Please be patient as the page loads
|
RPO3G_MOUSE
|
||||||
| SwissProt Accessions | Q6NXY9, Q8K0W5, Q9CV05 | Gene names | Polr3g | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA-directed RNA polymerase III subunit G (EC 2.7.7.6) (DNA-directed RNA polymerase III 32 kDa polypeptide) (RNA polymerase III C32 subunit). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
RPO3G_MOUSE
|
||||||
| θ value | 1.01059e-59 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6NXY9, Q8K0W5, Q9CV05 | Gene names | Polr3g | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA-directed RNA polymerase III subunit G (EC 2.7.7.6) (DNA-directed RNA polymerase III 32 kDa polypeptide) (RNA polymerase III C32 subunit). | |||||
|
RPO3G_HUMAN
|
||||||
| θ value | 2.94719e-51 (rank : 2) | NC score | 0.961382 (rank : 2) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O15318 | Gene names | POLR3G, RPC32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA-directed RNA polymerase III subunit G (EC 2.7.7.6) (DNA-directed RNA polymerase III 32 kDa polypeptide) (RNA polymerase III C32 subunit). | |||||
|
LEO1_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 3) | NC score | 0.144590 (rank : 5) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
SEC63_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 4) | NC score | 0.145938 (rank : 4) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UGP8, O95380, Q86VS9, Q8IWL0, Q9NTE0 | Gene names | SEC63, SEC63L | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
TCAL5_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 5) | NC score | 0.107825 (rank : 14) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
SEC63_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 6) | NC score | 0.127366 (rank : 9) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 601 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VHE0, Q8VEB9 | Gene names | Sec63, Sec63l | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
LEO1_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 7) | NC score | 0.137520 (rank : 7) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
NSBP1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 8) | NC score | 0.114643 (rank : 11) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
NALP5_MOUSE
|
||||||
| θ value | 0.279714 (rank : 9) | NC score | 0.064129 (rank : 40) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9R1M5, Q9JLR2 | Gene names | Nalp5, Mater | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Maternal antigen that embryos require) (Mater protein) (Ooplasm-specific protein 1) (OP1). | |||||
|
STK10_MOUSE
|
||||||
| θ value | 0.279714 (rank : 10) | NC score | 0.018300 (rank : 84) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 2132 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O55098 | Gene names | Stk10, Lok | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
ADNP_MOUSE
|
||||||
| θ value | 0.365318 (rank : 11) | NC score | 0.073341 (rank : 26) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z103 | Gene names | Adnp | |||
|
Domain Architecture |

|
|||||
| Description | Activity-dependent neuroprotector (Activity-dependent neuroprotective protein). | |||||
|
TCAL3_HUMAN
|
||||||
| θ value | 0.365318 (rank : 12) | NC score | 0.113132 (rank : 12) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q969E4 | Gene names | TCEAL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 3 (TCEA-like protein 3) (Transcription elongation factor S-II protein-like 3). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 13) | NC score | 0.092418 (rank : 21) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
SCG2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 14) | NC score | 0.066746 (rank : 37) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P13521, Q8TBH3 | Gene names | SCG2, CHGC | |||
|
Domain Architecture |

|
|||||
| Description | Secretogranin-2 precursor (Secretogranin II) (SgII) (Chromogranin C) [Contains: Secretoneurin (SN)]. | |||||
|
CC47_MOUSE
|
||||||
| θ value | 1.06291 (rank : 15) | NC score | 0.095226 (rank : 19) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9D024, Q3TSB5, Q3V1S7, Q8C5D9, Q920S6 | Gene names | Ccdc47, Asp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 47 precursor (Adipocyte-specific protein 4). | |||||
|
CT096_HUMAN
|
||||||
| θ value | 1.06291 (rank : 16) | NC score | 0.049163 (rank : 76) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NUD7, Q8N840, Q8NAX5 | Gene names | C20orf96 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf96. | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 17) | NC score | 0.085880 (rank : 22) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
STABP_HUMAN
|
||||||
| θ value | 1.81305 (rank : 18) | NC score | 0.039594 (rank : 79) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95630, Q3MJE7 | Gene names | STAMBP, AMSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STAM-binding protein (EC 3.1.2.15) (Associated molecule with the SH3 domain of STAM). | |||||
|
STK10_HUMAN
|
||||||
| θ value | 1.81305 (rank : 19) | NC score | 0.016964 (rank : 86) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O94804, Q9UIW4 | Gene names | STK10, LOK | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
CJ078_MOUSE
|
||||||
| θ value | 2.36792 (rank : 20) | NC score | 0.045276 (rank : 77) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
DIAP1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 21) | NC score | 0.016731 (rank : 87) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 839 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O08808 | Gene names | Diaph1, Diap1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 1 (Diaphanous-related formin-1) (DRF1) (mDIA1) (p140mDIA). | |||||
|
EEA1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 22) | NC score | 0.010100 (rank : 89) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
MK10_MOUSE
|
||||||
| θ value | 4.03905 (rank : 23) | NC score | 0.006313 (rank : 92) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 849 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61831, Q9R0U6 | Gene names | Mapk10, Jnk3, Prkm10, Serk2 | |||
|
Domain Architecture |
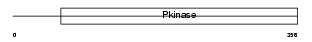
|
|||||
| Description | Mitogen-activated protein kinase 10 (EC 2.7.11.24) (Stress-activated protein kinase JNK3) (c-Jun N-terminal kinase 3) (MAP kinase p49 3F12). | |||||
|
BRD8_MOUSE
|
||||||
| θ value | 5.27518 (rank : 24) | NC score | 0.055945 (rank : 60) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8R3B7, Q8C049, Q8R583, Q8VDP0 | Gene names | Brd8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8. | |||||
|
CCD47_HUMAN
|
||||||
| θ value | 5.27518 (rank : 25) | NC score | 0.072205 (rank : 27) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96A33, Q96D00, Q96JZ7, Q9H3E4, Q9NRG3 | Gene names | CCDC47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 47 precursor. | |||||
|
ZN451_MOUSE
|
||||||
| θ value | 5.27518 (rank : 26) | NC score | 0.011229 (rank : 88) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C0P7 | Gene names | Znf451, Zfp451 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 451. | |||||
|
ANC5_MOUSE
|
||||||
| θ value | 6.88961 (rank : 27) | NC score | 0.024424 (rank : 82) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BTZ4, Q3TGA7, Q80UJ6, Q80W43, Q8BWW7, Q8C0F7, Q9CRX0, Q9CSL1, Q9JJB8 | Gene names | Anapc5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Anaphase-promoting complex subunit 5 (APC5) (Cyclosome subunit 5). | |||||
|
BRD8_HUMAN
|
||||||
| θ value | 6.88961 (rank : 28) | NC score | 0.050199 (rank : 75) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H0E9, O43178, Q15355, Q59GN0, Q969M9 | Gene names | BRD8, SMAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8 (p120) (Skeletal muscle abundant protein) (Thyroid hormone receptor coactivating protein 120kDa) (TrCP120). | |||||
|
EPN4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 29) | NC score | 0.043998 (rank : 78) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q14677, Q8NAF1, Q96E05 | Gene names | CLINT1, ENTH, EPN4, EPNR, KIAA0171 | |||
|
Domain Architecture |
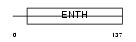
|
|||||
| Description | Clathrin interactor 1 (Epsin-4) (Epsin-related protein) (EpsinR) (Enthoprotin) (Clathrin-interacting protein localized in the trans- Golgi region) (Clint). | |||||
|
MATR3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 30) | NC score | 0.050748 (rank : 71) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8K310 | Gene names | Matr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Matrin-3. | |||||
|
RPGR_MOUSE
|
||||||
| θ value | 6.88961 (rank : 31) | NC score | 0.028599 (rank : 81) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9R0X5, O88408, Q9CU92 | Gene names | Rpgr | |||
|
Domain Architecture |

