, Q9UFE6 | Gene names | SPAG5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 5 (Astrin) (Mitotic spindle-associated protein p126) (MAP126) (Deepest). | |||||
|
SYCP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.053300 (rank : 62) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q15431, O14963 | Gene names | SYCP1, SCP1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
TAXB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.057449 (rank : 34) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 829 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q86VP1, O60398, O95770, Q13311, Q9BQG5, Q9UI88 | Gene names | TAX1BP1, T6BP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tax1-binding protein 1 (TRAF6-binding protein). | |||||
|
TPR_HUMAN
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.053198 (rank : 65) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
TRIPB_HUMAN
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.055825 (rank : 42) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
ZN291_HUMAN
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.052983 (rank : 68) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 647 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9BY12 | Gene names | ZNF291 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 291. | |||||
|
TANK_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q92844, Q92885 | Gene names | TANK, ITRAF | |||
|
Domain Architecture |

|
|||||
| Description | TRAF family member-associated NF-kappa-B activator (TRAF-interacting protein) (I-TRAF). | |||||
|
TANK_MOUSE
|
||||||
| NC score | 0.958225 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P70347, Q61178 | Gene names | Tank, Itraf | |||
|
Domain Architecture |

|
|||||
| Description | TRAF family member-associated NF-kappa-B activator (TRAF-interacting protein) (I-TRAF). | |||||
|
ZC313_HUMAN
|
||||||
| NC score | 0.086271 (rank : 3) | θ value | 0.47712 (rank : 14) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q5T200, O94936, Q5T1Z9, Q7Z7J3, Q8NDT6, Q9H0L6 | Gene names | ZC3H13, KIAA0853 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 13. | |||||
|
AKAP9_HUMAN
|
||||||
| NC score | 0.084723 (rank : 4) | θ value | 0.0563607 (rank : 3) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
SPAG5_MOUSE
|
||||||
| NC score | 0.083336 (rank : 5) | θ value | 0.0736092 (rank : 4) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 612 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q7TME2, Q8CH61 | Gene names | Spag5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 5 (Mastrin) (Mitotic spindle-associated protein p126) (MAP126). | |||||
|
TEX9_HUMAN
|
||||||
| NC score | 0.081517 (rank : 6) | θ value | 0.365318 (rank : 12) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 541 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8N6V9 | Gene names | TEX9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed sequence 9 protein. | |||||
|
ERC2_HUMAN
|
||||||
| NC score | 0.076936 (rank : 7) | θ value | 1.06291 (rank : 17) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1156 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O15083, Q86TK4 | Gene names | ERC2, KIAA0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2. | |||||
|
NDEL1_HUMAN
|
||||||
| NC score | 0.076376 (rank : 8) | θ value | 2.36792 (rank : 33) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9GZM8, Q86T80, Q8TAR7, Q9UH50 | Gene names | NDEL1, EOPA, MITAP1, NUDEL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear distribution protein nudE-like 1 (Protein Nudel) (Mitosin- associated protein 1). | |||||
|
NDEL1_MOUSE
|
||||||
| NC score | 0.076272 (rank : 9) | θ value | 2.36792 (rank : 34) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9ERR1, Q9D0Q4, Q9EPT6 | Gene names | Ndel1, Nudel | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear distribution protein nudE-like 1 (Protein Nudel) (Protein mNudE-like) (mNudE-L). | |||||
|
TEX9_MOUSE
|
||||||
| NC score | 0.076054 (rank : 10) | θ value | 1.38821 (rank : 24) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9D845, O54764 | Gene names | Tex9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed sequence 9 protein (Testis-specifically expressed protein 1) (Tsec-1). | |||||
|
RB6I2_MOUSE
|
||||||
| NC score | 0.075196 (rank : 11) | θ value | 2.36792 (rank : 36) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q99MI1, Q80TK7, Q8BPL1, Q8C7Y1, Q99MI2 | Gene names | Erc1, Cast2, Kiaa1081, Rab6ip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1) (ERC1) (CAZ-associated structural protein 2) (CAST2). | |||||
|
ERC2_MOUSE
|
||||||
| NC score | 0.074272 (rank : 12) | θ value | 2.36792 (rank : 30) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1166 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q6PH08, Q80U20, Q8CCP1 | Gene names | Erc2, Cast1, D14Ertd171e, Kiaa0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2 (CAZ-associated structural protein 1) (CAST1). | |||||
