Please be patient as the page loads
|
RIB1_HUMAN
|
||||||
| SwissProt Accessions | P04843 | Gene names | RPN1 | |||
|
Domain Architecture |

|
|||||
| Description | Dolichyl-diphosphooligosaccharide--protein glycosyltransferase 67 kDa subunit precursor (EC 2.4.1.119) (Ribophorin I) (RPN-I). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
RIB1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P04843 | Gene names | RPN1 | |||
|
Domain Architecture |

|
|||||
| Description | Dolichyl-diphosphooligosaccharide--protein glycosyltransferase 67 kDa subunit precursor (EC 2.4.1.119) (Ribophorin I) (RPN-I). | |||||
|
RIB1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.995425 (rank : 2) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q91YQ5, Q3U985 | Gene names | Rpn1 | |||
|
Domain Architecture |

|
|||||
| Description | Dolichyl-diphosphooligosaccharide--protein glycosyltransferase 67 kDa subunit precursor (EC 2.4.1.119) (Ribophorin I) (RPN-I). | |||||
|
MYH10_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 3) | NC score | 0.074670 (rank : 16) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P35580, Q16087 | Gene names | MYH10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
MYH10_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 4) | NC score | 0.074564 (rank : 17) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1612 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q61879, Q5SV63 | Gene names | Myh10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
TPM1_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 5) | NC score | 0.111280 (rank : 4) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 716 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P09493, P09494, P10469, Q86W64, Q96IK2, Q9UCY9 | Gene names | TPM1, TMSA | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin 1 alpha chain (Alpha-tropomyosin). | |||||
|
TPM1_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 6) | NC score | 0.111817 (rank : 3) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 729 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P58771, P02558, P19354, P46902, P99034 | Gene names | Tpm1, Tpm-1, Tpma | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin 1 alpha chain (Alpha-tropomyosin). | |||||
|
TPM3_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 7) | NC score | 0.106034 (rank : 6) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 689 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P06753, P12324, Q969Q2, Q9NQH8 | Gene names | TPM3 | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin alpha-3 chain (Tropomyosin-3) (Tropomyosin gamma) (hTM5). | |||||
|
TPM3_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 8) | NC score | 0.106092 (rank : 5) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P21107, Q09021, Q60606, Q80SW6, Q9EPW3 | Gene names | Tpm3, Tpm-5, Tpm5 | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin alpha-3 chain (Tropomyosin-3) (Tropomyosin gamma). | |||||
|
BICD2_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 9) | NC score | 0.096402 (rank : 8) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8TD16, O75181, Q5TBQ2, Q5TBQ3, Q96LH2, Q9BT84, Q9H561 | Gene names | BICD2, KIAA0699 | |||
|
Domain Architecture |

|
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 2. | |||||
|
EEA1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 10) | NC score | 0.073009 (rank : 18) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1637 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8BL66, Q6DIC2 | Gene names | Eea1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early endosome antigen 1. | |||||
|
BICD2_MOUSE
|
||||||
| θ value | 0.279714 (rank : 11) | NC score | 0.091697 (rank : 9) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 864 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q921C5, Q80TU1, Q8BTE3, Q9DCL3 | Gene names | Bicd2, Kiaa0699 | |||
|
Domain Architecture |

|
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 2. | |||||
|
ROCK2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 12) | NC score | 0.036610 (rank : 176) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 2073 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O75116, Q9UQN5 | Gene names | ROCK2, KIAA0619 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2) (Rho kinase 2). | |||||
|
ROCK2_MOUSE
|
||||||
| θ value | 0.279714 (rank : 13) | NC score | 0.037701 (rank : 174) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 2072 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P70336, Q8BR64, Q8CBR0 | Gene names | Rock2 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2). | |||||
|
CKAP4_MOUSE
|
||||||
| θ value | 0.365318 (rank : 14) | NC score | 0.099080 (rank : 7) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8BMK4, Q8BTK8, Q8R3F2 | Gene names | Ckap4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-associated protein 4 (63 kDa membrane protein) (p63). | |||||
|
MYH9_MOUSE
|
||||||
| θ value | 0.365318 (rank : 15) | NC score | 0.069187 (rank : 24) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
TRIPB_HUMAN
|
||||||
| θ value | 0.47712 (rank : 16) | NC score | 0.065979 (rank : 30) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
ZN148_HUMAN
|
||||||
| θ value | 0.62314 (rank : 17) | NC score | 0.013566 (rank : 181) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UQR1, O00389, O43591 | Gene names | ZNF148, ZBP89 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 148 (Zinc finger DNA-binding protein 89) (Transcription factor ZBP-89). | |||||
|
ZN148_MOUSE
|
||||||
| θ value | 0.62314 (rank : 18) | NC score | 0.013564 (rank : 182) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 755 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61624, P97475 | Gene names | Znf148, Zbp89, Zfp148 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 148 (Zinc finger DNA-binding protein 89) (Transcription factor ZBP-89) (G-rich box-binding protein) (Beta enolase repressor factor 1) (Transcription factor BFCOL1). | |||||
|
CF060_HUMAN
|
||||||
| θ value | 0.813845 (rank : 19) | NC score | 0.076737 (rank : 15) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1270 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8NB25, Q96GY8, Q9H851 | Gene names | C6orf60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf60. | |||||
|
GRAP1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 20) | NC score | 0.070115 (rank : 23) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q4V328, Q4V327, Q4V330, Q5HYG1, Q6N046, Q96DH8, Q9NQ43, Q9ULQ3 | Gene names | GRIPAP1, KIAA1167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP1-associated protein 1 (GRASP-1). | |||||
|
AKAP9_HUMAN
|
||||||
| θ value | 1.06291 (rank : 21) | NC score | 0.066869 (rank : 28) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
MYH9_HUMAN
|
||||||
| θ value | 1.06291 (rank : 22) | NC score | 0.067114 (rank : 27) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
HIP1R_HUMAN
|
||||||
| θ value | 1.81305 (rank : 23) | NC score | 0.070452 (rank : 22) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 809 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O75146, Q9UED9 | Gene names | HIP1R, HIP12, KIAA0655 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1-related protein (Hip1-related) (Hip 12). | |||||
|
KIF3B_HUMAN
|
||||||
| θ value | 1.81305 (rank : 24) | NC score | 0.033352 (rank : 177) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 540 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O15066 | Gene names | KIF3B, KIAA0359 | |||
|
Domain Architecture |
|
|||||
| Description | Kinesin-like protein KIF3B (Microtubule plus end-directed kinesin motor 3B) (HH0048). | |||||
|
KIF3B_MOUSE
|
||||||
| θ value | 1.81305 (rank : 25) | NC score | 0.033149 (rank : 178) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 533 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q61771 | Gene names | Kif3b | |||
|
Domain Architecture |
|
|||||
| Description | Kinesin-like protein KIF3B (Microtubule plus end-directed kinesin motor 3B). | |||||
|
MYH3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 26) | NC score | 0.063413 (rank : 35) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1660 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P11055, Q15492 | Gene names | MYH3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, fast skeletal muscle, embryonic (Muscle embryonic myosin heavy chain) (SMHCE). | |||||
|
SYNE1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 27) | NC score | 0.057272 (rank : 52) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
HIP1R_MOUSE
|
||||||
| θ value | 2.36792 (rank : 28) | NC score | 0.070490 (rank : 21) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9JKY5 | Gene names | Hip1r | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1-related protein (Hip1-related). | |||||
|
TPR_HUMAN
|
||||||
| θ value | 2.36792 (rank : 29) | NC score | 0.063084 (rank : 37) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
BAZ1A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 30) | NC score | 0.037386 (rank : 175) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9NRL2, Q9NZ15, Q9P065, Q9UIG1, Q9Y3V3 | Gene names | BAZ1A, ACF1, WCRF180 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1A (ATP-utilizing chromatin assembly and remodeling factor 1) (hACF1) (ATP-dependent chromatin remodelling protein) (Williams syndrome transcription factor-related chromatin remodeling factor 180) (WCRF180) (hWALp1) (CHRAC subunit ACF1). | |||||
|
CING_HUMAN
|
||||||
| θ value | 3.0926 (rank : 31) | NC score | 0.066682 (rank : 29) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9P2M7, Q5T386, Q9NR25 | Gene names | CGN, KIAA1319 | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
DZIP1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 32) | NC score | 0.051594 (rank : 143) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 675 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q86YF9, Q8WY45, Q8WY46, Q9UGA5, Q9Y2K0 | Gene names | DZIP1, DZIP, DZIP2, KIAA0996 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein DZIP1 (DAZ-interacting protein 1/2). | |||||
|
K0555_HUMAN
|
||||||
| θ value | 3.0926 (rank : 33) | NC score | 0.063171 (rank : 36) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 880 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q96AA8, O60302 | Gene names | KIAA0555 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0555. | |||||
|
MYH11_HUMAN
|
||||||
| θ value | 3.0926 (rank : 34) | NC score | 0.064623 (rank : 33) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
SAS6_HUMAN
|
||||||
| θ value | 3.0926 (rank : 35) | NC score | 0.067711 (rank : 26) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 791 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q6UVJ0, Q8N3K0 | Gene names | SASS6, SAS6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spindle assembly abnormal protein 6 homolog (HsSAS-6). | |||||
|
SAS6_MOUSE
|
||||||
| θ value | 3.0926 (rank : 36) | NC score | 0.068755 (rank : 25) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q80UK7, Q9CYT4 | Gene names | Sass6, Sas6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spindle assembly abnormal protein 6 homolog. | |||||
|
CCD41_HUMAN
|
||||||
| θ value | 4.03905 (rank : 37) | NC score | 0.064699 (rank : 32) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1139 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9Y592 | Gene names | CCDC41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41 (NY-REN-58 antigen). | |||||
|
CKAP4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 38) | NC score | 0.077165 (rank : 13) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q07065, Q504S5, Q53ES6 | Gene names | CKAP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-associated protein 4 (63 kDa membrane protein) (p63). | |||||
|
DAAM1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 39) | NC score | 0.031251 (rank : 179) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BPM0, Q80Y68, Q9CQQ2 | Gene names | Daam1 | |||
|
Domain Architecture |