|
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator (mRpgr). | |||||
|
CARD6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 32) | NC score | 0.023468 (rank : 83) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BX69 | Gene names | CARD6 | |||
|
Domain Architecture |
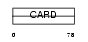
|
|||||
| Description | Caspase recruitment domain-containing protein 6. | |||||
|
NF2L3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 33) | NC score | 0.035171 (rank : 80) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y4A8, Q6NUS0, Q7Z498, Q86UJ4, Q86VR5, Q9UQA4 | Gene names | NFE2L3, NRF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor erythroid 2-related factor 3 (NF-E2-related factor 3) (NFE2-related factor 3) (Nuclear factor, erythroid derived 2, like 3). | |||||
|
P52K_HUMAN
|
||||||
| θ value | 8.99809 (rank : 34) | NC score | 0.017371 (rank : 85) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O43422, Q8WTW1, Q9Y3Z4 | Gene names | PRKRIR, DAP4, P52RIPK, THAP0 | |||
|
Domain Architecture |

|
|||||
| Description | 52 kDa repressor of the inhibitor of the protein kinase (p58IPK- interacting protein) (58 kDa interferon-induced protein kinase- interacting protein) (P52rIPK) (Death-associated protein 4) (THAP domain-containing protein 0). | |||||
|
TBA1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 35) | NC score | 0.010067 (rank : 90) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P68366, P05215 | Gene names | TUBA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulin alpha-1 chain (Alpha-tubulin 1) (Testis-specific alpha- tubulin) (Tubulin H2-alpha). | |||||
|
TBA4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 36) | NC score | 0.010067 (rank : 91) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P68368, P05215 | Gene names | Tuba4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulin alpha-4 chain (Alpha-tubulin 4) (Alpha-tubulin isotype M- alpha-4). | |||||
|
ABTAP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 37) | NC score | 0.057201 (rank : 57) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9H501, Q86X92, Q9H9Q5, Q9HA35, Q9NX93, Q9P1S6 | Gene names | ABTAP, C20orf6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ABT1-associated protein. | |||||
|
ABTAP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 38) | NC score | 0.059198 (rank : 53) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q3V1V3 | Gene names | Abtap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ABT1-associated protein. | |||||
|
ADNP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 39) | NC score | 0.050680 (rank : 73) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 265 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H2P0, O94881, Q9UG34 | Gene names | ADNP, KIAA0784 | |||
|
Domain Architecture |

|
|||||
| Description | Activity-dependent neuroprotector (Activity-dependent neuroprotective protein). | |||||
|
AN32B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 40) | NC score | 0.056172 (rank : 59) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q92688, O00655, P78458, P78459 | Gene names | ANP32B, APRIL, PHAPI2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member B (PHAPI2 protein) (Silver-stainable protein SSP29) (Acidic protein rich in leucines). | |||||
|
AN32B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 41) | NC score | 0.066758 (rank : 36) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9EST5 | Gene names | Anp32b, Pal31 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member B (Proliferation-related acidic leucine-rich protein PAL31). | |||||
|
ARS2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 42) | NC score | 0.055894 (rank : 62) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9BXP5, O95808, Q9BWP6, Q9BXP4 | Gene names | ARS2, ASR2 | |||
|
Domain Architecture |

|
|||||
| Description | Arsenite-resistance protein 2. | |||||
|
ARS2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 43) | NC score | 0.057754 (rank : 55) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99MR6, Q99MR4, Q99MR5, Q99MR7 | Gene names | Ars2, Asr2 | |||
|
Domain Architecture |