|
NDE1_MOUSE
|
||||||
| NC score | 0.074096 (rank : 13) | θ value | 3.0926 (rank : 42) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 602 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9CZA6, Q3UBS6, Q3UIC1, Q9ERR0 | Gene names | Nde1, Nude | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear distribution protein nudE homolog 1 (NudE) (mNudE). | |||||
|
RB6I2_HUMAN
|
||||||
| NC score | 0.073625 (rank : 14) | θ value | 2.36792 (rank : 35) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1584 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8IUD2, Q6NVK2, Q8IUD3, Q8IUD4, Q8IUD5, Q8NAS1, Q9NXN5, Q9UIK7, Q9UPS1 | Gene names | ERC1, ELKS, KIAA1081, RAB6IP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1). | |||||
|
NDE1_HUMAN
|
||||||
| NC score | 0.072986 (rank : 15) | θ value | 1.06291 (rank : 18) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 823 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9NXR1, Q49AQ2 | Gene names | NDE1, NUDE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear distribution protein nudE homolog 1 (NudE). | |||||
|
GOGA4_MOUSE
|
||||||
| NC score | 0.072555 (rank : 16) | θ value | 0.365318 (rank : 10) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
GCC2_MOUSE
|
||||||
| NC score | 0.070480 (rank : 17) | θ value | 0.279714 (rank : 7) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
NEMO_MOUSE
|
||||||
| NC score | 0.070237 (rank : 18) | θ value | 1.38821 (rank : 21) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O88522 | Gene names | Ikbkg, Nemo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NF-kappa-B essential modulator (NEMO) (NF-kappa-B essential modifier) (Inhibitor of nuclear factor kappa-B kinase subunit gamma) (IkB kinase subunit gamma) (I-kappa-B kinase gamma) (IKK-gamma) (IKKG) (IkB kinase-associated protein 1) (IKKAP1) (mFIP-3). | |||||
|
MD1L1_HUMAN
|
||||||
| NC score | 0.068222 (rank : 19) | θ value | 0.47712 (rank : 13) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9Y6D9, Q13312, Q75MI0, Q86UM4, Q9UNH0 | Gene names | MAD1L1, MAD1, TXBP181 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1) (Mitotic checkpoint MAD1 protein-homolog) (HsMAD1) (hMAD1) (Tax-binding protein 181). | |||||
|
TAXB1_MOUSE
|
||||||
| NC score | 0.066247 (rank : 20) | θ value | 6.88961 (rank : 58) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q3UKC1, Q91YT6, Q9CVF0, Q9DC45 | Gene names | Tax1bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tax1-binding protein 1 homolog. | |||||
|
TRHY_HUMAN
|
||||||
| NC score | 0.064391 (rank : 21) | θ value | 2.36792 (rank : 38) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
GOGA4_HUMAN
|
||||||
| NC score | 0.064260 (rank : 22) | θ value | 8.99809 (rank : 62) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
CF060_HUMAN
|
||||||
| NC score | 0.063118 (rank : 23) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1270 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8NB25, Q96GY8, Q9H851 | Gene names | C6orf60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf60. | |||||
|
EEA1_HUMAN
|
||||||
| NC score | 0.062853 (rank : 24) | θ value | 2.36792 (rank : 29) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
SPAG5_HUMAN
|
||||||
| NC score | 0.062188 (rank : 25) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 625 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q96R06, O95213, Q9BWE8, Q9NT17, Q9UFE6 | Gene names | SPAG5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 5 (Astrin) (Mitotic spindle-associated protein p126) (MAP126) (Deepest). | |||||
|
EEA1_MOUSE
|
||||||
| NC score | 0.061902 (rank : 26) | θ value | 6.88961 (rank : 55) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1637 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8BL66, Q6DIC2 | Gene names | Eea1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early endosome antigen 1. | |||||
|
CE290_HUMAN
|
||||||
| NC score | 0.061508 (rank : 27) | θ value | 6.88961 (rank : 52) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.061090 (rank : 28) | θ value | 1.81305 (rank : 25) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
REST_HUMAN
|
||||||
| NC score | 0.060044 (rank : 29) | θ value | 1.38821 (rank : 22) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1605 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P30622 | Gene names | RSN, CYLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Restin (Cytoplasmic linker protein 170 alpha-2) (CLIP-170) (Reed- Sternberg intermediate filament-associated protein) (Cytoplasmic linker protein 1). | |||||
|
REST_MOUSE
|
||||||