|
|||||
| Description | Disheveled-associated activator of morphogenesis 1. | |||||
|
KIF5A_MOUSE
|
||||||
| θ value | 5.27518 (rank : 40) | NC score | 0.042823 (rank : 173) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 859 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P33175, Q5DTP1, Q6PDY7, Q9Z2F9 | Gene names | Kif5a, Kiaa4086, Kif5, Nkhc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin heavy chain isoform 5A (Neuronal kinesin heavy chain) (NKHC) (Kinesin heavy chain neuron-specific 1). | |||||
|
VDP_MOUSE
|
||||||
| θ value | 5.27518 (rank : 41) | NC score | 0.047175 (rank : 172) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 469 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Z1Z0, Q3T9L9, Q3TH58, Q3U1C7, Q3UMW6, Q91WE7, Q99JZ5 | Gene names | Vdp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | General vesicular transport factor p115 (Transcytosis-associated protein) (TAP) (Vesicle docking protein). | |||||
|
CROCC_HUMAN
|
||||||
| θ value | 6.88961 (rank : 42) | NC score | 0.055931 (rank : 63) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1409 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q5TZA2, Q2VHY3, Q66GT7, Q7Z2L4, Q7Z5D7 | Gene names | CROCC, KIAA0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
CROCC_MOUSE
|
||||||
| θ value | 6.88961 (rank : 43) | NC score | 0.061981 (rank : 40) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1460 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8CJ40, Q7TQL2, Q80U01, Q8R0B9 | Gene names | Crocc, Kiaa0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
GRAP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 44) | NC score | 0.060507 (rank : 42) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1234 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8VD04, O35693, Q3T9C3, Q69ZP9 | Gene names | Gripap1, DXImx47e, Kiaa1167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP1-associated protein 1 (GRASP-1) (HCMV-interacting protein). | |||||
|
IFT74_MOUSE
|
||||||
| θ value | 6.88961 (rank : 45) | NC score | 0.064098 (rank : 34) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 939 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8BKE9, Q80ZI3, Q9CSY1, Q9CUS0, Q9D9T5 | Gene names | Ift74, Ccdc2, Cmg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 74 homolog (Coiled-coil domain-containing protein 2) (Capillary morphogenesis protein 1) (CMG-1). | |||||
|
SMC4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 46) | NC score | 0.054394 (rank : 98) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9NTJ3, O95752, Q8NDL4, Q9UNT9 | Gene names | SMC4L1, CAPC, SMC4 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 4-like 1 protein (Chromosome- associated polypeptide C) (hCAP-C) (XCAP-C homolog). | |||||
|
TARA_MOUSE
|
||||||
| θ value | 6.88961 (rank : 47) | NC score | 0.029694 (rank : 180) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
COPG_HUMAN
|
||||||
| θ value | 8.99809 (rank : 48) | NC score | 0.011152 (rank : 183) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y678 | Gene names | COPG | |||
|
Domain Architecture |