|
|||||
| Description | Arsenite-resistance protein 2. | |||||
|
CCDC9_MOUSE
|
||||||
| θ value | θ > 10 (rank : 44) | NC score | 0.055667 (rank : 63) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8VC31 | Gene names | Ccdc9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 9. | |||||
|
CENPB_HUMAN
|
||||||
| θ value | θ > 10 (rank : 45) | NC score | 0.063881 (rank : 41) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P07199 | Gene names | CENPB | |||
|
Domain Architecture |

|
|||||
| Description | Major centromere autoantigen B (Centromere protein B) (CENP-B). | |||||
|
CENPB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 46) | NC score | 0.074687 (rank : 25) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P27790 | Gene names | Cenpb, Cenp-b | |||
|
Domain Architecture |

|
|||||
| Description | Major centromere autoantigen B (Centromere protein B) (CENP-B). | |||||
|
CK051_HUMAN
|
||||||
| θ value | θ > 10 (rank : 47) | NC score | 0.052719 (rank : 68) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P60006, Q9CXK2, Q9Y269 | Gene names | C11orf51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf51. | |||||
|
CK051_MOUSE
|
||||||
| θ value | θ > 10 (rank : 48) | NC score | 0.050902 (rank : 70) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P60007, Q3UXV5, Q9CXK2, Q9Y269 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf51 homolog. | |||||
|
CYLC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 49) | NC score | 0.061084 (rank : 47) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P35663, Q5JQQ9 | Gene names | CYLC1, CYL, CYL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-1 (Cylicin I) (Multiple-band polypeptide I). | |||||
|
CYLC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 50) | NC score | 0.067875 (rank : 34) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q14093 | Gene names | CYLC2, CYL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-2 (Cylicin II) (Multiple-band polypeptide II). | |||||
|
DAXX_HUMAN
|
||||||
| θ value | θ > 10 (rank : 51) | NC score | 0.069730 (rank : 31) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UER7, O14747, O15141, O15208, Q9BWI3 | Gene names | DAXX, BING2, DAP6 | |||
|
Domain Architecture |

|
|||||
| Description | Death domain-associated protein 6 (Daxx) (hDaxx) (Fas death domain- associated protein) (ETS1-associated protein 1) (EAP1). | |||||
|
DAXX_MOUSE
|
||||||
| θ value | θ > 10 (rank : 52) | NC score | 0.061451 (rank : 46) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O35613, Q9QWT8, Q9QWV3 | Gene names | Daxx | |||
|
Domain Architecture |

|
|||||
| Description | Death domain-associated protein 6 (Daxx). | |||||
|
DMP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 53) | NC score | 0.069382 (rank : 32) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
DMP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 54) | NC score | 0.068293 (rank : 33) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
DSPP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 55) | NC score | 0.061737 (rank : 45) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
DSPP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 56) | NC score | 0.065104 (rank : 39) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
FAM9A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 57) | NC score | 0.053624 (rank : 67) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8IZU1 | Gene names | FAM9A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM9A. | |||||
|
GNAS3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 58) | NC score | 0.055070 (rank : 64) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Z0F1, Q9QXW5, Q9Z0H2 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuroendocrine secretory protein 55 (NESP55) [Contains: LHAL tetrapeptide; GPIPIRRH peptide]. | |||||
|
HIRP3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 59) | NC score | 0.059358 (rank : 52) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9BW71, O75707, O75708 | Gene names | HIRIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIRA-interacting protein 3. | |||||
|
HTSF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 60) | NC score | 0.065848 (rank : 38) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O43719, Q59G06, Q99730 | Gene names | HTATSF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 (Tat-SF1). | |||||
|
HTSF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 61) | NC score | 0.077551 (rank : 24) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BGC0, Q1WWK0, Q9CT41, Q9DAU3 | Gene names | Htatsf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 homolog. | |||||
|
MDN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 62) | NC score | 0.055902 (rank : 61) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
NCKX1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 63) | NC score | 0.063725 (rank : 42) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O60721, O43485, O75184 | Gene names | SLC24A1, KIAA0702, NCKX1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium/calcium exchanger 1 (Na(+)/K(+)/Ca(2+)-exchange protein 1) (Retinal rod Na-Ca+K exchanger). | |||||
|
NSBP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 64) | NC score | 0.113029 (rank : 13) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P82970 | Gene names | NSBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1. | |||||
|
NUCKS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.099757 (rank : 16) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9H1E3, Q9H1D6, Q9H723 | Gene names | NUCKS1, NUCKS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (P1). | |||||
|
NUCKS_MOUSE
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.102517 (rank : 15) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q80XU3, Q8BVD8 | Gene names | Nucks1, Nucks | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (JC7). | |||||
|
PELP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.053656 (rank : 66) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8IZL8, O15450, Q5EGN3, Q6NTE6, Q96FT1, Q9BU60 | Gene names | PELP1, HMX3, MNAR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor) (Transcription factor HMX3). | |||||
|
PELP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.051556 (rank : 69) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
PTMA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.146193 (rank : 3) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P06454, Q15249, Q15592 | Gene names | PTMA, TMSA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha-1]. | |||||
|
PTMA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.142111 (rank : 6) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P26350, Q3UQV6 | Gene names | Ptma | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha]. | |||||
|
PTMS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.126144 (rank : 10) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P20962 | Gene names | PTMS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parathymosin. | |||||
|
PTMS_MOUSE
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.129492 (rank : 8) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9D0J8 | Gene names | Ptms | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parathymosin. | |||||
|
RENT2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.050403 (rank : 74) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9HAU5, Q8N8U1, Q9H1J2, Q9NWL1, Q9P2D9, Q9Y4M9 | Gene names | UPF2, KIAA1408, RENT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of nonsense transcripts 2 (Nonsense mRNA reducing factor 2) (Up-frameshift suppressor 2 homolog) (hUpf2). | |||||
|
RGP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.060679 (rank : 49) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P46061, Q60801 | Gene names | Rangap1, Fug1 | |||
|
Domain Architecture |