| NC score | 0.058865 (rank : 30) | θ value | 4.03905 (rank : 47) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1506 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q922J3, Q571L7 | Gene names | Rsn, Kiaa4046 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin. | |||||
|
MD1L1_MOUSE
|
||||||
| NC score | 0.057809 (rank : 31) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1001 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9WTX8, Q9WTX9 | Gene names | Mad1l1, Mad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1). | |||||
|
CP250_MOUSE
|
||||||
| NC score | 0.057680 (rank : 32) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
SYNE1_HUMAN
|
||||||
| NC score | 0.057570 (rank : 33) | θ value | 3.0926 (rank : 44) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
TAXB1_HUMAN
|
||||||
| NC score | 0.057449 (rank : 34) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 829 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q86VP1, O60398, O95770, Q13311, Q9BQG5, Q9UI88 | Gene names | TAX1BP1, T6BP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tax1-binding protein 1 (TRAF6-binding protein). | |||||
|
DMD_HUMAN
|
||||||
| NC score | 0.057196 (rank : 35) | θ value | 0.365318 (rank : 9) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P11532, Q02295, Q14169, Q14170, Q5JYU0, Q7KZ48 | Gene names | DMD | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
CENPF_HUMAN
|
||||||
| NC score | 0.056971 (rank : 36) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
CE152_HUMAN
|
||||||
| NC score | 0.056873 (rank : 37) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 976 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O94986, Q6NTA0 | Gene names | CEP152, KIAA0912 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 152 kDa (Cep152 protein). | |||||
|
RBP24_HUMAN
|
||||||
| NC score | 0.056724 (rank : 38) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
GCC2_HUMAN
|
||||||
| NC score | 0.056617 (rank : 39) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
CCD46_HUMAN
|
||||||
| NC score | 0.056603 (rank : 40) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1430 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8N8E3, Q6PIB5, Q8NCR4, Q8NFR4 | Gene names | CCDC46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 46. | |||||
|
CP250_HUMAN
|
||||||
| NC score | 0.056076 (rank : 41) | θ value | 6.88961 (rank : 54) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
TRIPB_HUMAN
|
||||||
| NC score | 0.055825 (rank : 42) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
SDCG8_MOUSE
|
||||||
| NC score | 0.055777 (rank : 43) | θ value | 5.27518 (rank : 51) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q80UF4 | Gene names | Sdccag8, Cccap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serologically defined colon cancer antigen 8 (Centrosomal colon cancer autoantigen protein) (mCCCAP). | |||||
|
IFT74_HUMAN
|
||||||
| NC score | 0.055568 (rank : 44) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q96LB3, Q6PGQ8, Q9H643, Q9H8G7 | Gene names | IFT74, CCDC2, CMG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 74 homolog (Coiled-coil domain-containing protein 2) (Capillary morphogenesis protein 1) (CMG-1). | |||||
|
PCNT_HUMAN
|
||||||
| NC score | 0.055211 (rank : 45) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
CI093_HUMAN
|
||||||
| NC score | 0.055125 (rank : 46) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q6TFL3, Q5SU58, Q6P1W1, Q6ZR13, Q8N1Z4, Q8N8L3, Q8TBR2 | Gene names | C9orf93 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf93. | |||||
|
SMC4_MOUSE
|
||||||
| NC score | 0.054933 (rank : 47) | θ value | 1.38821 (rank : 23) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 830 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8CG47, Q8BTS7, Q8BTY9, Q99K21 | Gene names | Smc4l1, Capc, Smc4 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 4-like 1 protein (Chromosome- associated polypeptide C) (XCAP-C homolog). | |||||
|
CCD46_MOUSE
|
||||||
| NC score | 0.054919 (rank : 48) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q5PR68, Q80ZN6, Q99JS4, Q9D9T1 | Gene names | Ccdc46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled coil domain-containing protein 46. | |||||
|
K0831_MOUSE
|
||||||
| NC score | 0.054917 (rank : 49) | θ value | 3.0926 (rank : 40) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8CDJ3, Q69ZY1, Q6PFY6, Q8C6N0, Q8R3M3 | Gene names | Kiaa0831, D14Ertd436e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0831. | |||||
|
CCD39_HUMAN
|
||||||
| NC score | 0.054910 (rank : 50) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 868 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9UFE4 | Gene names | CCDC39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 39. | |||||
|
PIBF1_HUMAN
|
||||||
| NC score | 0.054787 (rank : 51) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8WXW3, O95664, Q6U9V2, Q6UG50, Q86V07, Q96SF4 | Gene names | PIBF1, C13orf24, PIBF | |||
