Please be patient as the page loads
|
TAB1_HUMAN
|
||||||
| SwissProt Accessions | Q15750, Q2PP09 | Gene names | MAP3K7IP1, TAB1 | |||
|
Domain Architecture |
|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 1 (TGF-beta-activated kinase 1-binding protein 1) (TAK1-binding protein 1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
TAB1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q15750, Q2PP09 | Gene names | MAP3K7IP1, TAB1 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 1 (TGF-beta-activated kinase 1-binding protein 1) (TAK1-binding protein 1). | |||||
|
TAB1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.996600 (rank : 2) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8CF89, Q7TQJ5, Q80V65, Q8R0D1 | Gene names | Map3k7ip1, Tab1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 1 (TGF-beta-activated kinase 1-binding protein 1) (TAK1-binding protein 1). | |||||
|
PP2CL_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 3) | NC score | 0.294846 (rank : 3) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5SGD2, Q96NM7 | Gene names | PPM1L, PP2CE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase 2C isoform epsilon (EC 3.1.3.16) (PP2C-epsilon) (Protein phosphatase 1L) (Protein phosphatase 1-like). | |||||
|
PP2CL_MOUSE
|
||||||
| θ value | 0.000121331 (rank : 4) | NC score | 0.293020 (rank : 4) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BHN0, Q3UGL2, Q810H0, Q8C021, Q8C1D5, Q9Z0T1 | Gene names | Ppm1l, Kiaa4175 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase 2C isoform epsilon (EC 3.1.3.16) (PP2C-epsilon) (Protein phosphatase 1L) (Protein phosphatase 1-like). | |||||
|
PDP1_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 5) | NC score | 0.234876 (rank : 12) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q3UV70 | Gene names | Pdp1, Ppm2c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | [Pyruvate dehydrogenase [lipoamide]]-phosphatase 1, mitochondrial precursor (EC 3.1.3.43) (PDP 1) (Pyruvate dehydrogenase phosphatase, catalytic subunit 1) (PDPC 1) (Protein phosphatase 2C). | |||||
|
PP2CA_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 6) | NC score | 0.246600 (rank : 8) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P35813, O75551 | Gene names | PPM1A, PPPM1A | |||
|
Domain Architecture |
|
|||||
| Description | Protein phosphatase 2C isoform alpha (EC 3.1.3.16) (PP2C-alpha) (IA) (Protein phosphatase 1A). | |||||
|
PP2CA_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 7) | NC score | 0.246447 (rank : 9) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P49443 | Gene names | Ppm1a, Pppm1a | |||
|
Domain Architecture |
|
|||||
| Description | Protein phosphatase 2C isoform alpha (EC 3.1.3.16) (PP2C-alpha) (IA) (Protein phosphatase 1A). | |||||
|
PP2CE_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 8) | NC score | 0.277877 (rank : 5) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BU27, Q9CSD6, Q9CU88 | Gene names | Ppm1m, Ppm1e | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 2C isoform eta (EC 3.1.3.16) (PP2C-eta) (PP2CE) (Protein phosphatase 1M). | |||||
|
PDP1_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 9) | NC score | 0.235193 (rank : 11) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9P0J1, Q5U5K1 | Gene names | PDP1, PDP, PPM2C | |||
|
Domain Architecture |

|
|||||
| Description | [Pyruvate dehydrogenase [lipoamide]]-phosphatase 1, mitochondrial precursor (EC 3.1.3.43) (PDP 1) (Pyruvate dehydrogenase phosphatase, catalytic subunit 1) (PDPC 1) (Protein phosphatase 2C). | |||||
|
PP2CG_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 10) | NC score | 0.251554 (rank : 7) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O15355 | Gene names | PPM1G, PPM1C | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 2C isoform gamma (EC 3.1.3.16) (PP2C-gamma) (Protein phosphatase magnesium-dependent 1 gamma) (Protein phosphatase 1C). | |||||
|
PP2CG_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 11) | NC score | 0.240903 (rank : 10) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q61074 | Gene names | Ppm1g, Fin13, Ppm1c | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 2C isoform gamma (EC 3.1.3.16) (PP2C-gamma) (Protein phosphatase magnesium-dependent 1 gamma) (Protein phosphatase 1C) (Fibroblast growth factor-inducible protein 13) (FIN13). | |||||