|
|||||
| Description | Ran GTPase-activating protein 1. | |||||
|
RPGR1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.062818 (rank : 44) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 713 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9EPQ2, Q8CAC2, Q8CDJ9, Q91WE0, Q9CUK6, Q9D5Q1 | Gene names | Rpgrip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
SFR11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.056416 (rank : 58) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q05519 | Gene names | SFRS11 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor arginine/serine-rich 11 (Arginine-rich 54 kDa nuclear protein) (p54). | |||||
|
SFR12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.061033 (rank : 48) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8WXA9, Q86X37 | Gene names | SFRS12, SRRP86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86) (Splicing regulatory protein 508) (SRrp508). | |||||
|
SFR12_MOUSE
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.054438 (rank : 65) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BZX4, Q8BLM8, Q8BWK7, Q8BYK0, Q8BYZ4 | Gene names | Sfrs12, Srrp86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86). | |||||
|
SRCH_HUMAN
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.078937 (rank : 23) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P23327, Q504Y6 | Gene names | HRC, HCP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcoplasmic reticulum histidine-rich calcium-binding protein precursor. | |||||
|
T2FA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.070209 (rank : 30) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P35269, Q9BWN0 | Gene names | GTF2F1, RAP74 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor IIF alpha subunit (EC 2.7.11.1) (TFIIF-alpha) (Transcription initiation factor RAP74) (General transcription factor IIF polypeptide 1 74 kDa subunit protein). | |||||
|
TCAL2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.067042 (rank : 35) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H3H9 | Gene names | TCEAL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 2 (TCEA-like protein 2) (Transcription elongation factor S-II protein-like 2). | |||||
|
TCAL3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.097254 (rank : 18) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8R0A5, Q9CYK5, Q9D1S4 | Gene names | Tceal3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 3 (TCEA-like protein 3) (Transcription elongation factor S-II protein-like 3). | |||||
|
TCAL4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.072074 (rank : 28) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96EI5, Q8WY12, Q9H2H1, Q9H775 | Gene names | TCEAL4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 4 (TCEA-like protein 4) (Transcription elongation factor S-II protein-like 4). | |||||
|
TCAL5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.093113 (rank : 20) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8CCT4, Q497R6 | Gene names | Tceal5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
TCAL6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.098257 (rank : 17) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6IPX3, Q5H9J8 | Gene names | TCEAL6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 6 (TCEA-like protein 6) (Transcription elongation factor S-II protein-like 6). | |||||
|
TRI44_HUMAN
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.060233 (rank : 51) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96DX7, Q96QY2, Q9UGK0 | Gene names | TRIM44, DIPB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB). | |||||
|
TRI44_MOUSE
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.057952 (rank : 54) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9QXA7, Q80SV9, Q9JHR8 | Gene names | Trim44, Dipb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB) (Protein Mc7). | |||||
|
TXD13_HUMAN
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.050712 (rank : 72) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9H1E5, Q8N4P7, Q8NCC1, Q9UJA1, Q9ULQ8 | Gene names | TXNDC13, KIAA1162 | |||
|
Domain Architecture |
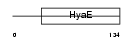
|
|||||
| Description | Thioredoxin domain-containing protein 13 precursor. | |||||
|
UBF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.063582 (rank : 43) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P17480 | Gene names | UBTF, UBF, UBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1) (Autoantigen NOR-90). | |||||
|
UBF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.060502 (rank : 50) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P25976 | Gene names | Ubtf, Tcfubf, Ubf-1, Ubf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1). | |||||
|
YTDC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.070822 (rank : 29) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q96MU7, Q7Z622, Q8TF35 | Gene names | YTHDC1, KIAA1966, YT521 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain-containing protein 1 (Putative splicing factor YT521). | |||||
|
ZC313_HUMAN
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.057505 (rank : 56) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5T200, O94936, Q5T1Z9, Q7Z7J3, Q8NDT6, Q9H0L6 | Gene names | ZC3H13, KIAA0853 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 13. | |||||
|
RPO3G_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 1.01059e-59 (rank : 1) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6NXY9, Q8K0W5, Q9CV05 | Gene names | Polr3g | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA-directed RNA polymerase III subunit G (EC 2.7.7.6) (DNA-directed RNA polymerase III 32 kDa polypeptide) (RNA polymerase III C32 subunit). | |||||
|
RPO3G_HUMAN
|
||||||
| NC score | 0.961382 (rank : 2) | θ value | 2.94719e-51 (rank : 2) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O15318 | Gene names | POLR3G, RPC32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA-directed RNA polymerase III subunit G (EC 2.7.7.6) (DNA-directed RNA polymerase III 32 kDa polypeptide) (RNA polymerase III C32 subunit). | |||||
|
PTMA_HUMAN
|
||||||
| NC score | 0.146193 (rank : 3) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P06454, Q15249, Q15592 | Gene names | PTMA, TMSA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha-1]. | |||||
|
SEC63_HUMAN
|
||||||
| NC score | 0.145938 (rank : 4) | θ value | 0.0148317 (rank : 4) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UGP8, O95380, Q86VS9, Q8IWL0, Q9NTE0 | Gene names | SEC63, SEC63L | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.144590 (rank : 5) | θ value | 0.00390308 (rank : 3) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
PTMA_MOUSE
|
||||||
| NC score | 0.142111 (rank : 6) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P26350, Q3UQV6 | Gene names | Ptma | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha]. | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.137520 (rank : 7) | θ value | 0.0736092 (rank : 7) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
PTMS_MOUSE
|
||||||
| NC score | 0.129492 (rank : 8) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9D0J8 | Gene names | Ptms | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parathymosin. | |||||
|
SEC63_MOUSE
|
||||||
| NC score | 0.127366 (rank : 9) | θ value | 0.0563607 (rank : 6) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 601 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VHE0, Q8VEB9 | Gene names | Sec63, Sec63l | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
PTMS_HUMAN
|
||||||
| NC score | 0.126144 (rank : 10) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P20962 | Gene names | PTMS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parathymosin. | |||||
|
NSBP1_MOUSE
|
||||||
| NC score | 0.114643 (rank : 11) | θ value | 0.21417 (rank : 8) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
TCAL3_HUMAN
|
||||||
| NC score | 0.113132 (rank : 12) | θ value | 0.365318 (rank : 12) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q969E4 | Gene names | TCEAL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 3 (TCEA-like protein 3) (Transcription elongation factor S-II protein-like 3). | |||||
|
NSBP1_HUMAN
|
||||||
| NC score | 0.113029 (rank : 13) | θ value | θ > 10 (rank : 64) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P82970 | Gene names | NSBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1. | |||||
|
TCAL5_HUMAN
|
||||||
| NC score | 0.107825 (rank : 14) | θ value | 0.0193708 (rank : 5) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
NUCKS_MOUSE
|
||||||
| NC score | 0.102517 (rank : 15) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q80XU3, Q8BVD8 | Gene names | Nucks1, Nucks | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (JC7). | |||||
|
NUCKS_HUMAN
|
||||||
| NC score | 0.099757 (rank : 16) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9H1E3, Q9H1D6, Q9H723 | Gene names | NUCKS1, NUCKS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (P1). | |||||
|
TCAL6_HUMAN
|
||||||
| NC score | 0.098257 (rank : 17) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6IPX3, Q5H9J8 | Gene names | TCEAL6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 6 (TCEA-like protein 6) (Transcription elongation factor S-II protein-like 6). | |||||
|
TCAL3_MOUSE
|
||||||
| NC score | 0.097254 (rank : 18) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8R0A5, Q9CYK5, Q9D1S4 | Gene names | Tceal3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 3 (TCEA-like protein 3) (Transcription elongation factor S-II protein-like 3). | |||||
|
CC47_MOUSE
|
||||||
| NC score | 0.095226 (rank : 19) | θ value | 1.06291 (rank : 15) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9D024, Q3TSB5, Q3V1S7, Q8C5D9, Q920S6 | Gene names | Ccdc47, Asp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 47 precursor (Adipocyte-specific protein 4). | |||||
|
TCAL5_MOUSE
|
||||||
| NC score | 0.093113 (rank : 20) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8CCT4, Q497R6 | Gene names | Tceal5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.092418 (rank : 21) | θ value | 0.47712 (rank : 13) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.085880 (rank : 22) | θ value | 1.06291 (rank : 17) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
SRCH_HUMAN
|
||||||
| NC score | 0.078937 (rank : 23) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P23327, Q504Y6 | Gene names | HRC, HCP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcoplasmic reticulum histidine-rich calcium-binding protein precursor. | |||||
|
HTSF1_MOUSE
|
||||||
| NC score | 0.077551 (rank : 24) | θ value | θ > 10 (rank : 61) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BGC0, Q1WWK0, Q9CT41, Q9DAU3 | Gene names | Htatsf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 homolog. | |||||
|
CENPB_MOUSE
|
||||||
| NC score | 0.074687 (rank : 25) | θ value | θ > 10 (rank : 46) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P27790 | Gene names | Cenpb, Cenp-b | |||
|
Domain Architecture |