|
Domain Architecture |

|
|||||
| Description | Progesterone-induced-blocking factor 1. | |||||
|
CEP63_HUMAN
|
||||||
| NC score | 0.054590 (rank : 52) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1005 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q96MT8, Q96CR0, Q9H8F5, Q9H8N0 | Gene names | CEP63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 63 kDa (Cep63 protein). | |||||
|
LRRF2_MOUSE
|
||||||
| NC score | 0.054587 (rank : 53) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q91WK0, Q8BVD1, Q9CU89 | Gene names | Lrrfip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 2 (LRR FLII- interacting protein 2). | |||||
|
CP135_MOUSE
|
||||||
| NC score | 0.054582 (rank : 54) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1278 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q6P5D4, Q6A030 | Gene names | Cep135, Cep4, Kiaa0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
KTN1_MOUSE
|
||||||
| NC score | 0.054168 (rank : 55) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1324 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q61595, Q8BG49, Q8BHF4, Q8BHM8, Q8C9Y5, Q8CG51, Q8CG52, Q8CG53, Q8CG54, Q8CG55, Q8CG56, Q8CG57, Q8CG58, Q8CG59, Q8CG60, Q8CG61, Q8CG62, Q8CG63 | Gene names | Ktn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin. | |||||
|
CYLN2_MOUSE
|
||||||
| NC score | 0.054120 (rank : 56) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1002 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9Z0H8, Q7TSI9, Q8CHU1, Q9EP81 | Gene names | Cyln2, Kiaa0291 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic linker protein 2 (Cytoplasmic linker protein 115) (CLIP- 115). | |||||
|
CP135_HUMAN
|
||||||
| NC score | 0.053894 (rank : 57) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1290 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q66GS9, O75130, Q9H8H7 | Gene names | CEP135, CEP4, KIAA0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
OPTN_MOUSE
|
||||||
| NC score | 0.053798 (rank : 58) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1120 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8K3K8 | Gene names | Optn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin. | |||||
|
CJ118_MOUSE
|
||||||
| NC score | 0.053782 (rank : 59) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1132 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8C9S4, Q6P3D6, Q8CJC0 | Gene names | Otg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf118 homolog (Oocyte-testis gene 1 protein). | |||||
|
HOOK3_HUMAN
|
||||||
| NC score | 0.053671 (rank : 60) | θ value | 2.36792 (rank : 31) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1070 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q86VS8, Q8NBH0, Q9BY13 | Gene names | HOOK3 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 3 (hHK3). | |||||
|
CI039_HUMAN
|
||||||
| NC score | 0.053435 (rank : 61) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1171 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9NXG0, Q5VYJ0, Q9HAJ5 | Gene names | C9orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf39. | |||||
|
SYCP1_HUMAN
|
||||||
| NC score | 0.053300 (rank : 62) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q15431, O14963 | Gene names | SYCP1, SCP1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
CN145_HUMAN
|
||||||
| NC score | 0.053263 (rank : 63) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1196 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q6ZU80, Q96ML4 | Gene names | C14orf145 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf145. | |||||
|
CCD39_MOUSE
|
||||||
| NC score | 0.053248 (rank : 64) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1026 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9D5Y1, Q8CDG6 | Gene names | Ccdc39, D3Ertd789e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 39. | |||||
|
TPR_HUMAN
|
||||||
| NC score | 0.053198 (rank : 65) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
NIN_HUMAN
|
||||||
| NC score | 0.053106 (rank : 66) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1416 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8N4C6, Q6P0P6, Q9BWU6, Q9C012, Q9C013, Q9C014, Q9H5I6, Q9HAT7, Q9HBY5, Q9HCK7, Q9UH61 | Gene names | NIN, KIAA1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein (hNinein) (Glycogen synthase kinase 3 beta-interacting protein) (GSK3B-interacting protein). | |||||
|
FYCO1_HUMAN
|
||||||
| NC score | 0.053040 (rank : 67) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1149 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9BQS8, Q3MJE6, Q86T41, Q86TB1, Q8TEF9, Q96IV5, Q9H8P9 | Gene names | FYCO1, ZFYVE7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1 (Zinc finger FYVE domain-containing protein 7). | |||||
|
ZN291_HUMAN
|
||||||