|
PP2CE_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 12) | NC score | 0.267632 (rank : 6) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96MI6, Q8N8J9, Q96DB8 | Gene names | PPM1M, PPM1E | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 2C isoform eta (EC 3.1.3.16) (PP2C-eta) (PP2CE) (Protein phosphatase 1M). | |||||
|
PP2CB_MOUSE
|
||||||
| θ value | 0.125558 (rank : 13) | NC score | 0.217611 (rank : 13) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P36993 | Gene names | Ppm1b, Pp2c2, Pppm1b | |||
|
Domain Architecture |
|
|||||
| Description | Protein phosphatase 2C isoform beta (EC 3.1.3.16) (PP2C-beta) (IA) (Protein phosphatase 1B). | |||||
|
PGBM_MOUSE
|
||||||
| θ value | 1.38821 (rank : 14) | NC score | 0.012809 (rank : 27) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 1073 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q05793 | Gene names | Hspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG) (Perlecan) (PLC). | |||||
|
CBL_MOUSE
|
||||||
| θ value | 1.81305 (rank : 15) | NC score | 0.036526 (rank : 21) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P22682, Q8CEA1 | Gene names | Cbl | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase CBL (EC 6.3.2.-) (Signal transduction protein CBL) (Proto-oncogene c-CBL) (Casitas B-lineage lymphoma proto- oncogene). | |||||
|
PERT_MOUSE
|
||||||
| θ value | 1.81305 (rank : 16) | NC score | 0.025149 (rank : 23) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P35419, Q8C8B1 | Gene names | Tpo | |||
|
Domain Architecture |
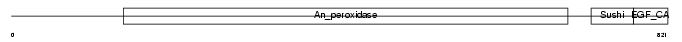
|
|||||
| Description | Thyroid peroxidase precursor (EC 1.11.1.8) (TPO). | |||||
|
HV2D_HUMAN
|
||||||
| θ value | 4.03905 (rank : 17) | NC score | 0.013570 (rank : 26) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P01817 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig heavy chain V-II region MCE. | |||||
|
OAZ1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 18) | NC score | 0.035516 (rank : 22) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P54369, O08610 | Gene names | Oaz1, Oaz | |||
|
Domain Architecture |
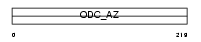
|
|||||
| Description | Ornithine decarboxylase antizyme (ODC-Az). | |||||
|
BAZ2A_MOUSE
|
||||||
| θ value | 5.27518 (rank : 19) | NC score | 0.010715 (rank : 28) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91YE5 | Gene names | Baz2a, Tip5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5). | |||||
|
PORIM_MOUSE
|
||||||
| θ value | 5.27518 (rank : 20) | NC score | 0.057883 (rank : 20) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91Z22, Q3T9F6, Q3TLL2, Q3U1R6, Q8CEX4 | Gene names | Tmem123 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Porimin precursor (Transmembrane protein 123). | |||||
|
TCF8_HUMAN
|
||||||
| θ value | 5.27518 (rank : 21) | NC score | 0.003523 (rank : 29) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 1057 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P37275, Q12924, Q13800 | Gene names | TCF8, AREB6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 8 (NIL-2-A zinc finger protein) (Negative regulator of IL2). | |||||
|
GA2L2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 22) | NC score | 0.019893 (rank : 24) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NHY3, Q8NHY4 | Gene names | GAS2L2, GAR17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 2 (Growth arrest-specific 2-like 2) (GAS2-related protein on chromosome 17) (GAR17 protein). | |||||
|
K1C17_MOUSE
|
||||||
| θ value | 8.99809 (rank : 23) | NC score | 0.002188 (rank : 30) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QWL7, Q61783 | Gene names | Krt17, Krt1-17 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 17 (Cytokeratin-17) (CK-17) (Keratin-17) (K17). | |||||
|
PPCKM_MOUSE
|
||||||
| θ value | 8.99809 (rank : 24) | NC score | 0.017606 (rank : 25) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BH04, Q8BMM9, Q91Z10 | Gene names | Pck2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphoenolpyruvate carboxykinase [GTP], mitochondrial precursor (EC 4.1.1.32) (Phosphoenolpyruvate carboxylase) (PEPCK-M). | |||||