|
|||||
| Description | Coatomer subunit gamma (Gamma-coat protein) (Gamma-COP). | |||||
|
MYLK_MOUSE
|
||||||
| θ value | 8.99809 (rank : 49) | NC score | 0.002552 (rank : 184) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1489 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6PDN3, Q3TSJ7, Q80UX0, Q80YN7, Q80YN8, Q8K026, Q924D2, Q9ERD3 | Gene names | Mylk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin light chain kinase, smooth muscle (EC 2.7.11.18) (MLCK) (Telokin) (Kinase-related protein) (KRP). | |||||
|
MYO5C_HUMAN
|
||||||
| θ value | 8.99809 (rank : 50) | NC score | 0.054091 (rank : 108) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1206 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9NQX4 | Gene names | MYO5C | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5C (Myosin Vc). | |||||
|
3BP5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 51) | NC score | 0.057122 (rank : 55) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O60239 | Gene names | SH3BP5, SAB | |||
|
Domain Architecture |
|
|||||
| Description | SH3 domain-binding protein 5 (SH3 domain-binding protein that preferentially associates with BTK). | |||||
|
3BP5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 52) | NC score | 0.054512 (rank : 95) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Z131, Q8C903, Q8VCZ0 | Gene names | Sh3bp5, Sab | |||
|
Domain Architecture |
|
|||||
| Description | SH3 domain-binding protein 5 (SH3 domain-binding protein that preferentially associates with BTK). | |||||
|
BICD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 53) | NC score | 0.071672 (rank : 20) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 844 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q96G01, O43892, O43893 | Gene names | BICD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 1. | |||||
|
BICD1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 54) | NC score | 0.071890 (rank : 19) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 837 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8BR07, O55206, Q8BQ18, Q8BRR8, Q8C4D3, Q8CAK6, Q8R2J4, Q8R2J5, Q8R2J6, Q91YP7 | Gene names | Bicd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 1. | |||||
|
BRE1B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 55) | NC score | 0.051745 (rank : 138) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O75150, Q6AHZ6, Q6N005, Q7L3T6, Q8N615, Q96T18, Q9BSV9, Q9HC82 | Gene names | RNF40, BRE1B, KIAA0661 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1B (EC 6.3.2.-) (BRE1-B) (RING finger protein 40) (95 kDa retinoblastoma-associated protein) (RBP95). | |||||
|
BRE1B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 56) | NC score | 0.052219 (rank : 131) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 970 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q3U319, Q6ZQ75, Q8BJA1, Q8BY03, Q8CHX4 | Gene names | Rnf40, Bre1b, Kiaa0661 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1B (EC 6.3.2.-) (BRE1-B) (RING finger protein 40). | |||||
|
CASP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 57) | NC score | 0.054092 (rank : 107) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q13948, Q53GU9, Q8TBS3 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
CASP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 58) | NC score | 0.054389 (rank : 100) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1152 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P70403, Q7TQJ4, Q91WN6 | Gene names | Cutl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
CCD13_HUMAN
|
||||||
| θ value | θ > 10 (rank : 59) | NC score | 0.065207 (rank : 31) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 721 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8IYE1 | Gene names | CCDC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 13. | |||||
|
CCD39_HUMAN
|
||||||
| θ value | θ > 10 (rank : 60) | NC score | 0.062518 (rank : 38) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 868 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9UFE4 | Gene names | CCDC39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 39. | |||||
|
CCD39_MOUSE
|
||||||
| θ value | θ > 10 (rank : 61) | NC score | 0.059823 (rank : 43) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1026 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9D5Y1, Q8CDG6 | Gene names | Ccdc39, D3Ertd789e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 39. | |||||
|
CCD41_MOUSE
|
||||||
| θ value | θ > 10 (rank : 62) | NC score | 0.056661 (rank : 60) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9D5R3, Q3U7X7, Q3UX57, Q80VF0 | Gene names | Ccdc41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41. | |||||
|
CCD42_HUMAN
|
||||||
| θ value | θ > 10 (rank : 63) | NC score | 0.054043 (rank : 109) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q96M95, Q8N6Q0 | Gene names | CCDC42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 42. | |||||
|
CCD46_HUMAN
|
||||||
| θ value | θ > 10 (rank : 64) | NC score | 0.054547 (rank : 93) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1430 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8N8E3, Q6PIB5, Q8NCR4, Q8NFR4 | Gene names | CCDC46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 46. | |||||
|
CCD46_MOUSE
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.054109 (rank : 106) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q5PR68, Q80ZN6, Q99JS4, Q9D9T1 | Gene names | Ccdc46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled coil domain-containing protein 46. | |||||
|
CCD62_HUMAN
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.052408 (rank : 129) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 631 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q6P9F0, Q6ZVF2, Q86VJ0, Q9BYZ5 | Gene names | CCDC62 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 60 (Aaa-protein) (Protein TSP- NY). | |||||
|
CCHCR_HUMAN
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.050906 (rank : 157) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 717 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8TD31, Q9NRK8, Q9NWY9, Q9NXJ4, Q9NXK3, Q9Y6W1, Q9Y6W2 | Gene names | CCHCR1, C6orf18, HCR | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil alpha-helical rod protein 1 (Alpha helical coiled-coil rod protein) (Putative gene 8 protein) (Pg8). | |||||
|
CDR2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.051461 (rank : 149) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 482 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P97817 | Gene names | Cdr2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cerebellar degeneration-related protein 2. | |||||
|
CE152_HUMAN
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.057224 (rank : 53) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 976 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O94986, Q6NTA0 | Gene names | CEP152, KIAA0912 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 152 kDa (Cep152 protein). | |||||
|
CE290_HUMAN
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.057320 (rank : 51) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
CENPE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.050656 (rank : 161) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
CENPF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.054923 (rank : 84) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
CENPH_HUMAN
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.050235 (rank : 166) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9H3R5 | Gene names | CENPH, ICEN35 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein H (CENP-H) (Interphase centromere complex protein 35). | |||||
|
CEP63_HUMAN
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.056948 (rank : 57) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1005 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q96MT8, Q96CR0, Q9H8F5, Q9H8N0 | Gene names | CEP63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 63 kDa (Cep63 protein). | |||||
|
CI039_HUMAN
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.054727 (rank : 87) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1171 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9NXG0, Q5VYJ0, Q9HAJ5 | Gene names | C9orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf39. | |||||
|
CI093_HUMAN
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.058152 (rank : 46) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q6TFL3, Q5SU58, Q6P1W1, Q6ZR13, Q8N1Z4, Q8N8L3, Q8TBR2 | Gene names | C9orf93 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf93. | |||||
|
CING_MOUSE
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.060793 (rank : 41) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1432 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P59242 | Gene names | Cgn | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
CJ118_MOUSE
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.051529 (rank : 146) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1132 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8C9S4, Q6P3D6, Q8CJC0 | Gene names | Otg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf118 homolog (Oocyte-testis gene 1 protein). | |||||
|
CK5P2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.051497 (rank : 147) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q96SN8, Q7Z3L4, Q7Z3U1, Q7Z7I6, Q9BSW0, Q9H6J6, Q9HCD9, Q9NV90, Q9UIW9 | Gene names | CDK5RAP2, CEP215, KIAA1633 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48) (Centrosome-associated protein 215). | |||||
|
CK5P2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.054632 (rank : 92) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1151 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8K389, Q6PCN1, Q6ZPL0 | Gene names | Cdk5rap2, Kiaa1633 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48). | |||||
|
CN145_HUMAN
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.055541 (rank : 71) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1196 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q6ZU80, Q96ML4 | Gene names | C14orf145 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf145. | |||||
|
CP135_HUMAN
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.055885 (rank : 66) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1290 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q66GS9, O75130, Q9H8H7 | Gene names | CEP135, CEP4, KIAA0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
CP135_MOUSE
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.056814 (rank : 58) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1278 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q6P5D4, Q6A030 | Gene names | Cep135, Cep4, Kiaa0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
CP250_HUMAN
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.052580 (rank : 128) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
CP250_MOUSE
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.052824 (rank : 124) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
CR034_MOUSE
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.055020 (rank : 83) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8CDV0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C18orf34 homolog. | |||||
|
CRCM_HUMAN
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.051276 (rank : 152) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 520 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P23508 | Gene names | MCC | |||
|
Domain Architecture |
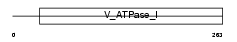
|
|||||
| Description | Colorectal mutant cancer protein (Protein MCC). | |||||
|
CT2NL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.050748 (rank : 160) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9P2B4, Q96B40 | Gene names | CTTNBP2NL, KIAA1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
CTGE5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.050475 (rank : 164) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 928 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8R311, Q8CIE3 | Gene names | Ctage5, Mea6, Mgea6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cutaneous T-cell lymphoma-associated antigen 5 homolog (cTAGE-5 protein) (Meningioma-expressed antigen 6). | |||||
|
CYLN2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.052770 (rank : 126) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1004 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UDT6, O14527, O43611 | Gene names | CYLN2, KIAA0291, WBSCR4, WSCR4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic linker protein 2 (Cytoplasmic linker protein 115) (CLIP- 115) (Williams-Beuren syndrome chromosome region 4 protein). | |||||
|
CYLN2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.053820 (rank : 112) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1002 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9Z0H8, Q7TSI9, Q8CHU1, Q9EP81 | Gene names | Cyln2, Kiaa0291 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic linker protein 2 (Cytoplasmic linker protein 115) (CLIP- 115). | |||||
|
CYTSA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.051377 (rank : 151) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1138 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q69YQ0, O15081 | Gene names | CYTSA, KIAA0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A (NY-REN-22 antigen). | |||||
|
CYTSA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.051258 (rank : 153) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q2KN98, Q8CHF9 | Gene names | Cytsa, Kiaa0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A. | |||||
|
EEA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.055888 (rank : 65) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
ERC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.051545 (rank : 145) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1156 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O15083, Q86TK4 | Gene names | ERC2, KIAA0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2. | |||||
|
ERC2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.052243 (rank : 130) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1166 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q6PH08, Q80U20, Q8CCP1 | Gene names | Erc2, Cast1, D14Ertd171e, Kiaa0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2 (CAZ-associated structural protein 1) (CAST1). | |||||
|
FLIP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.050558 (rank : 163) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q7Z7B0, Q5VUL6, Q86TC3, Q8N8B9, Q96SK6, Q9NVI8, Q9ULE5 | Gene names | FILIP1, KIAA1275 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
GAS8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.055294 (rank : 74) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 819 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O95995 | Gene names | GAS8, GAS11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Growth-arrest-specific protein 8 (Growth arrest-specific 11). | |||||
|
GCC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.055733 (rank : 69) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
GCC2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.053464 (rank : 119) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
GOGA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.052777 (rank : 125) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1222 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q92805, Q8IYZ9 | Gene names | GOLGA1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
GOGA1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.055801 (rank : 68) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 994 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9CW79, Q80YB0 | Gene names | Golga1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
GOGA2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.050114 (rank : 171) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q921M4 | Gene names | Golga2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgin subfamily A member 2 (Cis-Golgi matrix protein GM130). | |||||
|
GOGA3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.053565 (rank : 116) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q08378, O43241, Q6P9C7, Q86XW3, Q8TDA9, Q8WZA3 | Gene names | GOLGA3 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Golgi complex-associated protein of 170 kDa) (GCP170). | |||||
|
GOGA3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.054256 (rank : 102) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1437 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P55937, Q80VF5, Q8CCK4, Q9QYT2, Q9QYT3 | Gene names | Golga3, Mea2 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Male-enhanced antigen 2) (MEA-2). | |||||
|
GOGA4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.050178 (rank : 168) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
GOGA5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.053523 (rank : 117) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1168 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8TBA6, O95287, Q03962, Q9UQQ7 | Gene names | GOLGA5, RETII, RFG5 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (RET-fused gene 5 protein) (Ret-II protein). | |||||
|
GOGA5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.051474 (rank : 148) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1434 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9QYE6, O88317, Q3TGE7, Q3U6S5, Q3UUF9 | Gene names | Golga5, Retii | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (Sumiko protein) (Ret-II protein). | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.053659 (rank : 114) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
HIP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.059553 (rank : 45) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1016 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O00291, O00328, Q8TDL4 | Gene names | HIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1 (HIP-I). | |||||
|
HMMR_MOUSE
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.057745 (rank : 49) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q00547 | Gene names | Hmmr, Ihabp, Rhamm | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility). | |||||
|
HOOK3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.050123 (rank : 170) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1070 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q86VS8, Q8NBH0, Q9BY13 | Gene names | HOOK3 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 3 (hHK3). | |||||
|
HOOK3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.050641 (rank : 162) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8BUK6, Q8BV48, Q8BW57, Q8BY41 | Gene names | Hook3 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 3. | |||||
|
IFT74_HUMAN
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.062429 (rank : 39) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q96LB3, Q6PGQ8, Q9H643, Q9H8G7 | Gene names | IFT74, CCDC2, CMG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 74 homolog (Coiled-coil domain-containing protein 2) (Capillary morphogenesis protein 1) (CMG-1). | |||||
|
IFT81_HUMAN
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.054542 (rank : 94) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8WYA0, Q8NB51, Q9BSV2, Q9UNY8 | Gene names | IFT81, CDV1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 81 (Carnitine deficiency-associated protein expressed in ventricle 1) (CDV-1 protein). | |||||
|
IFT81_MOUSE
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.055950 (rank : 62) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O35594, Q8R4W2, Q9CSA1, Q9EQM1 | Gene names | Ift81, Cdv-1, Cdv1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 81 (Carnitine deficiency-associated protein expressed in ventricle 1) (CDV-1 protein). | |||||
|
KMHN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.054636 (rank : 90) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 950 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8TBZ0, Q86YI9, Q8N7W0 | Gene names | KMHN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cancer/testis antigen KM-HN-1. | |||||
|
KMHN1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.057024 (rank : 56) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q3V125, Q3TTQ8 | Gene names | Gm172 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cancer/testis antigen KM-HN-1 homolog. | |||||
|
KNTC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.055713 (rank : 70) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 673 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O14777, Q6PJX2 | Gene names | KNTC2, HEC, HEC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Hec1 (HsHec1) (Kinetochore-associated protein 2) (Highly expressed in cancer protein) (Retinoblastoma-associated protein HEC). | |||||
|
KTN1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.056126 (rank : 61) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1324 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q61595, Q8BG49, Q8BHF4, Q8BHM8, Q8C9Y5, Q8CG51, Q8CG52, Q8CG53, Q8CG54, Q8CG55, Q8CG56, Q8CG57, Q8CG58, Q8CG59, Q8CG60, Q8CG61, Q8CG62, Q8CG63 | Gene names | Ktn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin. | |||||
|
LRC45_HUMAN
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.050190 (rank : 167) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 933 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q96CN5 | Gene names | LRRC45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 45. | |||||
|
LRC45_MOUSE
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.053681 (rank : 113) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8CIM1, Q3U5Z2 | Gene names | Lrrc45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 45. | |||||
|
LRRF2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.053473 (rank : 118) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 791 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9Y608, Q68CV3, Q9NXH5 | Gene names | LRRFIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 2 (LRR FLII- interacting protein 2). | |||||
|
MD1L1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.053858 (rank : 110) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1001 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9WTX8, Q9WTX9 | Gene names | Mad1l1, Mad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1). | |||||
|
MPP9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.054487 (rank : 97) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q99550, Q9H976 | Gene names | MPHOSPH9, MPP9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | M-phase phosphoprotein 9. | |||||
|
MY18A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.054703 (rank : 88) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
MY18A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.055529 (rank : 72) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
MY18B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.050806 (rank : 158) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1299 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8IUG5, Q8NDI8, Q8TE65, Q8WWS0, Q96KH2, Q96KR8, Q96KR9 | Gene names | MYO18B | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18B (Myosin XVIIIb). | |||||
|
MYH11_MOUSE
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.059771 (rank : 44) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
MYH13_HUMAN
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.054234 (rank : 103) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1504 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UKX3, O95252 | Gene names | MYH13 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-13 (Myosin heavy chain, skeletal muscle, extraocular) (MyHC- eo). | |||||
|
MYH14_HUMAN
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.057189 (rank : 54) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 995 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q7Z406, Q5CZ75, Q6XYE4, Q76B62, Q8WV23, Q96I22, Q9BT27, Q9BW35, Q9H882 | Gene names | MYH14, KIAA2034 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
MYH14_MOUSE
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.058124 (rank : 47) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1225 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q6URW6, Q80V64, Q80ZE6 | Gene names | Myh14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
MYH1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.054690 (rank : 89) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1478 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P12882, Q9Y622 | Gene names | MYH1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1) (Myosin heavy chain IIx/d) (MyHC-IIx/d). | |||||
|
MYH1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.055076 (rank : 81) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1461 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q5SX40 | Gene names | Myh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1). | |||||
|
MYH2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.055223 (rank : 78) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1491 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UKX2, Q14322, Q16229 | Gene names | MYH2, MYHSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, skeletal muscle, adult 2 (Myosin heavy chain IIa) (MyHC-IIa). | |||||
|
MYH4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.053438 (rank : 122) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1377 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9Y623 | Gene names | MYH4 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal) (Myosin heavy chain IIb) (MyHC-IIb). | |||||
|
MYH4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.055239 (rank : 76) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
MYH6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.054222 (rank : 105) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1426 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P13533, Q13943, Q14906, Q14907 | Gene names | MYH6, MYHCA | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
MYH6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.054364 (rank : 101) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1501 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q02566, Q64258, Q64738 | Gene names | Myh6, Myhca | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
MYH7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.055499 (rank : 73) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P12883, Q14836, Q14837, Q14904, Q16579, Q92679, Q9H1D5, Q9UMM8 | Gene names | MYH7, MYHCB | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle beta isoform (MyHC-beta). | |||||
|
MYH8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.055053 (rank : 82) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1610 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P13535, Q14910 | Gene names | MYH8 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
MYH8_MOUSE
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.054770 (rank : 86) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1600 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P13542, Q5SX36 | Gene names | Myh8, Myhsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
MYO5A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.051956 (rank : 136) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1092 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9Y4I1, O60653, Q07902, Q16249, Q9UE30, Q9UE31 | Gene names | MYO5A, MYH12 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5A (Myosin Va) (Dilute myosin heavy chain, non-muscle) (Myosin heavy chain 12) (Myoxin). | |||||
|
MYO5A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.051024 (rank : 156) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1117 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q99104 | Gene names | Myo5a, Dilute | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5A (Myosin Va) (Dilute myosin heavy chain, non-muscle). | |||||
|
NDE1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.054391 (rank : 99) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 823 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9NXR1, Q49AQ2 | Gene names | NDE1, NUDE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear distribution protein nudE homolog 1 (NudE). | |||||
|
NIN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.050155 (rank : 169) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1416 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8N4C6, Q6P0P6, Q9BWU6, Q9C012, Q9C013, Q9C014, Q9H5I6, Q9HAT7, Q9HBY5, Q9HCK7, Q9UH61 | Gene names | NIN, KIAA1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein (hNinein) (Glycogen synthase kinase 3 beta-interacting protein) (GSK3B-interacting protein). | |||||
|
NIN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.051029 (rank : 155) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q61043, Q6ZPM7 | Gene names | Nin, Kiaa1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein. | |||||
|
NUF2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.055258 (rank : 75) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 661 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9BZD4, Q8WU69, Q96HJ4, Q96Q78 | Gene names | CDCA1, NUF2, NUF2R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Nuf2 (hNuf2) (hNuf2R) (hsNuf2) (Cell division cycle-associated protein 1). | |||||
|
OPTN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.053574 (rank : 115) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 827 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q96CV9, Q5T672, Q5T673, Q5T674, Q5T675, Q7LDL9, Q8N562, Q9UET9, Q9UEV4, Q9Y218 | Gene names | OPTN, FIP2, GLC1E, NRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin (Optic neuropathy-inducing protein) (E3-14.7K-interacting protein) (FIP-2) (Huntingtin-interacting protein HYPL) (NEMO-related protein) (Transcription factor IIIA-interacting protein) (TFIIIA- IntP). | |||||
|
OPTN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.054918 (rank : 85) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1120 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8K3K8 | Gene names | Optn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin. | |||||
|
PCNT_HUMAN
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.051682 (rank : 141) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
PIBF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.057575 (rank : 50) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8WXW3, O95664, Q6U9V2, Q6UG50, Q86V07, Q96SF4 | Gene names | PIBF1, C13orf24, PIBF | |||
|
Domain Architecture |