| NC score | 0.052983 (rank : 68) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 647 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9BY12 | Gene names | ZNF291 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 291. | |||||
|
NIN_MOUSE
|
||||||
| NC score | 0.052962 (rank : 69) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q61043, Q6ZPM7 | Gene names | Nin, Kiaa1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein. | |||||
|
CJ118_HUMAN
|
||||||
| NC score | 0.052933 (rank : 70) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q7Z3E2, Q2M2V6, Q6NS91, Q7RTP1, Q8N117, Q8N3G3, Q8N6C2, Q9NWA3 | Gene names | C10orf118 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf118 (CTCL tumor antigen HD-CL-01/L14-2). | |||||
|
CCD41_HUMAN
|
||||||
| NC score | 0.052704 (rank : 71) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1139 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9Y592 | Gene names | CCDC41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41 (NY-REN-58 antigen). | |||||
|
SDCG8_HUMAN
|
||||||
| NC score | 0.052693 (rank : 72) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1002 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q86SQ7, O60527, Q3ZCR6, Q8N5F2, Q9P0F1 | Gene names | SDCCAG8, CCCAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serologically defined colon cancer antigen 8 (Centrosomal colon cancer autoantigen protein) (hCCCAP) (Antigen NY-CO-8). | |||||
|
CAR10_MOUSE
|
||||||
| NC score | 0.052548 (rank : 73) | θ value | 2.36792 (rank : 28) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 407 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P58660 | Gene names | Card10, Bimp1 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 10 (Bcl10-interacting MAGUK protein 1) (Bimp1). | |||||
|
UACA_MOUSE
|
||||||
| NC score | 0.052373 (rank : 74) | θ value | 2.36792 (rank : 39) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8CGB3, Q69ZG3, Q7TN77, Q8BJC8 | Gene names | Uaca, Kiaa1561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uveal autoantigen with coiled-coil domains and ankyrin repeats protein (Nucling) (Nuclear membrane-binding protein). | |||||
|
KMHN1_HUMAN
|
||||||
| NC score | 0.052262 (rank : 75) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 950 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8TBZ0, Q86YI9, Q8N7W0 | Gene names | KMHN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cancer/testis antigen KM-HN-1. | |||||
|
IFT81_MOUSE
|
||||||
| NC score | 0.052148 (rank : 76) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O35594, Q8R4W2, Q9CSA1, Q9EQM1 | Gene names | Ift81, Cdv-1, Cdv1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 81 (Carnitine deficiency-associated protein expressed in ventricle 1) (CDV-1 protein). | |||||
|
NUMA1_HUMAN
|
||||||
| NC score | 0.052139 (rank : 77) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
HMMR_MOUSE
|
||||||
| NC score | 0.052000 (rank : 78) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q00547 | Gene names | Hmmr, Ihabp, Rhamm | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility). | |||||
|
RABE1_HUMAN
|
||||||
| NC score | 0.051949 (rank : 79) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q15276, O95369, Q8IVX3 | Gene names | RABEP1, RABPT5, RABPT5A | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha) (Rabaptin-4) (NY-REN-17 antigen). | |||||
|
GOGA3_MOUSE
|
||||||
| NC score | 0.051904 (rank : 80) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1437 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P55937, Q80VF5, Q8CCK4, Q9QYT2, Q9QYT3 | Gene names | Golga3, Mea2 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Male-enhanced antigen 2) (MEA-2). | |||||
|
RABE1_MOUSE
|
||||||
| NC score | 0.051783 (rank : 81) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1105 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O35551, Q99L08, Q9CRP3, Q9EQF9, Q9JI94 | Gene names | Rabep1, Rab5ep, Rabpt5, Rabpt5a | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha). | |||||
|
HMMR_HUMAN
|
||||||
| NC score | 0.051735 (rank : 82) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O75330, Q92767 | Gene names | HMMR, IHABP, RHAMM | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility) (CD168 antigen). | |||||
|
K0555_HUMAN
|
||||||
| NC score | 0.051447 (rank : 83) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 880 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q96AA8, O60302 | Gene names | KIAA0555 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0555. | |||||
|
CYLN2_HUMAN
|
||||||
| NC score | 0.051376 (rank : 84) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1004 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9UDT6, O14527, O43611 | Gene names | CYLN2, KIAA0291, WBSCR4, WSCR4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic linker protein 2 (Cytoplasmic linker protein 115) (CLIP- 115) (Williams-Beuren syndrome chromosome region 4 protein). | |||||