|
PDP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 25) | NC score | 0.182075 (rank : 14) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9P2J9 | Gene names | PDP2, KIAA1348 | |||
|
Domain Architecture |

|
|||||
| Description | [Pyruvate dehydrogenase [lipoamide]]-phosphatase 2, mitochondrial precursor (EC 3.1.3.43) (PDP 2) (Pyruvate dehydrogenase phosphatase, catalytic subunit 2) (PDPC 2). | |||||
|
PP2CB_HUMAN
|
||||||
| θ value | θ > 10 (rank : 26) | NC score | 0.169872 (rank : 15) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O75688, Q9HAY8 | Gene names | PPM1B, PP2CB | |||
|
Domain Architecture |
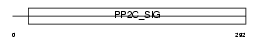
|
|||||
| Description | Protein phosphatase 2C isoform beta (EC 3.1.3.16) (PP2C-beta). | |||||
|
PP2CD_HUMAN
|
||||||
| θ value | θ > 10 (rank : 27) | NC score | 0.109216 (rank : 18) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O15297 | Gene names | PPM1D, WIP1 | |||
|
Domain Architecture |
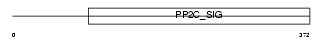
|
|||||
| Description | Protein phosphatase 2C isoform delta (EC 3.1.3.16) (PP2C-delta) (p53- induced protein phosphatase 1) (Protein phosphatase magnesium- dependent 1 delta). | |||||
|
PP2CD_MOUSE
|
||||||
| θ value | θ > 10 (rank : 28) | NC score | 0.107577 (rank : 19) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9QZ67 | Gene names | Ppm1d, Wip1 | |||
|
Domain Architecture |
|
|||||
| Description | Protein phosphatase 2C isoform delta (EC 3.1.3.16) (PP2C-delta) (p53- induced protein phosphatase 1) (Protein phosphatase magnesium- dependent 1 delta). | |||||
|
PPM1F_HUMAN
|
||||||
| θ value | θ > 10 (rank : 29) | NC score | 0.137624 (rank : 16) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P49593, Q96PM2 | Gene names | PPM1F, KIAA0015, POPX2 | |||
|
Domain Architecture |

|
|||||
| Description | Ca(2+)/calmodulin-dependent protein kinase phosphatase (EC 3.1.3.16) (CaM-kinase phosphatase) (CaMKPase) (Partner of PIX 2) (hFEM-2) (Protein phosphatase 1F). | |||||
|
PPM1F_MOUSE
|
||||||
| θ value | θ > 10 (rank : 30) | NC score | 0.127970 (rank : 17) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8CGA0 | Gene names | Ppm1f | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ca(2+)/calmodulin-dependent protein kinase phosphatase (EC 3.1.3.16) (CaM-kinase phosphatase) (CaMKPase) (Protein phosphatase 1F). | |||||
|
TAB1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q15750, Q2PP09 | Gene names | MAP3K7IP1, TAB1 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 1 (TGF-beta-activated kinase 1-binding protein 1) (TAK1-binding protein 1). | |||||
|
TAB1_MOUSE
|
||||||
| NC score | 0.996600 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8CF89, Q7TQJ5, Q80V65, Q8R0D1 | Gene names | Map3k7ip1, Tab1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 1 (TGF-beta-activated kinase 1-binding protein 1) (TAK1-binding protein 1). | |||||
|
PP2CL_HUMAN
|
||||||
| NC score | 0.294846 (rank : 3) | θ value | 9.29e-05 (rank : 3) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5SGD2, Q96NM7 | Gene names | PPM1L, PP2CE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase 2C isoform epsilon (EC 3.1.3.16) (PP2C-epsilon) (Protein phosphatase 1L) (Protein phosphatase 1-like). | |||||
|
PP2CL_MOUSE
|
||||||
| NC score | 0.293020 (rank : 4) | θ value | 0.000121331 (rank : 4) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BHN0, Q3UGL2, Q810H0, Q8C021, Q8C1D5, Q9Z0T1 | Gene names | Ppm1l, Kiaa4175 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase 2C isoform epsilon (EC 3.1.3.16) (PP2C-epsilon) (Protein phosphatase 1L) (Protein phosphatase 1-like). | |||||
|
PP2CE_MOUSE
|
||||||
| NC score | 0.277877 (rank : 5) | θ value | 0.00228821 (rank : 8) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BU27, Q9CSD6, Q9CU88 | Gene names | Ppm1m, Ppm1e | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 2C isoform eta (EC 3.1.3.16) (PP2C-eta) (PP2CE) (Protein phosphatase 1M). | |||||
|
PP2CE_HUMAN
|
||||||