|
|||||
| Description | Progesterone-induced-blocking factor 1. | |||||
|
POF1B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.051816 (rank : 137) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8K4L4, Q8C3A2 | Gene names | Pof1b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein POF1B. | |||||
|
RABE1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.053458 (rank : 120) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q15276, O95369, Q8IVX3 | Gene names | RABEP1, RABPT5, RABPT5A | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha) (Rabaptin-4) (NY-REN-17 antigen). | |||||
|
RABE1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.055180 (rank : 79) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1105 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O35551, Q99L08, Q9CRP3, Q9EQF9, Q9JI94 | Gene names | Rabep1, Rab5ep, Rabpt5, Rabpt5a | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha). | |||||
|
RAD50_HUMAN
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.058029 (rank : 48) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q92878, O43254, Q6GMT7, Q6P5X3, Q9UP86 | Gene names | RAD50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (hRAD50). | |||||
|
RAD50_MOUSE
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.055930 (rank : 64) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1366 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P70388, Q6PEB0, Q8C2T7, Q9CU59 | Gene names | Rad50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (mRad50). | |||||
|
RB6I2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.052189 (rank : 132) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1584 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8IUD2, Q6NVK2, Q8IUD3, Q8IUD4, Q8IUD5, Q8NAS1, Q9NXN5, Q9UIK7, Q9UPS1 | Gene names | ERC1, ELKS, KIAA1081, RAB6IP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1). | |||||
|
RB6I2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.052119 (rank : 135) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q99MI1, Q80TK7, Q8BPL1, Q8C7Y1, Q99MI2 | Gene names | Erc1, Cast2, Kiaa1081, Rab6ip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1) (ERC1) (CAZ-associated structural protein 2) (CAST2). | |||||
|
RBP24_HUMAN
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.055840 (rank : 67) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
REST_HUMAN
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.053072 (rank : 123) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1605 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P30622 | Gene names | RSN, CYLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Restin (Cytoplasmic linker protein 170 alpha-2) (CLIP-170) (Reed- Sternberg intermediate filament-associated protein) (Cytoplasmic linker protein 1). | |||||
|
REST_MOUSE
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.053825 (rank : 111) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1506 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q922J3, Q571L7 | Gene names | Rsn, Kiaa4046 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin. | |||||
|
SDCG8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.051732 (rank : 139) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1002 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q86SQ7, O60527, Q3ZCR6, Q8N5F2, Q9P0F1 | Gene names | SDCCAG8, CCCAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serologically defined colon cancer antigen 8 (Centrosomal colon cancer autoantigen protein) (hCCCAP) (Antigen NY-CO-8). | |||||
|
SDCG8_MOUSE
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.051116 (rank : 154) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q80UF4 | Gene names | Sdccag8, Cccap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serologically defined colon cancer antigen 8 (Centrosomal colon cancer autoantigen protein) (mCCCAP). | |||||
|
SM1L2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.052652 (rank : 127) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1019 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8NDV3, Q5TIC3, Q9Y3G5 | Gene names | SMC1L2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 2 protein (SMC1beta protein). | |||||
|
SM1L2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.051607 (rank : 142) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 918 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q920F6 | Gene names | Smc1l2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 1-like 2 protein (SMC1beta protein). | |||||
|
SMC1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.050790 (rank : 159) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1346 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q14683, O14995, Q16351 | Gene names | SMC1L1, DXS423E, KIAA0178, SB1.8, SMC1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Sb1.8). | |||||
|
SMC1A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.051385 (rank : 150) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1384 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9CU62, Q3V480, Q9CUX9, Q9D959, Q9WTU1 | Gene names | Smc1l1, Sb1.8, Smc1, Smcb | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Chromosome segregation protein SmcB) (Sb1.8). | |||||
|
SMC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.055101 (rank : 80) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O95347, Q9P1P2 | Gene names | SMC2L1, CAPE, SMC2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (hCAP-E) (XCAP-E homolog). | |||||
|
SYCP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.051705 (rank : 140) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q15431, O14963 | Gene names | SYCP1, SCP1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
SYCP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.052184 (rank : 133) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1456 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q62209, O09205, P70192, Q62329 | Gene names | Sycp1, Scp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
T3JAM_HUMAN
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.054635 (rank : 91) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 533 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9Y228, Q2YDB5, Q4VY06, Q7Z706 | Gene names | TRAF3IP3, T3JAM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRAF3-interacting JNK-activating modulator (TRAF3-interacting protein 3). | |||||
|
TAXB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.053442 (rank : 121) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 829 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q86VP1, O60398, O95770, Q13311, Q9BQG5, Q9UI88 | Gene names | TAX1BP1, T6BP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tax1-binding protein 1 (TRAF6-binding protein). | |||||
|
TAXB1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.055229 (rank : 77) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q3UKC1, Q91YT6, Q9CVF0, Q9DC45 | Gene names | Tax1bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tax1-binding protein 1 homolog. | |||||
|
TEX9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.054224 (rank : 104) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 541 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8N6V9 | Gene names | TEX9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed sequence 9 protein. | |||||
|
TEX9_MOUSE
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.056799 (rank : 59) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9D845, O54764 | Gene names | Tex9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed sequence 9 protein (Testis-specifically expressed protein 1) (Tsec-1). | |||||
|
TPM2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.080734 (rank : 11) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 677 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P07951, P06468, Q13894, Q53FM4, Q5TCU4, Q5TCU7, Q9UH67 | Gene names | TPM2, TMSB | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin beta chain (Tropomyosin 2) (Beta-tropomyosin). | |||||
|
TPM2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.080280 (rank : 12) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P58774, P02560, P46901 | Gene names | Tpm2, Tpm-2 | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin beta chain (Tropomyosin 2) (Beta-tropomyosin). | |||||
|
TPM4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.083058 (rank : 10) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 525 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P67936, P07226, Q15659, Q9BU85, Q9H8Q3 | Gene names | TPM4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tropomyosin alpha-4 chain (Tropomyosin-4) (TM30p1). | |||||
|
TPM4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.077057 (rank : 14) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 703 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q6IRU2 | Gene names | Tpm4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tropomyosin alpha-4 chain (Tropomyosin-4). | |||||
|
TRAIP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 181) | NC score | 0.050339 (rank : 165) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9BWF2, O00467 | Gene names | TRAIP, TRIP | |||
|
Domain Architecture |

|
|||||
| Description | TRAF-interacting protein. | |||||
|
TRAIP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 182) | NC score | 0.051555 (rank : 144) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 745 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8VIG6, O08854, Q922M8, Q9CPP4 | Gene names | Traip, Trip | |||
|
Domain Architecture |

|
|||||
| Description | TRAF-interacting protein. | |||||
|
TUFT1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 183) | NC score | 0.054492 (rank : 96) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 686 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9NNX1, Q5T384, Q5T385, Q9BU28, Q9H5L1 | Gene names | TUFT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tuftelin. | |||||
|
YO11_MOUSE
|
||||||
| θ value | θ > 10 (rank : 184) | NC score | 0.052170 (rank : 134) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P11260 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Hypothetical protein ORF-1137. | |||||
|
RIB1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P04843 | Gene names | RPN1 | |||
|
Domain Architecture |

|
|||||
| Description | Dolichyl-diphosphooligosaccharide--protein glycosyltransferase 67 kDa subunit precursor (EC 2.4.1.119) (Ribophorin I) (RPN-I). | |||||
|
RIB1_MOUSE
|
||||||
| NC score | 0.995425 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q91YQ5, Q3U985 | Gene names | Rpn1 | |||
|
Domain Architecture |

|
|||||
| Description | Dolichyl-diphosphooligosaccharide--protein glycosyltransferase 67 kDa subunit precursor (EC 2.4.1.119) (Ribophorin I) (RPN-I). | |||||
|
TPM1_MOUSE
|
||||||
| NC score | 0.111817 (rank : 3) | θ value | 0.0431538 (rank : 6) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 729 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P58771, P02558, P19354, P46902, P99034 | Gene names | Tpm1, Tpm-1, Tpma | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin 1 alpha chain (Alpha-tropomyosin). | |||||
|
TPM1_HUMAN
|
||||||
| NC score | 0.111280 (rank : 4) | θ value | 0.0431538 (rank : 5) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 716 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P09493, P09494, P10469, Q86W64, Q96IK2, Q9UCY9 | Gene names | TPM1, TMSA | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin 1 alpha chain (Alpha-tropomyosin). | |||||
|
TPM3_MOUSE
|
||||||
| NC score | 0.106092 (rank : 5) | θ value | 0.0563607 (rank : 8) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P21107, Q09021, Q60606, Q80SW6, Q9EPW3 | Gene names | Tpm3, Tpm-5, Tpm5 | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin alpha-3 chain (Tropomyosin-3) (Tropomyosin gamma). | |||||
|
TPM3_HUMAN
|
||||||
| NC score | 0.106034 (rank : 6) | θ value | 0.0563607 (rank : 7) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 689 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P06753, P12324, Q969Q2, Q9NQH8 | Gene names | TPM3 | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin alpha-3 chain (Tropomyosin-3) (Tropomyosin gamma) (hTM5). | |||||
|
CKAP4_MOUSE
|
||||||
| NC score | 0.099080 (rank : 7) | θ value | 0.365318 (rank : 14) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8BMK4, Q8BTK8, Q8R3F2 | Gene names | Ckap4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-associated protein 4 (63 kDa membrane protein) (p63). | |||||
|
BICD2_HUMAN
|
||||||
| NC score | 0.096402 (rank : 8) | θ value | 0.0736092 (rank : 9) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8TD16, O75181, Q5TBQ2, Q5TBQ3, Q96LH2, Q9BT84, Q9H561 | Gene names | BICD2, KIAA0699 | |||
|
Domain Architecture |

|
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 2. | |||||
|
BICD2_MOUSE
|
||||||
| NC score | 0.091697 (rank : 9) | θ value | 0.279714 (rank : 11) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 864 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q921C5, Q80TU1, Q8BTE3, Q9DCL3 | Gene names | Bicd2, Kiaa0699 | |||
|
Domain Architecture |