|
MPP9_HUMAN
|
||||||
| NC score | 0.051365 (rank : 85) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q99550, Q9H976 | Gene names | MPHOSPH9, MPP9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | M-phase phosphoprotein 9. | |||||
|
CCD41_MOUSE
|
||||||
| NC score | 0.051342 (rank : 86) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9D5R3, Q3U7X7, Q3UX57, Q80VF0 | Gene names | Ccdc41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41. | |||||
|
RAD50_HUMAN
|
||||||
| NC score | 0.051294 (rank : 87) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q92878, O43254, Q6GMT7, Q6P5X3, Q9UP86 | Gene names | RAD50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (hRAD50). | |||||
|
CAR10_HUMAN
|
||||||
| NC score | 0.051222 (rank : 88) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 569 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9BWT7, Q14CQ8, Q5TFG6, Q9UGR5, Q9UGR6, Q9Y3H0 | Gene names | CARD10, CARMA3 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 10 (CARD-containing MAGUK protein 3) (Carma 3). | |||||
|
PCNT_MOUSE
|
||||||
| NC score | 0.051068 (rank : 89) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1207 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P48725 | Gene names | Pcnt, Pcnt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pericentrin. | |||||
|
IFT74_MOUSE
|
||||||
| NC score | 0.051035 (rank : 90) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 939 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8BKE9, Q80ZI3, Q9CSY1, Q9CUS0, Q9D9T5 | Gene names | Ift74, Ccdc2, Cmg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 74 homolog (Coiled-coil domain-containing protein 2) (Capillary morphogenesis protein 1) (CMG-1). | |||||
|
CENPE_HUMAN
|
||||||
| NC score | 0.050984 (rank : 91) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
RAD50_MOUSE
|
||||||
| NC score | 0.050963 (rank : 92) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1366 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P70388, Q6PEB0, Q8C2T7, Q9CU59 | Gene names | Rad50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (mRad50). | |||||
|
NUF2_HUMAN
|
||||||
| NC score | 0.050940 (rank : 93) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 661 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9BZD4, Q8WU69, Q96HJ4, Q96Q78 | Gene names | CDCA1, NUF2, NUF2R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Nuf2 (hNuf2) (hNuf2R) (hsNuf2) (Cell division cycle-associated protein 1). | |||||
|
FLIP1_HUMAN
|
||||||
| NC score | 0.050826 (rank : 94) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q7Z7B0, Q5VUL6, Q86TC3, Q8N8B9, Q96SK6, Q9NVI8, Q9ULE5 | Gene names | FILIP1, KIAA1275 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
CROCC_HUMAN
|
||||||
| NC score | 0.050732 (rank : 95) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1409 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q5TZA2, Q2VHY3, Q66GT7, Q7Z2L4, Q7Z5D7 | Gene names | CROCC, KIAA0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
GOGA3_HUMAN
|
||||||
| NC score | 0.050573 (rank : 96) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q08378, O43241, Q6P9C7, Q86XW3, Q8TDA9, Q8WZA3 | Gene names | GOLGA3 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Golgi complex-associated protein of 170 kDa) (GCP170). | |||||
|
SAS6_MOUSE
|
||||||
| NC score | 0.050484 (rank : 97) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q80UK7, Q9CYT4 | Gene names | Sass6, Sas6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spindle assembly abnormal protein 6 homolog. | |||||
|
CARD9_HUMAN
|
||||||
| NC score | 0.050367 (rank : 98) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 602 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9H257, Q5SXM6, Q9H854 | Gene names | CARD9 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 9 (hCARD9). | |||||
|
CK5P2_MOUSE
|
||||||
| NC score | 0.050331 (rank : 99) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1151 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8K389, Q6PCN1, Q6ZPL0 | Gene names | Cdk5rap2, Kiaa1633 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48). | |||||
|
ANR26_MOUSE
|
||||||
| NC score | 0.050071 (rank : 100) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1554 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q811D2, Q69ZS2, Q9CS61 | Gene names | Ankrd26, Kiaa1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
SM1L2_MOUSE
|
||||||
| NC score | 0.050040 (rank : 101) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 918 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q920F6 | Gene names | Smc1l2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 1-like 2 protein (SMC1beta protein). | |||||
|
GOGA1_MOUSE
|
||||||