| NC score | 0.267632 (rank : 6) | θ value | 0.0252991 (rank : 12) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96MI6, Q8N8J9, Q96DB8 | Gene names | PPM1M, PPM1E | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 2C isoform eta (EC 3.1.3.16) (PP2C-eta) (PP2CE) (Protein phosphatase 1M). | |||||
|
PP2CG_HUMAN
|
||||||
| NC score | 0.251554 (rank : 7) | θ value | 0.00509761 (rank : 10) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O15355 | Gene names | PPM1G, PPM1C | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 2C isoform gamma (EC 3.1.3.16) (PP2C-gamma) (Protein phosphatase magnesium-dependent 1 gamma) (Protein phosphatase 1C). | |||||
|
PP2CA_HUMAN
|
||||||
| NC score | 0.246600 (rank : 8) | θ value | 0.00228821 (rank : 6) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P35813, O75551 | Gene names | PPM1A, PPPM1A | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 2C isoform alpha (EC 3.1.3.16) (PP2C-alpha) (IA) (Protein phosphatase 1A). | |||||
|
PP2CA_MOUSE
|
||||||
| NC score | 0.246447 (rank : 9) | θ value | 0.00228821 (rank : 7) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P49443 | Gene names | Ppm1a, Pppm1a | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 2C isoform alpha (EC 3.1.3.16) (PP2C-alpha) (IA) (Protein phosphatase 1A). | |||||
|
PP2CG_MOUSE
|
||||||
| NC score | 0.240903 (rank : 10) | θ value | 0.00869519 (rank : 11) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q61074 | Gene names | Ppm1g, Fin13, Ppm1c | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 2C isoform gamma (EC 3.1.3.16) (PP2C-gamma) (Protein phosphatase magnesium-dependent 1 gamma) (Protein phosphatase 1C) (Fibroblast growth factor-inducible protein 13) (FIN13). | |||||
|
PDP1_HUMAN
|
||||||
| NC score | 0.235193 (rank : 11) | θ value | 0.00509761 (rank : 9) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9P0J1, Q5U5K1 | Gene names | PDP1, PDP, PPM2C | |||
|
Domain Architecture |

|
|||||
| Description | [Pyruvate dehydrogenase [lipoamide]]-phosphatase 1, mitochondrial precursor (EC 3.1.3.43) (PDP 1) (Pyruvate dehydrogenase phosphatase, catalytic subunit 1) (PDPC 1) (Protein phosphatase 2C). | |||||
|
PDP1_MOUSE
|
||||||
| NC score | 0.234876 (rank : 12) | θ value | 0.00228821 (rank : 5) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q3UV70 | Gene names | Pdp1, Ppm2c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | [Pyruvate dehydrogenase [lipoamide]]-phosphatase 1, mitochondrial precursor (EC 3.1.3.43) (PDP 1) (Pyruvate dehydrogenase phosphatase, catalytic subunit 1) (PDPC 1) (Protein phosphatase 2C). | |||||
|
PP2CB_MOUSE
|
||||||
| NC score | 0.217611 (rank : 13) | θ value | 0.125558 (rank : 13) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P36993 | Gene names | Ppm1b, Pp2c2, Pppm1b | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 2C isoform beta (EC 3.1.3.16) (PP2C-beta) (IA) (Protein phosphatase 1B). | |||||
|
PDP2_HUMAN
|
||||||
| NC score | 0.182075 (rank : 14) | θ value | θ > 10 (rank : 25) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9P2J9 | Gene names | PDP2, KIAA1348 | |||
|
Domain Architecture |

|
|||||
| Description | [Pyruvate dehydrogenase [lipoamide]]-phosphatase 2, mitochondrial precursor (EC 3.1.3.43) (PDP 2) (Pyruvate dehydrogenase phosphatase, catalytic subunit 2) (PDPC 2). | |||||
|
PP2CB_HUMAN
|
||||||
| NC score | 0.169872 (rank : 15) | θ value | θ > 10 (rank : 26) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O75688, Q9HAY8 | Gene names | PPM1B, PP2CB | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 2C isoform beta (EC 3.1.3.16) (PP2C-beta). | |||||
|
PPM1F_HUMAN
|
||||||
| NC score | 0.137624 (rank : 16) | θ value | θ > 10 (rank : 29) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P49593, Q96PM2 | Gene names | PPM1F, KIAA0015, POPX2 | |||
|
Domain Architecture |

|
|||||
| Description | Ca(2+)/calmodulin-dependent protein kinase phosphatase (EC 3.1.3.16) (CaM-kinase phosphatase) (CaMKPase) (Partner of PIX 2) (hFEM-2) (Protein phosphatase 1F). | |||||
|
PPM1F_MOUSE
|
||||||
| NC score | 0.127970 (rank : 17) | θ value | θ > 10 (rank : 30) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8CGA0 | Gene names | Ppm1f | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ca(2+)/calmodulin-dependent protein kinase phosphatase (EC 3.1.3.16) (CaM-kinase phosphatase) (CaMKPase) (Protein phosphatase 1F). | |||||