|
|||||
| Description | Major centromere autoantigen B (Centromere protein B) (CENP-B). | |||||
|
ADNP_MOUSE
|
||||||
| NC score | 0.073341 (rank : 26) | θ value | 0.365318 (rank : 11) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z103 | Gene names | Adnp | |||
|
Domain Architecture |

|
|||||
| Description | Activity-dependent neuroprotector (Activity-dependent neuroprotective protein). | |||||
|
CCD47_HUMAN
|
||||||
| NC score | 0.072205 (rank : 27) | θ value | 5.27518 (rank : 25) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96A33, Q96D00, Q96JZ7, Q9H3E4, Q9NRG3 | Gene names | CCDC47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 47 precursor. | |||||
|
TCAL4_HUMAN
|
||||||
| NC score | 0.072074 (rank : 28) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96EI5, Q8WY12, Q9H2H1, Q9H775 | Gene names | TCEAL4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 4 (TCEA-like protein 4) (Transcription elongation factor S-II protein-like 4). | |||||
|
YTDC1_HUMAN
|
||||||
| NC score | 0.070822 (rank : 29) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q96MU7, Q7Z622, Q8TF35 | Gene names | YTHDC1, KIAA1966, YT521 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain-containing protein 1 (Putative splicing factor YT521). | |||||
|
T2FA_HUMAN
|
||||||
| NC score | 0.070209 (rank : 30) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P35269, Q9BWN0 | Gene names | GTF2F1, RAP74 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor IIF alpha subunit (EC 2.7.11.1) (TFIIF-alpha) (Transcription initiation factor RAP74) (General transcription factor IIF polypeptide 1 74 kDa subunit protein). | |||||
|
DAXX_HUMAN
|
||||||
| NC score | 0.069730 (rank : 31) | θ value | θ > 10 (rank : 51) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UER7, O14747, O15141, O15208, Q9BWI3 | Gene names | DAXX, BING2, DAP6 | |||
|
Domain Architecture |