|
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 2. | |||||
|
TPM4_HUMAN
|
||||||
| NC score | 0.083058 (rank : 10) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 525 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P67936, P07226, Q15659, Q9BU85, Q9H8Q3 | Gene names | TPM4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tropomyosin alpha-4 chain (Tropomyosin-4) (TM30p1). | |||||
|
TPM2_HUMAN
|
||||||
| NC score | 0.080734 (rank : 11) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 677 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P07951, P06468, Q13894, Q53FM4, Q5TCU4, Q5TCU7, Q9UH67 | Gene names | TPM2, TMSB | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin beta chain (Tropomyosin 2) (Beta-tropomyosin). | |||||
|
TPM2_MOUSE
|
||||||
| NC score | 0.080280 (rank : 12) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P58774, P02560, P46901 | Gene names | Tpm2, Tpm-2 | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin beta chain (Tropomyosin 2) (Beta-tropomyosin). | |||||
|
CKAP4_HUMAN
|
||||||
| NC score | 0.077165 (rank : 13) | θ value | 5.27518 (rank : 38) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q07065, Q504S5, Q53ES6 | Gene names | CKAP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-associated protein 4 (63 kDa membrane protein) (p63). | |||||
|
TPM4_MOUSE
|
||||||
| NC score | 0.077057 (rank : 14) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 703 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q6IRU2 | Gene names | Tpm4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tropomyosin alpha-4 chain (Tropomyosin-4). | |||||
|
CF060_HUMAN
|
||||||
| NC score | 0.076737 (rank : 15) | θ value | 0.813845 (rank : 19) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1270 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8NB25, Q96GY8, Q9H851 | Gene names | C6orf60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf60. | |||||
|
MYH10_HUMAN
|
||||||
| NC score | 0.074670 (rank : 16) | θ value | 0.0193708 (rank : 3) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P35580, Q16087 | Gene names | MYH10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
MYH10_MOUSE
|
||||||
| NC score | 0.074564 (rank : 17) | θ value | 0.0252991 (rank : 4) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1612 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q61879, Q5SV63 | Gene names | Myh10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
EEA1_MOUSE
|
||||||
| NC score | 0.073009 (rank : 18) | θ value | 0.21417 (rank : 10) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1637 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8BL66, Q6DIC2 | Gene names | Eea1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early endosome antigen 1. | |||||
|
BICD1_MOUSE
|
||||||
| NC score | 0.071890 (rank : 19) | θ value | θ > 10 (rank : 54) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 837 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8BR07, O55206, Q8BQ18, Q8BRR8, Q8C4D3, Q8CAK6, Q8R2J4, Q8R2J5, Q8R2J6, Q91YP7 | Gene names | Bicd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 1. | |||||
|
BICD1_HUMAN
|
||||||
| NC score | 0.071672 (rank : 20) | θ value | θ > 10 (rank : 53) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 844 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q96G01, O43892, O43893 | Gene names | BICD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 1. | |||||
|
HIP1R_MOUSE
|
||||||
| NC score | 0.070490 (rank : 21) | θ value | 2.36792 (rank : 28) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9JKY5 | Gene names | Hip1r | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1-related protein (Hip1-related). | |||||
|
HIP1R_HUMAN
|
||||||
| NC score | 0.070452 (rank : 22) | θ value | 1.81305 (rank : 23) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 809 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O75146, Q9UED9 | Gene names | HIP1R, HIP12, KIAA0655 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1-related protein (Hip1-related) (Hip 12). | |||||
|
GRAP1_HUMAN
|
||||||
| NC score | 0.070115 (rank : 23) | θ value | 0.813845 (rank : 20) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q4V328, Q4V327, Q4V330, Q5HYG1, Q6N046, Q96DH8, Q9NQ43, Q9ULQ3 | Gene names | GRIPAP1, KIAA1167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP1-associated protein 1 (GRASP-1). | |||||
|
MYH9_MOUSE
|
||||||
| NC score | 0.069187 (rank : 24) | θ value | 0.365318 (rank : 15) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
SAS6_MOUSE
|
||||||
| NC score | 0.068755 (rank : 25) | θ value | 3.0926 (rank : 36) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q80UK7, Q9CYT4 | Gene names | Sass6, Sas6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spindle assembly abnormal protein 6 homolog. | |||||
|
SAS6_HUMAN
|
||||||
| NC score | 0.067711 (rank : 26) | θ value | 3.0926 (rank : 35) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 791 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q6UVJ0, Q8N3K0 | Gene names | SASS6, SAS6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spindle assembly abnormal protein 6 homolog (HsSAS-6). | |||||
|
MYH9_HUMAN
|
||||||
| NC score | 0.067114 (rank : 27) | θ value | 1.06291 (rank : 22) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
AKAP9_HUMAN
|
||||||
| NC score | 0.066869 (rank : 28) | θ value | 1.06291 (rank : 21) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
CING_HUMAN
|
||||||
| NC score | 0.066682 (rank : 29) | θ value | 3.0926 (rank : 31) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9P2M7, Q5T386, Q9NR25 | Gene names | CGN, KIAA1319 | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
TRIPB_HUMAN
|
||||||
| NC score | 0.065979 (rank : 30) | θ value | 0.47712 (rank : 16) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
CCD13_HUMAN
|
||||||
| NC score | 0.065207 (rank : 31) | θ value | θ > 10 (rank : 59) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 721 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8IYE1 | Gene names | CCDC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 13. | |||||
|
CCD41_HUMAN
|
||||||
| NC score | 0.064699 (rank : 32) | θ value | 4.03905 (rank : 37) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1139 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9Y592 | Gene names | CCDC41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41 (NY-REN-58 antigen). | |||||
|
MYH11_HUMAN
|
||||||
| NC score | 0.064623 (rank : 33) | θ value | 3.0926 (rank : 34) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
IFT74_MOUSE
|
||||||
| NC score | 0.064098 (rank : 34) | θ value | 6.88961 (rank : 45) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 939 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8BKE9, Q80ZI3, Q9CSY1, Q9CUS0, Q9D9T5 | Gene names | Ift74, Ccdc2, Cmg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 74 homolog (Coiled-coil domain-containing protein 2) (Capillary morphogenesis protein 1) (CMG-1). | |||||
|
MYH3_HUMAN
|
||||||
| NC score | 0.063413 (rank : 35) | θ value | 1.81305 (rank : 26) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1660 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P11055, Q15492 | Gene names | MYH3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, fast skeletal muscle, embryonic (Muscle embryonic myosin heavy chain) (SMHCE). | |||||
|
K0555_HUMAN
|
||||||
| NC score | 0.063171 (rank : 36) | θ value | 3.0926 (rank : 33) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 880 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q96AA8, O60302 | Gene names | KIAA0555 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0555. | |||||
|
TPR_HUMAN
|
||||||
| NC score | 0.063084 (rank : 37) | θ value | 2.36792 (rank : 29) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
CCD39_HUMAN
|
||||||
| NC score | 0.062518 (rank : 38) | θ value | θ > 10 (rank : 60) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 868 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9UFE4 | Gene names | CCDC39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 39. | |||||
|
IFT74_HUMAN
|
||||||
| NC score | 0.062429 (rank : 39) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q96LB3, Q6PGQ8, Q9H643, Q9H8G7 | Gene names | IFT74, CCDC2, CMG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 74 homolog (Coiled-coil domain-containing protein 2) (Capillary morphogenesis protein 1) (CMG-1). | |||||
|
CROCC_MOUSE
|
||||||
| NC score | 0.061981 (rank : 40) | θ value | 6.88961 (rank : 43) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1460 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8CJ40, Q7TQL2, Q80U01, Q8R0B9 | Gene names | Crocc, Kiaa0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
CING_MOUSE
|
||||||
| NC score | 0.060793 (rank : 41) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1432 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P59242 | Gene names | Cgn | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
GRAP1_MOUSE
|
||||||
| NC score | 0.060507 (rank : 42) | θ value | 6.88961 (rank : 44) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1234 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8VD04, O35693, Q3T9C3, Q69ZP9 | Gene names | Gripap1, DXImx47e, Kiaa1167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP1-associated protein 1 (GRASP-1) (HCMV-interacting protein). | |||||
|
CCD39_MOUSE
|
||||||
| NC score | 0.059823 (rank : 43) | θ value | θ > 10 (rank : 61) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1026 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9D5Y1, Q8CDG6 | Gene names | Ccdc39, D3Ertd789e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 39. | |||||
|
MYH11_MOUSE
|
||||||
| NC score | 0.059771 (rank : 44) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
HIP1_HUMAN
|
||||||
| NC score | 0.059553 (rank : 45) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1016 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O00291, O00328, Q8TDL4 | Gene names | HIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1 (HIP-I). | |||||
|
CI093_HUMAN
|
||||||
| NC score | 0.058152 (rank : 46) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q6TFL3, Q5SU58, Q6P1W1, Q6ZR13, Q8N1Z4, Q8N8L3, Q8TBR2 | Gene names | C9orf93 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf93. | |||||
|
MYH14_MOUSE
|
||||||
| NC score | 0.058124 (rank : 47) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1225 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q6URW6, Q80V64, Q80ZE6 | Gene names | Myh14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
RAD50_HUMAN
|
||||||
| NC score | 0.058029 (rank : 48) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q92878, O43254, Q6GMT7, Q6P5X3, Q9UP86 | Gene names | RAD50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (hRAD50). | |||||
|
HMMR_MOUSE
|
||||||
| NC score | 0.057745 (rank : 49) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q00547 | Gene names | Hmmr, Ihabp, Rhamm | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility). | |||||
|
PIBF1_HUMAN
|
||||||
| NC score | 0.057575 (rank : 50) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8WXW3, O95664, Q6U9V2, Q6UG50, Q86V07, Q96SF4 | Gene names | PIBF1, C13orf24, PIBF | |||
|
Domain Architecture |