| NC score | 0.050003 (rank : 102) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 994 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9CW79, Q80YB0 | Gene names | Golga1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
MYH10_MOUSE
|
||||||
| NC score | 0.046559 (rank : 103) | θ value | 5.27518 (rank : 49) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1612 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q61879, Q5SV63 | Gene names | Myh10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
MYH10_HUMAN
|
||||||
| NC score | 0.046075 (rank : 104) | θ value | 6.88961 (rank : 56) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P35580, Q16087 | Gene names | MYH10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
MYH14_HUMAN
|
||||||
| NC score | 0.043670 (rank : 105) | θ value | 1.38821 (rank : 20) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 995 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q7Z406, Q5CZ75, Q6XYE4, Q76B62, Q8WV23, Q96I22, Q9BT27, Q9BW35, Q9H882 | Gene names | MYH14, KIAA2034 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
ANR12_HUMAN
|
||||||
| NC score | 0.041358 (rank : 106) | θ value | 2.36792 (rank : 27) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1081 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q6UB98, O94951, Q658K1, Q6QMF7, Q9H231, Q9H784 | Gene names | ANKRD12, ANCO2, KIAA0874 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 12 (Ankyrin repeat-containing cofactor 2) (GAC-1 protein). | |||||
|
KINH_MOUSE
|
||||||
| NC score | 0.040027 (rank : 107) | θ value | 5.27518 (rank : 48) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q61768, O08711, Q61580 | Gene names | Kif5b, Khcs, Kns1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain (Ubiquitous kinesin heavy chain) (UKHC). | |||||
|
GUC2D_HUMAN
|
||||||
| NC score | 0.039170 (rank : 108) | θ value | 0.365318 (rank : 11) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 715 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q02846, Q6LEA7 | Gene names | GUCY2D, CORD6, GUC1A4, GUC2D, RETGC, RETGC1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinal guanylyl cyclase 1 precursor (EC 4.6.1.2) (Guanylate cyclase 2D, retinal) (RETGC-1) (Rod outer segment membrane guanylate cyclase) (ROS-GC). | |||||
|
TXLNG_HUMAN
|
||||||
| NC score | 0.038883 (rank : 109) | θ value | 8.99809 (rank : 65) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 568 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9NUQ3, Q9P0X1 | Gene names | TXLNG, CXorf15, LSR5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gamma-taxilin (Lipopolysaccharide-specific response protein 5). | |||||
|
EP15_HUMAN
|
||||||
| NC score | 0.038517 (rank : 110) | θ value | 8.99809 (rank : 60) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1021 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P42566 | Gene names | EPS15, AF1P | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15 (Protein Eps15) (AF-1p protein). | |||||
|
MOES_MOUSE
|
||||||
| NC score | 0.037798 (rank : 111) | θ value | 2.36792 (rank : 32) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 923 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P26041, Q3UL28, Q8BSN4 | Gene names | Msn | |||
|
Domain Architecture |

|
|||||
| Description | Moesin (Membrane-organizing extension spike protein). | |||||
|
RSF1_HUMAN
|
||||||
| NC score | 0.036294 (rank : 112) | θ value | 3.0926 (rank : 43) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q96T23, Q86X86, Q9H3L8, Q9NVZ8, Q9NYU0 | Gene names | RSF1, HBXAP, XAP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Remodeling and spacing factor 1 (Rsf-1) (Hepatitis B virus X- associated protein) (HBV pX-associated protein 8) (p325 subunit of RSF chromatin remodelling complex). | |||||
|
GUC2E_MOUSE
|
||||||
| NC score | 0.034083 (rank : 113) | θ value | 1.81305 (rank : 26) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 692 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P52785 | Gene names | Gucy2e, Guc2e | |||
|
Domain Architecture |

|
|||||
| Description | Guanylyl cyclase GC-E precursor (EC 4.6.1.2) (Guanylate cyclase 2E). | |||||
|
ATN1_HUMAN
|
||||||
| NC score | 0.033955 (rank : 114) | θ value | 0.62314 (rank : 15) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
TRAF5_HUMAN
|
||||||
| NC score | 0.032740 (rank : 115) | θ value | 3.0926 (rank : 45) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O00463 | Gene names | TRAF5, RNF84 | |||
|
Domain Architecture |

|
|||||
| Description | TNF receptor-associated factor 5 (RING finger protein 84). | |||||
|
TBCD2_HUMAN
|
||||||
| NC score | 0.031953 (rank : 116) | θ value | 6.88961 (rank : 59) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9BYX2 | Gene names | TBC1D2, PARIS1 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 2 (Prostate antigen recognized and identified by SEREX) (PARIS-1). | |||||