|
PP2CD_HUMAN
|
||||||
| NC score | 0.109216 (rank : 18) | θ value | θ > 10 (rank : 27) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O15297 | Gene names | PPM1D, WIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 2C isoform delta (EC 3.1.3.16) (PP2C-delta) (p53- induced protein phosphatase 1) (Protein phosphatase magnesium- dependent 1 delta). | |||||
|
PP2CD_MOUSE
|
||||||
| NC score | 0.107577 (rank : 19) | θ value | θ > 10 (rank : 28) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9QZ67 | Gene names | Ppm1d, Wip1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 2C isoform delta (EC 3.1.3.16) (PP2C-delta) (p53- induced protein phosphatase 1) (Protein phosphatase magnesium- dependent 1 delta). | |||||
|
PORIM_MOUSE
|
||||||
| NC score | 0.057883 (rank : 20) | θ value | 5.27518 (rank : 20) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91Z22, Q3T9F6, Q3TLL2, Q3U1R6, Q8CEX4 | Gene names | Tmem123 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Porimin precursor (Transmembrane protein 123). | |||||
|
CBL_MOUSE
|
||||||
| NC score | 0.036526 (rank : 21) | θ value | 1.81305 (rank : 15) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P22682, Q8CEA1 | Gene names | Cbl | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase CBL (EC 6.3.2.-) (Signal transduction protein CBL) (Proto-oncogene c-CBL) (Casitas B-lineage lymphoma proto- oncogene). | |||||
|
OAZ1_MOUSE
|
||||||
| NC score | 0.035516 (rank : 22) | θ value | 4.03905 (rank : 18) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P54369, O08610 | Gene names | Oaz1, Oaz | |||
|
Domain Architecture |

|
|||||
| Description | Ornithine decarboxylase antizyme (ODC-Az). | |||||
|
PERT_MOUSE
|
||||||
| NC score | 0.025149 (rank : 23) | θ value | 1.81305 (rank : 16) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P35419, Q8C8B1 | Gene names | Tpo | |||
|
Domain Architecture |
|
|||||
| Description | Thyroid peroxidase precursor (EC 1.11.1.8) (TPO). | |||||
|
GA2L2_HUMAN
|
||||||
| NC score | 0.019893 (rank : 24) | θ value | 6.88961 (rank : 22) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NHY3, Q8NHY4 | Gene names | GAS2L2, GAR17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 2 (Growth arrest-specific 2-like 2) (GAS2-related protein on chromosome 17) (GAR17 protein). | |||||
|
PPCKM_MOUSE
|
||||||
| NC score | 0.017606 (rank : 25) | θ value | 8.99809 (rank : 24) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BH04, Q8BMM9, Q91Z10 | Gene names | Pck2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphoenolpyruvate carboxykinase [GTP], mitochondrial precursor (EC 4.1.1.32) (Phosphoenolpyruvate carboxylase) (PEPCK-M). | |||||
|
HV2D_HUMAN
|
||||||
| NC score | 0.013570 (rank : 26) | θ value | 4.03905 (rank : 17) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P01817 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig heavy chain V-II region MCE. | |||||
|
PGBM_MOUSE
|
||||||
| NC score | 0.012809 (rank : 27) | θ value | 1.38821 (rank : 14) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 1073 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q05793 | Gene names | Hspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG) (Perlecan) (PLC). | |||||
|
BAZ2A_MOUSE
|
||||||
| NC score | 0.010715 (rank : 28) | θ value | 5.27518 (rank : 19) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91YE5 | Gene names | Baz2a, Tip5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5). | |||||
|
TCF8_HUMAN
|
||||||
| NC score | 0.003523 (rank : 29) | θ value | 5.27518 (rank : 21) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 1057 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P37275, Q12924, Q13800 | Gene names | TCF8, AREB6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 8 (NIL-2-A zinc finger protein) (Negative regulator of IL2). | |||||
|
K1C17_MOUSE
|
||||||
| NC score | 0.002188 (rank : 30) | θ value | 8.99809 (rank : 23) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QWL7, Q61783 | Gene names | Krt17, Krt1-17 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 17 (Cytokeratin-17) (CK-17) (Keratin-17) (K17). | |||||