|
|||||
| Description | Progesterone-induced-blocking factor 1. | |||||
|
CE290_HUMAN
|
||||||
| NC score | 0.057320 (rank : 51) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
SYNE1_HUMAN
|
||||||
| NC score | 0.057272 (rank : 52) | θ value | 1.81305 (rank : 27) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
CE152_HUMAN
|
||||||
| NC score | 0.057224 (rank : 53) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 976 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O94986, Q6NTA0 | Gene names | CEP152, KIAA0912 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 152 kDa (Cep152 protein). | |||||
|
MYH14_HUMAN
|
||||||
| NC score | 0.057189 (rank : 54) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 995 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q7Z406, Q5CZ75, Q6XYE4, Q76B62, Q8WV23, Q96I22, Q9BT27, Q9BW35, Q9H882 | Gene names | MYH14, KIAA2034 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
3BP5_HUMAN
|
||||||
| NC score | 0.057122 (rank : 55) | θ value | θ > 10 (rank : 51) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O60239 | Gene names | SH3BP5, SAB | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 5 (SH3 domain-binding protein that preferentially associates with BTK). | |||||
|
KMHN1_MOUSE
|
||||||
| NC score | 0.057024 (rank : 56) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q3V125, Q3TTQ8 | Gene names | Gm172 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cancer/testis antigen KM-HN-1 homolog. | |||||
|
CEP63_HUMAN
|
||||||
| NC score | 0.056948 (rank : 57) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1005 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q96MT8, Q96CR0, Q9H8F5, Q9H8N0 | Gene names | CEP63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 63 kDa (Cep63 protein). | |||||
|
CP135_MOUSE
|
||||||
| NC score | 0.056814 (rank : 58) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1278 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q6P5D4, Q6A030 | Gene names | Cep135, Cep4, Kiaa0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
TEX9_MOUSE
|
||||||
| NC score | 0.056799 (rank : 59) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9D845, O54764 | Gene names | Tex9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed sequence 9 protein (Testis-specifically expressed protein 1) (Tsec-1). | |||||
|
CCD41_MOUSE
|
||||||
| NC score | 0.056661 (rank : 60) | θ value | θ > 10 (rank : 62) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9D5R3, Q3U7X7, Q3UX57, Q80VF0 | Gene names | Ccdc41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41. | |||||
|
KTN1_MOUSE
|
||||||
| NC score | 0.056126 (rank : 61) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1324 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q61595, Q8BG49, Q8BHF4, Q8BHM8, Q8C9Y5, Q8CG51, Q8CG52, Q8CG53, Q8CG54, Q8CG55, Q8CG56, Q8CG57, Q8CG58, Q8CG59, Q8CG60, Q8CG61, Q8CG62, Q8CG63 | Gene names | Ktn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin. | |||||
|
IFT81_MOUSE
|
||||||
| NC score | 0.055950 (rank : 62) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O35594, Q8R4W2, Q9CSA1, Q9EQM1 | Gene names | Ift81, Cdv-1, Cdv1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 81 (Carnitine deficiency-associated protein expressed in ventricle 1) (CDV-1 protein). | |||||
|
CROCC_HUMAN
|
||||||
| NC score | 0.055931 (rank : 63) | θ value | 6.88961 (rank : 42) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1409 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q5TZA2, Q2VHY3, Q66GT7, Q7Z2L4, Q7Z5D7 | Gene names | CROCC, KIAA0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
RAD50_MOUSE
|
||||||
| NC score | 0.055930 (rank : 64) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1366 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P70388, Q6PEB0, Q8C2T7, Q9CU59 | Gene names | Rad50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (mRad50). | |||||
|
EEA1_HUMAN
|
||||||
| NC score | 0.055888 (rank : 65) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
CP135_HUMAN
|
||||||
| NC score | 0.055885 (rank : 66) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1290 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q66GS9, O75130, Q9H8H7 | Gene names | CEP135, CEP4, KIAA0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
RBP24_HUMAN
|
||||||
| NC score | 0.055840 (rank : 67) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
GOGA1_MOUSE
|
||||||
| NC score | 0.055801 (rank : 68) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 994 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9CW79, Q80YB0 | Gene names | Golga1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
GCC2_HUMAN
|
||||||
| NC score | 0.055733 (rank : 69) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
KNTC2_HUMAN
|
||||||
| NC score | 0.055713 (rank : 70) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 673 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O14777, Q6PJX2 | Gene names | KNTC2, HEC, HEC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Hec1 (HsHec1) (Kinetochore-associated protein 2) (Highly expressed in cancer protein) (Retinoblastoma-associated protein HEC). | |||||
|
CN145_HUMAN
|
||||||
| NC score | 0.055541 (rank : 71) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1196 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q6ZU80, Q96ML4 | Gene names | C14orf145 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf145. | |||||
|
MY18A_MOUSE
|
||||||
| NC score | 0.055529 (rank : 72) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
MYH7_HUMAN
|
||||||
| NC score | 0.055499 (rank : 73) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P12883, Q14836, Q14837, Q14904, Q16579, Q92679, Q9H1D5, Q9UMM8 | Gene names | MYH7, MYHCB | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle beta isoform (MyHC-beta). | |||||
|
GAS8_HUMAN
|
||||||
| NC score | 0.055294 (rank : 74) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 819 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O95995 | Gene names | GAS8, GAS11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Growth-arrest-specific protein 8 (Growth arrest-specific 11). | |||||
|
NUF2_HUMAN
|
||||||
| NC score | 0.055258 (rank : 75) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 661 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9BZD4, Q8WU69, Q96HJ4, Q96Q78 | Gene names | CDCA1, NUF2, NUF2R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Nuf2 (hNuf2) (hNuf2R) (hsNuf2) (Cell division cycle-associated protein 1). | |||||
|
MYH4_MOUSE
|
||||||
| NC score | 0.055239 (rank : 76) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
TAXB1_MOUSE
|
||||||
| NC score | 0.055229 (rank : 77) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q3UKC1, Q91YT6, Q9CVF0, Q9DC45 | Gene names | Tax1bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tax1-binding protein 1 homolog. | |||||
|
MYH2_HUMAN
|
||||||
| NC score | 0.055223 (rank : 78) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1491 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UKX2, Q14322, Q16229 | Gene names | MYH2, MYHSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, skeletal muscle, adult 2 (Myosin heavy chain IIa) (MyHC-IIa). | |||||
|
RABE1_MOUSE
|
||||||
| NC score | 0.055180 (rank : 79) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1105 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O35551, Q99L08, Q9CRP3, Q9EQF9, Q9JI94 | Gene names | Rabep1, Rab5ep, Rabpt5, Rabpt5a | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha). | |||||
|
SMC2_HUMAN
|
||||||
| NC score | 0.055101 (rank : 80) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O95347, Q9P1P2 | Gene names | SMC2L1, CAPE, SMC2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (hCAP-E) (XCAP-E homolog). | |||||
|
MYH1_MOUSE
|
||||||
| NC score | 0.055076 (rank : 81) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1461 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q5SX40 | Gene names | Myh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1). | |||||
|
MYH8_HUMAN
|
||||||
| NC score | 0.055053 (rank : 82) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1610 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P13535, Q14910 | Gene names | MYH8 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
CR034_MOUSE
|
||||||
| NC score | 0.055020 (rank : 83) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8CDV0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C18orf34 homolog. | |||||
|
CENPF_HUMAN
|
||||||
| NC score | 0.054923 (rank : 84) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
OPTN_MOUSE
|
||||||
| NC score | 0.054918 (rank : 85) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1120 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8K3K8 | Gene names | Optn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin. | |||||
|
MYH8_MOUSE
|
||||||
| NC score | 0.054770 (rank : 86) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1600 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P13542, Q5SX36 | Gene names | Myh8, Myhsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
CI039_HUMAN
|
||||||
| NC score | 0.054727 (rank : 87) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1171 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9NXG0, Q5VYJ0, Q9HAJ5 | Gene names | C9orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf39. | |||||
|
MY18A_HUMAN
|
||||||
| NC score | 0.054703 (rank : 88) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
MYH1_HUMAN
|
||||||
| NC score | 0.054690 (rank : 89) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1478 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P12882, Q9Y622 | Gene names | MYH1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1) (Myosin heavy chain IIx/d) (MyHC-IIx/d). | |||||
|
KMHN1_HUMAN
|
||||||
| NC score | 0.054636 (rank : 90) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 950 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8TBZ0, Q86YI9, Q8N7W0 | Gene names | KMHN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cancer/testis antigen KM-HN-1. | |||||
|
T3JAM_HUMAN
|
||||||
| NC score | 0.054635 (rank : 91) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 533 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9Y228, Q2YDB5, Q4VY06, Q7Z706 | Gene names | TRAF3IP3, T3JAM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRAF3-interacting JNK-activating modulator (TRAF3-interacting protein 3). | |||||
|
CK5P2_MOUSE
|
||||||
| NC score | 0.054632 (rank : 92) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1151 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8K389, Q6PCN1, Q6ZPL0 | Gene names | Cdk5rap2, Kiaa1633 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48). | |||||
|
CCD46_HUMAN
|
||||||
| NC score | 0.054547 (rank : 93) | θ value | θ > 10 (rank : 64) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1430 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8N8E3, Q6PIB5, Q8NCR4, Q8NFR4 | Gene names | CCDC46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 46. | |||||
|
IFT81_HUMAN
|
||||||
| NC score | 0.054542 (rank : 94) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8WYA0, Q8NB51, Q9BSV2, Q9UNY8 | Gene names | IFT81, CDV1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 81 (Carnitine deficiency-associated protein expressed in ventricle 1) (CDV-1 protein). | |||||
|
3BP5_MOUSE
|
||||||
| NC score | 0.054512 (rank : 95) | θ value | θ > 10 (rank : 52) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Z131, Q8C903, Q8VCZ0 | Gene names | Sh3bp5, Sab | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 5 (SH3 domain-binding protein that preferentially associates with BTK). | |||||
|
TUFT1_HUMAN
|
||||||
| NC score | 0.054492 (rank : 96) | θ value | θ > 10 (rank : 183) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 686 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9NNX1, Q5T384, Q5T385, Q9BU28, Q9H5L1 | Gene names | TUFT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tuftelin. | |||||
|
MPP9_HUMAN
|
||||||
| NC score | 0.054487 (rank : 97) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q99550, Q9H976 | Gene names | MPHOSPH9, MPP9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | M-phase phosphoprotein 9. | |||||
|
SMC4_HUMAN
|
||||||
| NC score | 0.054394 (rank : 98) | θ value | 6.88961 (rank : 46) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9NTJ3, O95752, Q8NDL4, Q9UNT9 | Gene names | SMC4L1, CAPC, SMC4 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 4-like 1 protein (Chromosome- associated polypeptide C) (hCAP-C) (XCAP-C homolog). | |||||
|
NDE1_HUMAN
|
||||||
| NC score | 0.054391 (rank : 99) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 823 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9NXR1, Q49AQ2 | Gene names | NDE1, NUDE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear distribution protein nudE homolog 1 (NudE). | |||||
|
CASP_MOUSE
|
||||||
| NC score | 0.054389 (rank : 100) | θ value | θ > 10 (rank : 58) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1152 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P70403, Q7TQJ4, Q91WN6 | Gene names | Cutl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
MYH6_MOUSE
|
||||||
| NC score | 0.054364 (rank : 101) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1501 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q02566, Q64258, Q64738 | Gene names | Myh6, Myhca | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
GOGA3_MOUSE
|
||||||
| NC score | 0.054256 (rank : 102) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1437 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P55937, Q80VF5, Q8CCK4, Q9QYT2, Q9QYT3 | Gene names | Golga3, Mea2 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Male-enhanced antigen 2) (MEA-2). | |||||
|
MYH13_HUMAN
|
||||||
| NC score | 0.054234 (rank : 103) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1504 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UKX3, O95252 | Gene names | MYH13 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-13 (Myosin heavy chain, skeletal muscle, extraocular) (MyHC- eo). | |||||
|
TEX9_HUMAN
|
||||||
| NC score | 0.054224 (rank : 104) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 541 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8N6V9 | Gene names | TEX9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed sequence 9 protein. | |||||
|
MYH6_HUMAN
|
||||||
| NC score | 0.054222 (rank : 105) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1426 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P13533, Q13943, Q14906, Q14907 | Gene names | MYH6, MYHCA | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
CCD46_MOUSE
|
||||||
| NC score | 0.054109 (rank : 106) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q5PR68, Q80ZN6, Q99JS4, Q9D9T1 | Gene names | Ccdc46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled coil domain-containing protein 46. | |||||
|
CASP_HUMAN
|
||||||
| NC score | 0.054092 (rank : 107) | θ value | θ > 10 (rank : 57) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q13948, Q53GU9, Q8TBS3 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
MYO5C_HUMAN
|
||||||
| NC score | 0.054091 (rank : 108) | θ value | 8.99809 (rank : 50) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1206 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9NQX4 | Gene names | MYO5C | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5C (Myosin Vc). | |||||
|
CCD42_HUMAN
|
||||||
| NC score | 0.054043 (rank : 109) | θ value | θ > 10 (rank : 63) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q96M95, Q8N6Q0 | Gene names | CCDC42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 42. | |||||
|
MD1L1_MOUSE
|
||||||
| NC score | 0.053858 (rank : 110) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1001 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9WTX8, Q9WTX9 | Gene names | Mad1l1, Mad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1). | |||||
|
REST_MOUSE
|
||||||
| NC score | 0.053825 (rank : 111) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1506 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q922J3, Q571L7 | Gene names | Rsn, Kiaa4046 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin. | |||||
|
CYLN2_MOUSE
|
||||||
| NC score | 0.053820 (rank : 112) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1002 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9Z0H8, Q7TSI9, Q8CHU1, Q9EP81 | Gene names | Cyln2, Kiaa0291 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic linker protein 2 (Cytoplasmic linker protein 115) (CLIP- 115). | |||||
|
LRC45_MOUSE
|
||||||
| NC score | 0.053681 (rank : 113) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8CIM1, Q3U5Z2 | Gene names | Lrrc45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 45. | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.053659 (rank : 114) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
OPTN_HUMAN
|
||||||
| NC score | 0.053574 (rank : 115) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 827 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q96CV9, Q5T672, Q5T673, Q5T674, Q5T675, Q7LDL9, Q8N562, Q9UET9, Q9UEV4, Q9Y218 | Gene names | OPTN, FIP2, GLC1E, NRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin (Optic neuropathy-inducing protein) (E3-14.7K-interacting protein) (FIP-2) (Huntingtin-interacting protein HYPL) (NEMO-related protein) (Transcription factor IIIA-interacting protein) (TFIIIA- IntP). | |||||
|
GOGA3_HUMAN
|
||||||
| NC score | 0.053565 (rank : 116) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q08378, O43241, Q6P9C7, Q86XW3, Q8TDA9, Q8WZA3 | Gene names | GOLGA3 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Golgi complex-associated protein of 170 kDa) (GCP170). | |||||
|
GOGA5_HUMAN
|
||||||
| NC score | 0.053523 (rank : 117) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1168 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8TBA6, O95287, Q03962, Q9UQQ7 | Gene names | GOLGA5, RETII, RFG5 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (RET-fused gene 5 protein) (Ret-II protein). | |||||
|
LRRF2_HUMAN
|
||||||
| NC score | 0.053473 (rank : 118) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 791 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9Y608, Q68CV3, Q9NXH5 | Gene names | LRRFIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 2 (LRR FLII- interacting protein 2). | |||||
|
GCC2_MOUSE
|
||||||
| NC score | 0.053464 (rank : 119) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
RABE1_HUMAN
|
||||||
| NC score | 0.053458 (rank : 120) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q15276, O95369, Q8IVX3 | Gene names | RABEP1, RABPT5, RABPT5A | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha) (Rabaptin-4) (NY-REN-17 antigen). | |||||
|
TAXB1_HUMAN
|
||||||
| NC score | 0.053442 (rank : 121) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 829 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q86VP1, O60398, O95770, Q13311, Q9BQG5, Q9UI88 | Gene names | TAX1BP1, T6BP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tax1-binding protein 1 (TRAF6-binding protein). | |||||
|
MYH4_HUMAN
|
||||||
| NC score | 0.053438 (rank : 122) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1377 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9Y623 | Gene names | MYH4 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal) (Myosin heavy chain IIb) (MyHC-IIb). | |||||
|
REST_HUMAN
|
||||||
| NC score | 0.053072 (rank : 123) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1605 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P30622 | Gene names | RSN, CYLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Restin (Cytoplasmic linker protein 170 alpha-2) (CLIP-170) (Reed- Sternberg intermediate filament-associated protein) (Cytoplasmic linker protein 1). | |||||
|
CP250_MOUSE
|
||||||
| NC score | 0.052824 (rank : 124) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
GOGA1_HUMAN
|
||||||
| NC score | 0.052777 (rank : 125) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1222 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q92805, Q8IYZ9 | Gene names | GOLGA1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
CYLN2_HUMAN
|
||||||
| NC score | 0.052770 (rank : 126) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1004 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UDT6, O14527, O43611 | Gene names | CYLN2, KIAA0291, WBSCR4, WSCR4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic linker protein 2 (Cytoplasmic linker protein 115) (CLIP- 115) (Williams-Beuren syndrome chromosome region 4 protein). | |||||
|
SM1L2_HUMAN
|
||||||
| NC score | 0.052652 (rank : 127) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1019 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8NDV3, Q5TIC3, Q9Y3G5 | Gene names | SMC1L2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 2 protein (SMC1beta protein). | |||||
|
CP250_HUMAN
|
||||||
| NC score | 0.052580 (rank : 128) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
CCD62_HUMAN
|
||||||
| NC score | 0.052408 (rank : 129) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 631 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q6P9F0, Q6ZVF2, Q86VJ0, Q9BYZ5 | Gene names | CCDC62 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 60 (Aaa-protein) (Protein TSP- NY). | |||||
|
ERC2_MOUSE
|
||||||
| NC score | 0.052243 (rank : 130) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1166 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q6PH08, Q80U20, Q8CCP1 | Gene names | Erc2, Cast1, D14Ertd171e, Kiaa0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2 (CAZ-associated structural protein 1) (CAST1). | |||||
|
BRE1B_MOUSE
|
||||||
| NC score | 0.052219 (rank : 131) | θ value | θ > 10 (rank : 56) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 970 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q3U319, Q6ZQ75, Q8BJA1, Q8BY03, Q8CHX4 | Gene names | Rnf40, Bre1b, Kiaa0661 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1B (EC 6.3.2.-) (BRE1-B) (RING finger protein 40). | |||||
|
RB6I2_HUMAN
|
||||||
| NC score | 0.052189 (rank : 132) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1584 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8IUD2, Q6NVK2, Q8IUD3, Q8IUD4, Q8IUD5, Q8NAS1, Q9NXN5, Q9UIK7, Q9UPS1 | Gene names | ERC1, ELKS, KIAA1081, RAB6IP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1). | |||||
|
SYCP1_MOUSE
|
||||||
| NC score | 0.052184 (rank : 133) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1456 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q62209, O09205, P70192, Q62329 | Gene names | Sycp1, Scp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
YO11_MOUSE
|
||||||
| NC score | 0.052170 (rank : 134) | θ value | θ > 10 (rank : 184) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P11260 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Hypothetical protein ORF-1137. | |||||
|
RB6I2_MOUSE
|
||||||
| NC score | 0.052119 (rank : 135) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q99MI1, Q80TK7, Q8BPL1, Q8C7Y1, Q99MI2 | Gene names | Erc1, Cast2, Kiaa1081, Rab6ip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1) (ERC1) (CAZ-associated structural protein 2) (CAST2). | |||||
|
MYO5A_HUMAN
|
||||||
| NC score | 0.051956 (rank : 136) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1092 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9Y4I1, O60653, Q07902, Q16249, Q9UE30, Q9UE31 | Gene names | MYO5A, MYH12 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5A (Myosin Va) (Dilute myosin heavy chain, non-muscle) (Myosin heavy chain 12) (Myoxin). | |||||
|
POF1B_MOUSE
|
||||||
| NC score | 0.051816 (rank : 137) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8K4L4, Q8C3A2 | Gene names | Pof1b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein POF1B. | |||||
|
BRE1B_HUMAN
|
||||||
| NC score | 0.051745 (rank : 138) | θ value | θ > 10 (rank : 55) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O75150, Q6AHZ6, Q6N005, Q7L3T6, Q8N615, Q96T18, Q9BSV9, Q9HC82 | Gene names | RNF40, BRE1B, KIAA0661 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1B (EC 6.3.2.-) (BRE1-B) (RING finger protein 40) (95 kDa retinoblastoma-associated protein) (RBP95). | |||||
|
SDCG8_HUMAN
|
||||||
| NC score | 0.051732 (rank : 139) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1002 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q86SQ7, O60527, Q3ZCR6, Q8N5F2, Q9P0F1 | Gene names | SDCCAG8, CCCAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serologically defined colon cancer antigen 8 (Centrosomal colon cancer autoantigen protein) (hCCCAP) (Antigen NY-CO-8). | |||||
|
SYCP1_HUMAN
|
||||||
| NC score | 0.051705 (rank : 140) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q15431, O14963 | Gene names | SYCP1, SCP1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
PCNT_HUMAN
|
||||||
| NC score | 0.051682 (rank : 141) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
SM1L2_MOUSE
|
||||||
| NC score | 0.051607 (rank : 142) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 918 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q920F6 | Gene names | Smc1l2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 1-like 2 protein (SMC1beta protein). | |||||
|
DZIP1_HUMAN
|
||||||
| NC score | 0.051594 (rank : 143) | θ value | 3.0926 (rank : 32) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 675 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q86YF9, Q8WY45, Q8WY46, Q9UGA5, Q9Y2K0 | Gene names | DZIP1, DZIP, DZIP2, KIAA0996 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein DZIP1 (DAZ-interacting protein 1/2). | |||||
|
TRAIP_MOUSE
|
||||||
| NC score | 0.051555 (rank : 144) | θ value | θ > 10 (rank : 182) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 745 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8VIG6, O08854, Q922M8, Q9CPP4 | Gene names | Traip, Trip | |||
|
Domain Architecture |