|
|||||
| Description | Death domain-associated protein 6 (Daxx) (hDaxx) (Fas death domain- associated protein) (ETS1-associated protein 1) (EAP1). | |||||
|
DMP1_HUMAN
|
||||||
| NC score | 0.069382 (rank : 32) | θ value | θ > 10 (rank : 53) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
DMP1_MOUSE
|
||||||
| NC score | 0.068293 (rank : 33) | θ value | θ > 10 (rank : 54) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
CYLC2_HUMAN
|
||||||
| NC score | 0.067875 (rank : 34) | θ value | θ > 10 (rank : 50) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q14093 | Gene names | CYLC2, CYL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-2 (Cylicin II) (Multiple-band polypeptide II). | |||||
|
TCAL2_HUMAN
|
||||||
| NC score | 0.067042 (rank : 35) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H3H9 | Gene names | TCEAL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 2 (TCEA-like protein 2) (Transcription elongation factor S-II protein-like 2). | |||||
|
AN32B_MOUSE
|
||||||
| NC score | 0.066758 (rank : 36) | θ value | θ > 10 (rank : 41) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9EST5 | Gene names | Anp32b, Pal31 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member B (Proliferation-related acidic leucine-rich protein PAL31). | |||||
|
SCG2_HUMAN
|
||||||
| NC score | 0.066746 (rank : 37) | θ value | 0.813845 (rank : 14) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P13521, Q8TBH3 | Gene names | SCG2, CHGC | |||
|
Domain Architecture |

|
|||||
| Description | Secretogranin-2 precursor (Secretogranin II) (SgII) (Chromogranin C) [Contains: Secretoneurin (SN)]. | |||||
|
HTSF1_HUMAN
|
||||||
| NC score | 0.065848 (rank : 38) | θ value | θ > 10 (rank : 60) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O43719, Q59G06, Q99730 | Gene names | HTATSF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 (Tat-SF1). | |||||
|
DSPP_MOUSE
|
||||||
| NC score | 0.065104 (rank : 39) | θ value | θ > 10 (rank : 56) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
NALP5_MOUSE
|
||||||
| NC score | 0.064129 (rank : 40) | θ value | 0.279714 (rank : 9) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9R1M5, Q9JLR2 | Gene names | Nalp5, Mater | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Maternal antigen that embryos require) (Mater protein) (Ooplasm-specific protein 1) (OP1). | |||||
|
CENPB_HUMAN
|
||||||
| NC score | 0.063881 (rank : 41) | θ value | θ > 10 (rank : 45) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P07199 | Gene names | CENPB | |||
|
Domain Architecture |

|
|||||
| Description | Major centromere autoantigen B (Centromere protein B) (CENP-B). | |||||
|
NCKX1_HUMAN
|
||||||
| NC score | 0.063725 (rank : 42) | θ value | θ > 10 (rank : 63) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O60721, O43485, O75184 | Gene names | SLC24A1, KIAA0702, NCKX1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium/calcium exchanger 1 (Na(+)/K(+)/Ca(2+)-exchange protein 1) (Retinal rod Na-Ca+K exchanger). | |||||
|
UBF1_HUMAN
|
||||||
| NC score | 0.063582 (rank : 43) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P17480 | Gene names | UBTF, UBF, UBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1) (Autoantigen NOR-90). | |||||
|
RPGR1_MOUSE
|
||||||
| NC score | 0.062818 (rank : 44) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 713 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9EPQ2, Q8CAC2, Q8CDJ9, Q91WE0, Q9CUK6, Q9D5Q1 | Gene names | Rpgrip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
DSPP_HUMAN
|
||||||
| NC score | 0.061737 (rank : 45) | θ value | θ > 10 (rank : 55) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
DAXX_MOUSE
|
||||||
| NC score | 0.061451 (rank : 46) | θ value | θ > 10 (rank : 52) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O35613, Q9QWT8, Q9QWV3 | Gene names | Daxx | |||
|
Domain Architecture |

|
|||||
| Description | Death domain-associated protein 6 (Daxx). | |||||
|
CYLC1_HUMAN
|
||||||
| NC score | 0.061084 (rank : 47) | θ value | θ > 10 (rank : 49) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P35663, Q5JQQ9 | Gene names | CYLC1, CYL, CYL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-1 (Cylicin I) (Multiple-band polypeptide I). | |||||
|
SFR12_HUMAN
|
||||||
| NC score | 0.061033 (rank : 48) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8WXA9, Q86X37 | Gene names | SFRS12, SRRP86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86) (Splicing regulatory protein 508) (SRrp508). | |||||
|
RGP1_MOUSE
|
||||||
| NC score | 0.060679 (rank : 49) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P46061, Q60801 | Gene names | Rangap1, Fug1 | |||
|
Domain Architecture |

|
|||||
| Description | Ran GTPase-activating protein 1. | |||||
|
UBF1_MOUSE
|
||||||
| NC score | 0.060502 (rank : 50) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P25976 | Gene names | Ubtf, Tcfubf, Ubf-1, Ubf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1). | |||||
|
TRI44_HUMAN
|
||||||
| NC score | 0.060233 (rank : 51) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96DX7, Q96QY2, Q9UGK0 | Gene names | TRIM44, DIPB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB). | |||||
|
HIRP3_HUMAN
|
||||||
| NC score | 0.059358 (rank : 52) | θ value | θ > 10 (rank : 59) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9BW71, O75707, O75708 | Gene names | HIRIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIRA-interacting protein 3. | |||||
|
ABTAP_MOUSE
|
||||||
| NC score | 0.059198 (rank : 53) | θ value | θ > 10 (rank : 38) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q3V1V3 | Gene names | Abtap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ABT1-associated protein. | |||||
|
TRI44_MOUSE
|
||||||
| NC score | 0.057952 (rank : 54) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9QXA7, Q80SV9, Q9JHR8 | Gene names | Trim44, Dipb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB) (Protein Mc7). | |||||
|
ARS2_MOUSE
|
||||||
| NC score | 0.057754 (rank : 55) | θ value | θ > 10 (rank : 43) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99MR6, Q99MR4, Q99MR5, Q99MR7 | Gene names | Ars2, Asr2 | |||
|
Domain Architecture |