|
GUC2F_HUMAN
|
||||||
| NC score | 0.030488 (rank : 117) | θ value | 0.163984 (rank : 6) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 740 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P51841, Q9UJF1 | Gene names | GUCY2F, GUC2F, RETGC2 | |||
|
Domain Architecture |

|
|||||
| Description | Retinal guanylyl cyclase 2 precursor (EC 4.6.1.2) (Guanylate cyclase 2F, retinal) (RETGC-2) (Rod outer segment membrane guanylate cyclase 2) (ROS-GC2) (Guanylate cyclase F) (GC-F). | |||||
|
KIF11_MOUSE
|
||||||
| NC score | 0.029131 (rank : 118) | θ value | 3.0926 (rank : 41) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 446 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6P9P6, Q9Z1J0 | Gene names | Kif11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin-like protein KIF11 (Kinesin-related motor protein Eg5). | |||||
|
SLK_HUMAN
|
||||||
| NC score | 0.029053 (rank : 119) | θ value | 0.125558 (rank : 5) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1775 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9H2G2, O00211, Q6P1Z4, Q86WU7, Q86WW1, Q92603, Q9NQL0, Q9NQL1 | Gene names | SLK, KIAA0204, STK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (hSLK) (Serine/threonine-protein kinase 2) (CTCL tumor antigen se20-9). | |||||
|
TMCC3_MOUSE
|
||||||
| NC score | 0.028844 (rank : 120) | θ value | 2.36792 (rank : 37) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8R310 | Gene names | Tmcc3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane and coiled-coil domains protein 3. | |||||
|
SLK_MOUSE
|
||||||
| NC score | 0.026948 (rank : 121) | θ value | 1.06291 (rank : 19) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1798 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O54988, Q80U65, Q8CAU2, Q8CDW2, Q9WU41 | Gene names | Slk, Kiaa0204, Stk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (mSLK) (Serine/threonine-protein kinase 2) (STE20-related kinase SMAK) (Etk4). | |||||
|
ANLN_HUMAN
|
||||||
| NC score | 0.026051 (rank : 122) | θ value | 4.03905 (rank : 46) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NQW6, Q5CZ78, Q6NSK5, Q9H8Y4, Q9NVN9, Q9NVP0 | Gene names | ANLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding protein anillin. | |||||
|
MASTL_HUMAN
|
||||||
| NC score | 0.024033 (rank : 123) | θ value | 0.279714 (rank : 8) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 894 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96GX5, Q5T8D5, Q5T8D7, Q8NCD6, Q96SJ5 | Gene names | MASTL, THC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase-like (EC 2.7.11.1). | |||||
|
MPP8_HUMAN
|
||||||
| NC score | 0.022912 (rank : 124) | θ value | 8.99809 (rank : 64) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 948 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q99549, Q86TK3, Q9BTP1 | Gene names | MPHOSPH8, MPP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | M-phase phosphoprotein 8. | |||||
|
PER3_MOUSE
|
||||||
| NC score | 0.017291 (rank : 125) | θ value | 5.27518 (rank : 50) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O70361 | Gene names | Per3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (mPER3). | |||||
|
SNPC4_MOUSE
|
||||||
| NC score | 0.017019 (rank : 126) | θ value | 6.88961 (rank : 57) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BP86, Q6PGG7, Q80UG9, Q810L1 | Gene names | Snapc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 4 (SNAPc subunit 4) (snRNA- activating protein complex 190 kDa subunit) (SNAPc 190 kDa subunit). | |||||
|
GLT14_HUMAN
|
||||||
| NC score | 0.016246 (rank : 127) | θ value | 0.62314 (rank : 16) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96FL9, Q8IVI4, Q9BRH1, Q9H827, Q9H9J8 | Gene names | GALNT14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 14 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 14) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 14) (Polypeptide GalNAc transferase 14) (GalNAc-T14) (pp-GaNTase 14). | |||||
|
EVI2B_HUMAN
|
||||||
| NC score | 0.016101 (rank : 128) | θ value | 8.99809 (rank : 61) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P34910 | Gene names | EVI2B, EVDB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EVI2B protein precursor (Ecotropic viral integration site 2B protein). | |||||
|
CLOCK_MOUSE
|
||||||
| NC score | 0.011725 (rank : 129) | θ value | 6.88961 (rank : 53) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O08785 | Gene names | Clock | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (mCLOCK). | |||||
|
IRAK1_HUMAN
|
||||||
| NC score | 0.008671 (rank : 130) | θ value | 8.99809 (rank : 63) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P51617, Q7Z5V4, Q96RL2 | Gene names | IRAK1, IRAK | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-1 receptor-associated kinase 1 (EC 2.7.11.1) (IRAK-1). | |||||