|
|||||
| Description | TRAF-interacting protein. | |||||
|
ERC2_HUMAN
|
||||||
| NC score | 0.051545 (rank : 145) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1156 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O15083, Q86TK4 | Gene names | ERC2, KIAA0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2. | |||||
|
CJ118_MOUSE
|
||||||
| NC score | 0.051529 (rank : 146) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1132 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8C9S4, Q6P3D6, Q8CJC0 | Gene names | Otg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf118 homolog (Oocyte-testis gene 1 protein). | |||||
|
CK5P2_HUMAN
|
||||||
| NC score | 0.051497 (rank : 147) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q96SN8, Q7Z3L4, Q7Z3U1, Q7Z7I6, Q9BSW0, Q9H6J6, Q9HCD9, Q9NV90, Q9UIW9 | Gene names | CDK5RAP2, CEP215, KIAA1633 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48) (Centrosome-associated protein 215). | |||||
|
GOGA5_MOUSE
|
||||||
| NC score | 0.051474 (rank : 148) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1434 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9QYE6, O88317, Q3TGE7, Q3U6S5, Q3UUF9 | Gene names | Golga5, Retii | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (Sumiko protein) (Ret-II protein). | |||||
|
CDR2_MOUSE
|
||||||
| NC score | 0.051461 (rank : 149) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 482 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P97817 | Gene names | Cdr2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cerebellar degeneration-related protein 2. | |||||
|
SMC1A_MOUSE
|
||||||
| NC score | 0.051385 (rank : 150) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1384 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9CU62, Q3V480, Q9CUX9, Q9D959, Q9WTU1 | Gene names | Smc1l1, Sb1.8, Smc1, Smcb | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Chromosome segregation protein SmcB) (Sb1.8). | |||||
|
CYTSA_HUMAN
|
||||||
| NC score | 0.051377 (rank : 151) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1138 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q69YQ0, O15081 | Gene names | CYTSA, KIAA0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A (NY-REN-22 antigen). | |||||
|
CRCM_HUMAN
|
||||||
| NC score | 0.051276 (rank : 152) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 520 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P23508 | Gene names | MCC | |||
|
Domain Architecture |