|
|||||
| Description | Arsenite-resistance protein 2. | |||||
|
ZC313_HUMAN
|
||||||
| NC score | 0.057505 (rank : 56) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5T200, O94936, Q5T1Z9, Q7Z7J3, Q8NDT6, Q9H0L6 | Gene names | ZC3H13, KIAA0853 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 13. | |||||
|
ABTAP_HUMAN
|
||||||
| NC score | 0.057201 (rank : 57) | θ value | θ > 10 (rank : 37) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9H501, Q86X92, Q9H9Q5, Q9HA35, Q9NX93, Q9P1S6 | Gene names | ABTAP, C20orf6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ABT1-associated protein. | |||||
|
SFR11_HUMAN
|
||||||
| NC score | 0.056416 (rank : 58) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q05519 | Gene names | SFRS11 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor arginine/serine-rich 11 (Arginine-rich 54 kDa nuclear protein) (p54). | |||||
|
AN32B_HUMAN
|
||||||
| NC score | 0.056172 (rank : 59) | θ value | θ > 10 (rank : 40) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q92688, O00655, P78458, P78459 | Gene names | ANP32B, APRIL, PHAPI2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member B (PHAPI2 protein) (Silver-stainable protein SSP29) (Acidic protein rich in leucines). | |||||
|
BRD8_MOUSE
|
||||||
| NC score | 0.055945 (rank : 60) | θ value | 5.27518 (rank : 24) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8R3B7, Q8C049, Q8R583, Q8VDP0 | Gene names | Brd8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8. | |||||
|
MDN1_HUMAN
|
||||||
| NC score | 0.055902 (rank : 61) | θ value | θ > 10 (rank : 62) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
ARS2_HUMAN
|
||||||
| NC score | 0.055894 (rank : 62) | θ value | θ > 10 (rank : 42) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9BXP5, O95808, Q9BWP6, Q9BXP4 | Gene names | ARS2, ASR2 | |||
|
Domain Architecture |

|
|||||
| Description | Arsenite-resistance protein 2. | |||||
|
CCDC9_MOUSE
|
||||||
| NC score | 0.055667 (rank : 63) | θ value | θ > 10 (rank : 44) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8VC31 | Gene names | Ccdc9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 9. | |||||
|
GNAS3_MOUSE
|
||||||
| NC score | 0.055070 (rank : 64) | θ value | θ > 10 (rank : 58) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Z0F1, Q9QXW5, Q9Z0H2 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuroendocrine secretory protein 55 (NESP55) [Contains: LHAL tetrapeptide; GPIPIRRH peptide]. | |||||
|
SFR12_MOUSE
|
||||||
| NC score | 0.054438 (rank : 65) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BZX4, Q8BLM8, Q8BWK7, Q8BYK0, Q8BYZ4 | Gene names | Sfrs12, Srrp86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86). | |||||
|
PELP1_HUMAN
|
||||||
| NC score | 0.053656 (rank : 66) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8IZL8, O15450, Q5EGN3, Q6NTE6, Q96FT1, Q9BU60 | Gene names | PELP1, HMX3, MNAR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor) (Transcription factor HMX3). | |||||
|
FAM9A_HUMAN
|
||||||
| NC score | 0.053624 (rank : 67) | θ value | θ > 10 (rank : 57) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8IZU1 | Gene names | FAM9A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM9A. | |||||
|
CK051_HUMAN
|
||||||
| NC score | 0.052719 (rank : 68) | θ value | θ > 10 (rank : 47) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P60006, Q9CXK2, Q9Y269 | Gene names | C11orf51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf51. | |||||
|
PELP1_MOUSE
|
||||||
| NC score | 0.051556 (rank : 69) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
CK051_MOUSE
|
||||||
| NC score | 0.050902 (rank : 70) | θ value | θ > 10 (rank : 48) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P60007, Q3UXV5, Q9CXK2, Q9Y269 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf51 homolog. | |||||
|
MATR3_MOUSE
|
||||||
| NC score | 0.050748 (rank : 71) | θ value | 6.88961 (rank : 30) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8K310 | Gene names | Matr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Matrin-3. | |||||
|
TXD13_HUMAN
|
||||||
| NC score | 0.050712 (rank : 72) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9H1E5, Q8N4P7, Q8NCC1, Q9UJA1, Q9ULQ8 | Gene names | TXNDC13, KIAA1162 | |||
|
Domain Architecture |

|
|||||
| Description | Thioredoxin domain-containing protein 13 precursor. | |||||
|
ADNP_HUMAN
|
||||||
| NC score | 0.050680 (rank : 73) | θ value | θ > 10 (rank : 39) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 265 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H2P0, O94881, Q9UG34 | Gene names | ADNP, KIAA0784 | |||
|
Domain Architecture |

|
|||||
| Description | Activity-dependent neuroprotector (Activity-dependent neuroprotective protein). | |||||
|
RENT2_HUMAN
|
||||||
| NC score | 0.050403 (rank : 74) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9HAU5, Q8N8U1, Q9H1J2, Q9NWL1, Q9P2D9, Q9Y4M9 | Gene names | UPF2, KIAA1408, RENT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of nonsense transcripts 2 (Nonsense mRNA reducing factor 2) (Up-frameshift suppressor 2 homolog) (hUpf2). | |||||
|
BRD8_HUMAN
|
||||||
| NC score | 0.050199 (rank : 75) | θ value | 6.88961 (rank : 28) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H0E9, O43178, Q15355, Q59GN0, Q969M9 | Gene names | BRD8, SMAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8 (p120) (Skeletal muscle abundant protein) (Thyroid hormone receptor coactivating protein 120kDa) (TrCP120). | |||||
|
CT096_HUMAN
|
||||||
| NC score | 0.049163 (rank : 76) | θ value | 1.06291 (rank : 16) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NUD7, Q8N840, Q8NAX5 | Gene names | C20orf96 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf96. | |||||
|
CJ078_MOUSE
|
||||||
| NC score | 0.045276 (rank : 77) | θ value | 2.36792 (rank : 20) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
EPN4_HUMAN
|
||||||
| NC score | 0.043998 (rank : 78) | θ value | 6.88961 (rank : 29) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q14677, Q8NAF1, Q96E05 | Gene names | CLINT1, ENTH, EPN4, EPNR, KIAA0171 | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin interactor 1 (Epsin-4) (Epsin-related protein) (EpsinR) (Enthoprotin) (Clathrin-interacting protein localized in the trans- Golgi region) (Clint). | |||||
|
STABP_HUMAN
|
||||||
| NC score | 0.039594 (rank : 79) | θ value | 1.81305 (rank : 18) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95630, Q3MJE7 | Gene names | STAMBP, AMSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STAM-binding protein (EC 3.1.2.15) (Associated molecule with the SH3 domain of STAM). | |||||
|
NF2L3_HUMAN
|
||||||
| NC score | 0.035171 (rank : 80) | θ value | 8.99809 (rank : 33) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y4A8, Q6NUS0, Q7Z498, Q86UJ4, Q86VR5, Q9UQA4 | Gene names | NFE2L3, NRF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor erythroid 2-related factor 3 (NF-E2-related factor 3) (NFE2-related factor 3) (Nuclear factor, erythroid derived 2, like 3). | |||||
|
RPGR_MOUSE
|
||||||
| NC score | 0.028599 (rank : 81) | θ value | 6.88961 (rank : 31) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9R0X5, O88408, Q9CU92 | Gene names | Rpgr | |||
|
Domain Architecture |

|
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator (mRpgr). | |||||
|
ANC5_MOUSE
|
||||||
| NC score | 0.024424 (rank : 82) | θ value | 6.88961 (rank : 27) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BTZ4, Q3TGA7, Q80UJ6, Q80W43, Q8BWW7, Q8C0F7, Q9CRX0, Q9CSL1, Q9JJB8 | Gene names | Anapc5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Anaphase-promoting complex subunit 5 (APC5) (Cyclosome subunit 5). | |||||
|
CARD6_HUMAN
|
||||||
| NC score | 0.023468 (rank : 83) | θ value | 8.99809 (rank : 32) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BX69 | Gene names | CARD6 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 6. | |||||
|
STK10_MOUSE
|
||||||
| NC score | 0.018300 (rank : 84) | θ value | 0.279714 (rank : 10) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 2132 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O55098 | Gene names | Stk10, Lok | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
P52K_HUMAN
|
||||||
| NC score | 0.017371 (rank : 85) | θ value | 8.99809 (rank : 34) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O43422, Q8WTW1, Q9Y3Z4 | Gene names | PRKRIR, DAP4, P52RIPK, THAP0 | |||
|
Domain Architecture |

|
|||||
| Description | 52 kDa repressor of the inhibitor of the protein kinase (p58IPK- interacting protein) (58 kDa interferon-induced protein kinase- interacting protein) (P52rIPK) (Death-associated protein 4) (THAP domain-containing protein 0). | |||||
|
STK10_HUMAN
|
||||||
| NC score | 0.016964 (rank : 86) | θ value | 1.81305 (rank : 19) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O94804, Q9UIW4 | Gene names | STK10, LOK | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
DIAP1_MOUSE
|
||||||
| NC score | 0.016731 (rank : 87) | θ value | 4.03905 (rank : 21) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 839 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O08808 | Gene names | Diaph1, Diap1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 1 (Diaphanous-related formin-1) (DRF1) (mDIA1) (p140mDIA). | |||||
|
ZN451_MOUSE
|
||||||
| NC score | 0.011229 (rank : 88) | θ value | 5.27518 (rank : 26) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C0P7 | Gene names | Znf451, Zfp451 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 451. | |||||
|
EEA1_HUMAN
|
||||||
| NC score | 0.010100 (rank : 89) | θ value | 4.03905 (rank : 22) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
TBA1_HUMAN
|
||||||
| NC score | 0.010067 (rank : 90) | θ value | 8.99809 (rank : 35) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P68366, P05215 | Gene names | TUBA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulin alpha-1 chain (Alpha-tubulin 1) (Testis-specific alpha- tubulin) (Tubulin H2-alpha). | |||||
|
TBA4_MOUSE
|
||||||
| NC score | 0.010067 (rank : 91) | θ value | 8.99809 (rank : 36) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P68368, P05215 | Gene names | Tuba4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulin alpha-4 chain (Alpha-tubulin 4) (Alpha-tubulin isotype M- alpha-4). | |||||
|
MK10_MOUSE
|
||||||
| NC score | 0.006313 (rank : 92) | θ value | 4.03905 (rank : 23) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 849 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61831, Q9R0U6 | Gene names | Mapk10, Jnk3, Prkm10, Serk2 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase 10 (EC 2.7.11.24) (Stress-activated protein kinase JNK3) (c-Jun N-terminal kinase 3) (MAP kinase p49 3F12). | |||||