|
|||||
| Description | Colorectal mutant cancer protein (Protein MCC). | |||||
|
CYTSA_MOUSE
|
||||||
| NC score | 0.051258 (rank : 153) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q2KN98, Q8CHF9 | Gene names | Cytsa, Kiaa0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A. | |||||
|
SDCG8_MOUSE
|
||||||
| NC score | 0.051116 (rank : 154) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q80UF4 | Gene names | Sdccag8, Cccap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serologically defined colon cancer antigen 8 (Centrosomal colon cancer autoantigen protein) (mCCCAP). | |||||
|
NIN_MOUSE
|
||||||
| NC score | 0.051029 (rank : 155) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q61043, Q6ZPM7 | Gene names | Nin, Kiaa1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein. | |||||
|
MYO5A_MOUSE
|
||||||
| NC score | 0.051024 (rank : 156) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1117 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q99104 | Gene names | Myo5a, Dilute | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5A (Myosin Va) (Dilute myosin heavy chain, non-muscle). | |||||
|
CCHCR_HUMAN
|
||||||
| NC score | 0.050906 (rank : 157) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 717 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8TD31, Q9NRK8, Q9NWY9, Q9NXJ4, Q9NXK3, Q9Y6W1, Q9Y6W2 | Gene names | CCHCR1, C6orf18, HCR | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil alpha-helical rod protein 1 (Alpha helical coiled-coil rod protein) (Putative gene 8 protein) (Pg8). | |||||
|
MY18B_HUMAN
|
||||||
| NC score | 0.050806 (rank : 158) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1299 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8IUG5, Q8NDI8, Q8TE65, Q8WWS0, Q96KH2, Q96KR8, Q96KR9 | Gene names | MYO18B | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18B (Myosin XVIIIb). | |||||
|
SMC1A_HUMAN
|
||||||
| NC score | 0.050790 (rank : 159) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1346 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q14683, O14995, Q16351 | Gene names | SMC1L1, DXS423E, KIAA0178, SB1.8, SMC1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Sb1.8). | |||||
|
CT2NL_HUMAN
|
||||||
| NC score | 0.050748 (rank : 160) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9P2B4, Q96B40 | Gene names | CTTNBP2NL, KIAA1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
CENPE_HUMAN
|
||||||
| NC score | 0.050656 (rank : 161) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
HOOK3_MOUSE
|
||||||
| NC score | 0.050641 (rank : 162) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8BUK6, Q8BV48, Q8BW57, Q8BY41 | Gene names | Hook3 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 3. | |||||
|
FLIP1_HUMAN
|
||||||
| NC score | 0.050558 (rank : 163) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q7Z7B0, Q5VUL6, Q86TC3, Q8N8B9, Q96SK6, Q9NVI8, Q9ULE5 | Gene names | FILIP1, KIAA1275 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
CTGE5_MOUSE
|
||||||
| NC score | 0.050475 (rank : 164) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 928 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8R311, Q8CIE3 | Gene names | Ctage5, Mea6, Mgea6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cutaneous T-cell lymphoma-associated antigen 5 homolog (cTAGE-5 protein) (Meningioma-expressed antigen 6). | |||||
|
TRAIP_HUMAN
|
||||||
| NC score | 0.050339 (rank : 165) | θ value | θ > 10 (rank : 181) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9BWF2, O00467 | Gene names | TRAIP, TRIP | |||
|
Domain Architecture |

|
|||||
| Description | TRAF-interacting protein. | |||||
|
CENPH_HUMAN
|
||||||
| NC score | 0.050235 (rank : 166) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9H3R5 | Gene names | CENPH, ICEN35 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein H (CENP-H) (Interphase centromere complex protein 35). | |||||
|
LRC45_HUMAN
|
||||||
| NC score | 0.050190 (rank : 167) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 933 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q96CN5 | Gene names | LRRC45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 45. | |||||
|
GOGA4_MOUSE
|
||||||
| NC score | 0.050178 (rank : 168) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
NIN_HUMAN
|
||||||
| NC score | 0.050155 (rank : 169) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1416 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8N4C6, Q6P0P6, Q9BWU6, Q9C012, Q9C013, Q9C014, Q9H5I6, Q9HAT7, Q9HBY5, Q9HCK7, Q9UH61 | Gene names | NIN, KIAA1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein (hNinein) (Glycogen synthase kinase 3 beta-interacting protein) (GSK3B-interacting protein). | |||||
|
HOOK3_HUMAN
|
||||||
| NC score | 0.050123 (rank : 170) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1070 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q86VS8, Q8NBH0, Q9BY13 | Gene names | HOOK3 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 3 (hHK3). | |||||
|
GOGA2_MOUSE
|
||||||
| NC score | 0.050114 (rank : 171) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q921M4 | Gene names | Golga2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgin subfamily A member 2 (Cis-Golgi matrix protein GM130). | |||||
|
VDP_MOUSE
|
||||||
| NC score | 0.047175 (rank : 172) | θ value | 5.27518 (rank : 41) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 469 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Z1Z0, Q3T9L9, Q3TH58, Q3U1C7, Q3UMW6, Q91WE7, Q99JZ5 | Gene names | Vdp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | General vesicular transport factor p115 (Transcytosis-associated protein) (TAP) (Vesicle docking protein). | |||||
|
KIF5A_MOUSE
|
||||||
| NC score | 0.042823 (rank : 173) | θ value | 5.27518 (rank : 40) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 859 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P33175, Q5DTP1, Q6PDY7, Q9Z2F9 | Gene names | Kif5a, Kiaa4086, Kif5, Nkhc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin heavy chain isoform 5A (Neuronal kinesin heavy chain) (NKHC) (Kinesin heavy chain neuron-specific 1). | |||||
|
ROCK2_MOUSE
|
||||||
| NC score | 0.037701 (rank : 174) | θ value | 0.279714 (rank : 13) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 2072 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P70336, Q8BR64, Q8CBR0 | Gene names | Rock2 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2). | |||||
|
BAZ1A_HUMAN
|
||||||
| NC score | 0.037386 (rank : 175) | θ value | 3.0926 (rank : 30) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9NRL2, Q9NZ15, Q9P065, Q9UIG1, Q9Y3V3 | Gene names | BAZ1A, ACF1, WCRF180 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1A (ATP-utilizing chromatin assembly and remodeling factor 1) (hACF1) (ATP-dependent chromatin remodelling protein) (Williams syndrome transcription factor-related chromatin remodeling factor 180) (WCRF180) (hWALp1) (CHRAC subunit ACF1). | |||||
|
ROCK2_HUMAN
|
||||||
| NC score | 0.036610 (rank : 176) | θ value | 0.279714 (rank : 12) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 2073 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O75116, Q9UQN5 | Gene names | ROCK2, KIAA0619 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2) (Rho kinase 2). | |||||
|
KIF3B_HUMAN
|
||||||
| NC score | 0.033352 (rank : 177) | θ value | 1.81305 (rank : 24) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 540 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O15066 | Gene names | KIF3B, KIAA0359 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF3B (Microtubule plus end-directed kinesin motor 3B) (HH0048). | |||||
|
KIF3B_MOUSE
|
||||||
| NC score | 0.033149 (rank : 178) | θ value | 1.81305 (rank : 25) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 533 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q61771 | Gene names | Kif3b | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF3B (Microtubule plus end-directed kinesin motor 3B). | |||||
|
DAAM1_MOUSE
|
||||||
| NC score | 0.031251 (rank : 179) | θ value | 5.27518 (rank : 39) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BPM0, Q80Y68, Q9CQQ2 | Gene names | Daam1 | |||
|
Domain Architecture |

|
|||||
| Description | Disheveled-associated activator of morphogenesis 1. | |||||
|
TARA_MOUSE
|
||||||
| NC score | 0.029694 (rank : 180) | θ value | 6.88961 (rank : 47) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
ZN148_HUMAN
|
||||||
| NC score | 0.013566 (rank : 181) | θ value | 0.62314 (rank : 17) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UQR1, O00389, O43591 | Gene names | ZNF148, ZBP89 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 148 (Zinc finger DNA-binding protein 89) (Transcription factor ZBP-89). | |||||
|
ZN148_MOUSE
|
||||||
| NC score | 0.013564 (rank : 182) | θ value | 0.62314 (rank : 18) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 755 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61624, P97475 | Gene names | Znf148, Zbp89, Zfp148 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 148 (Zinc finger DNA-binding protein 89) (Transcription factor ZBP-89) (G-rich box-binding protein) (Beta enolase repressor factor 1) (Transcription factor BFCOL1). | |||||
|
COPG_HUMAN
|
||||||
| NC score | 0.011152 (rank : 183) | θ value | 8.99809 (rank : 48) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y678 | Gene names | COPG | |||
|
Domain Architecture |

|
|||||
| Description | Coatomer subunit gamma (Gamma-coat protein) (Gamma-COP). | |||||
|
MYLK_MOUSE
|
||||||
| NC score | 0.002552 (rank : 184) | θ value | 8.99809 (rank : 49) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1489 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6PDN3, Q3TSJ7, Q80UX0, Q80YN7, Q80YN8, Q8K026, Q924D2, Q9ERD3 | Gene names | Mylk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin light chain kinase, smooth muscle (EC 2.7.11.18) (MLCK) (Telokin) (Kinase-related protein) (KRP). | |||||